Abstract
Leucine-rich amelogenin peptide (LRAP), an alternatively spliced amelogenin protein, possesses a signaling property shown to induce osteogenic differentiation. In the current study, we detected LRAP expression during osteogenesis of wild-type (WT) embryonic stem (ES) cells and observed the absence of LRAP expression in amelogenin-null (KO) ES cells. We explored the signaling effect of LRAP on wild-type ES cells, and the ability of LRAP to rescue the impaired osteogenesis phenotype observed in KO ES cells. Our data indicate that LRAP treatment of WT and KO ES cells induces a significant increase in mineral matrix formation, and significant increases in bone sialoprotein and osterix gene expression. In addition, the amelogenin KO phenotype is partially rescued by the addition of exogenous LRAP. These data suggest a unique function of LRAP during ES cell differentiation along osteogenic lineage.
Keywords: Leucine-rich amelogenin peptide, LRAP, osteogenesis, ES cell differentiation
INTRODUCTION
The formation of hard tissues such as bone, cartilage and tooth occurs by a process of matrix-mediated biomineralization in which intracellular and extracellular organic proteins regulate the initiation, growth and deposition of mineral crystals [1]. In developing enamel matrix, the majority of the organic extracellular matrix proteins are comprised by a group of highly conserved, structural proteins called amelogenins [2]. Amelogenins have been identified to function as cell adhesion molecules and to facilitate nucleation and growth of hydroxyapatite crystals during the mineralization phase of amelogenesis [3]. As a result of amelogenin mRNA alternative splicing, different amelogenin isoforms are detected in the developing enamel matrix [4]. Although the function of amelogenin has been proven indispensable for proper development of enamel [5], the function of various amelogenin isoforms is not known.
Leucine-rich Amelogenin Peptide (LRAP) is a 59-residue protein translated from exon 2, 3, 5, 6D and 7 of amelogenin mRNA [6]. Failure of LRAP to rescue the amelogenin-null enamel phenotype indicates that LRAP may have distinctive function(s) outside guiding mineral crystallite formation [7]. The commercial porcine enamel matrix derivative, known as Emdogain, is composed predominantly of amelogenins. Emdogain has the ability to induce tissue repair by promoting periodontal tissue regeneration. Periodontal tissue repair relies upon pathways involving enhanced proliferation and differentiation of precursor cells [8], and the upregulation of bone marker genes [9]. Analyses of the amelogenin constituents of Emdogain suggest that LRAP functions as a signaling molecule [10]. In the previous studies, LRAP has been found to induce osteogenesis in various cell types, including rat muscle fibroblasts [10], mouse cementoblasts [11], and mouse oral mucosal cells [12].
Amelogenin null mice develop hypomineralized enamel lacking normal prism structure, but are healthy and fertile [5]. However, the amelogenin null mice are smaller than wild-type mice prior to weaning. The smaller size of amelogenin null mice could potentially be due to the lack of LRAP expression in some of these tissues, leading to a delay in development [13].
In the current experiment, we studied the effect of exogenous LRAP on ES cell differentiation to the osteogenic lineage by monitoring changes in gene expression and changes in mineral deposition in the presence of exogenous LRAP. In addition, we explore the effect of LRAP to rescue the impaired phenotype observed in osteogenic differentiation of amelogenin-null ES cells. We propose that LRAP functions as a moonlighting protein [14] outside its original location in enamel matrix to serve as a signaling molecule in other tissues.
MATERIALS AND METHODS
Mouse embryonic stem cell culture
Mouse ES cells (RW4) were maintained on the irradiated MEF feeder layers in Knockout D-MEM (Invitrogen) containing 15% FBS (Hyclone) supplemented with murine leukemia inhibitory factor (LIF; 1000 U/mL; Chemicon), 10 mM HEPES buffer solution, 0.1 mM MEM non-essential amino acids solution, 0.05 mM 2-mercaptoethanol, 2 mM L-glutamine, penicillin (50 U/mL) and streptomycin (50 μg/mL). Cells were grown at 37°C in 7.5% CO2 with daily media changed. Cell passage was achieved by treatment of the cells with 0.05% Trypsin and reseeded onto a fresh MEF feeder layer. Amelogenin-null (KO) ES cells were generated as previously described [15] (See supplement and Fig S1) and were maintained in the same condition as the wild-type ES cells.
Induction of cell differentiation
Mouse ES cells were induced to form embryoid bodies (EBs) in a hanging drop culture according to a standard protocol [16]. After 2 days, EBs were collected and resuspended in 100 mm petri dishes in knockout D-MEM supplemented with 10-7M all-trans retinoic acids for additional 3 days with medium change every day.
After 3 days, the EBs were collected, and transferred to gelatinized 6-well tissue culture plates for osteogenic induction experiments using one of the three media each with different supplements. The “basal media” was prepared by using MEM-alpha media, supplemented with 15% batch-tested FBS, 10 mM HEPES buffer, 0.1 mM MEM non-essential amino acids solution, 0.05 mM 2-mercaptoethanol, 2 mM L-glutamine, penicillin (50 U/mL) and streptomycin (50 μg/mL). The “control media” was prepared by supplementing the basal media with ascorbic acid (50 μg/mL) and β-glycerophosphate (10mM). The “LRAP media” was prepared by adding chemically synthesized and purified murine LRAP protein (10ng/mL) to the control media. The EBs were cultured in one of these media at 37°C in 5.0% CO2 for 15 days with the media changed every other day.
RNA extraction, cDNA synthesis, and quantitative real-time PCR analysis
RNA was isolated from EB cultures at day-10, and at day-20 (day 0=EB formation) by using RNAqueous®-4PCR Kit (Ambion) following the manufacturer’s instructions. Synthesis of cDNA was performed by using RETROscript® Kit (Ambion). For cDNA template preparation, 2 μg of total RNA from each EB sample was used in a 20 μL reaction. For quantitative real-time PCR analysis, a 25 μL reaction was prepared for each sample. Included in this reaction volume was 1 μL of the resulting cDNA, iQ™ SYBR® Green Supermix (Bio-Rad) containing dNTP and iTaq DNA polymerase, and the appropriate primers (Table S1). The resulting threshold cycle (CT) value from each primer pair was normalized with the CT value for Gapdh, which served as an internal control.
Analysis of mineral deposition
EBs at day-20 were stained with Alizarin Red. Quantification of calcium concentration was measured by means of the intensity of blue color at 612 nm (QuantiChrom™ Calcium Assay kit; BioAssay Systems). The total amount of protein in each sample was used as a standard to normalize the calcium data.
Statistical analysis
For gene expression analysis and calcium concentration analysis, independent measurements were performed in triplicate and averaged for control of internal error. Each experiment was repeated at least three times. Mean and standard deviation from each experiment were used to statistically analyze the difference between each pair of samples. A P-value less than 0.05 was considered significant.
RESULTS
Different amelogenin isoforms are expressed during osteogenic differentiation of ES cells
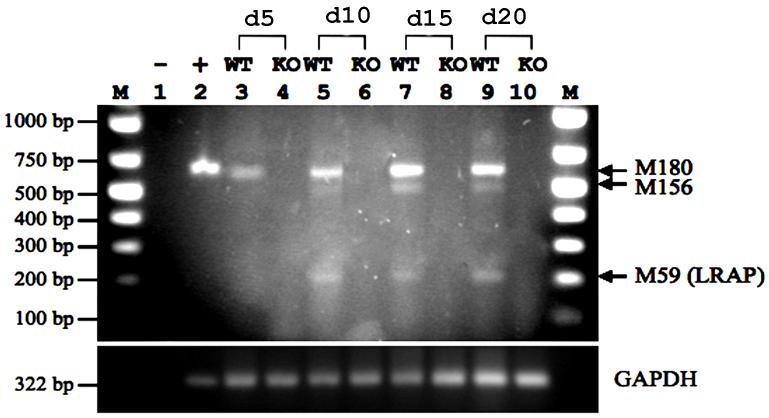
To understand the function of LRAP during the differentiation of mouse ES cells, we first determined the expression of LRAP during differentiation in both wild-type ES cells and amelogenin-null ES cells. Mouse ES cells were induced to form EBs (day-0), followed by subsequent osteogenic induction for 20 days according to an established protocol [16]. At selected time points, RNA samples were extracted for PCR analyses using a forward primer in exon 1 and a reverse primer in exon 6D (Supplemental Figure S1) of the amelogenin gene to detect all possible splicing variants. Our results showed that amelogenin transcripts were detected only in wild-type ES cells but not in amelogenin-null ES cells (Fig. 1). Only the full-length M180 isoform (598 bp) was expressed in EBs prior to osteogenic differentiation (Fig. 1 lane 3). LRAP (aka M59; 235 bp) was detected in differentiating cells (Fig. 1 lanes 5, 7, 9), suggesting its functional implication during osteogenic differentiation. An additional isoform, M156 (520 bp) was also detected in differentiating cells (Fig. 1 lanes 5, 7, 9).
Figure 1. Identification of amelogenin splicing isoforms during osteogenic differentiation of mouse embryonic stem cells.
Mouse wild-type RW4 (WT) and amelogenin knock-out-ES cells (KO) were induced to form EBs, and subsequently induced to osteogenic differentiation [16]. RNA samples were extracted from EB-derived cells at day-5, day-10, day-15, and day-20 (day-0=EB formation). Reverse transcription products were subject to PCR analyses using a forward primer in exon 1 (5′-ATCAAGCATCCCTGAGCTTCAGAC-3′) and a reverse primer in exon 6D (5′-GCTCAGGAAGAATGGGGGACAG-3′) of the amelogenin gene to detect all possible splicing variants. GAPDH was used as an internal control. Top panel: amelogenin; Bottom panel: GAPDH. Lane 1, negative control; Lane 2, positive control; Lane 3, RW4 day-5; Lane 4, KO day-5; Lane 5, RW4 day-10; Lane 6, KO day-10; Lane 7, RW4 day-15; Lane 8, KO day-15; Lane 9, RW4 day-20; Lane 10, KO day-20; M, marker.
LRAP enhanced mineral deposition in osteogenic-induced mouse ES cells
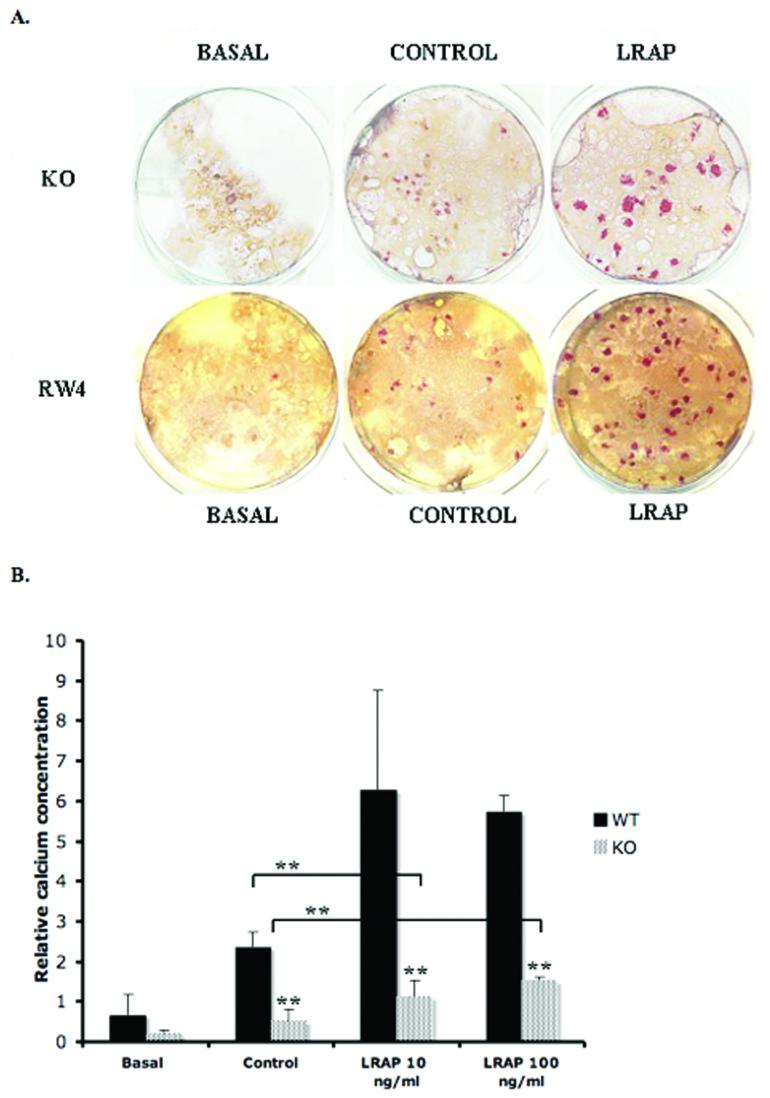
To analyze the terminal phenotype, EBs at day-20 were stained with Alizarin red to detect mineral deposits in the matrix. In both RW4 and KO ES cell differentiation, increased dye-stained area was evidenced in control group, suggesting that both RW4 and KO ES cells underwent proper osteogenic cell differentiation (Fig. 2A). The addition of exogenous LRAP to the control media resulted in a marked increase in mineral deposition, when compared to outcomes from control media alone, suggesting enhanced mineral formation with LRAP treatment (Fig. 2A).
Figure 2. ES cell differentiation and mineral nodule formation.
(A) Alizarin red staining. RW4 ES cells, and amelogenin-KO ES cells were induced to osteogenic differentiation in the basal media, control media, or LRAP media. At day-20, the cells were analyzed for mineral nodule formation by Alizarin red staining. Both RW4- and KO-ES cells underwent osteogenic differentiation as indicated by the dye accumulation in the synthesized matrix. Marked increased in mineral formation, as indicated by the increased numbers of nodules and the increased intensity of red-stained areas, was observed in LRAP-treated culture. KO = amelogenin null ES cells, RW4 = wild-type ES cells, Basal = basal media, Control = control media, LRAP = LRAP media. (B) Quantification of calcium concentration in osteogenic-induced ES cells. Significant increase of calcium concentration was observed in the LRAP-treated group in both RW4 and KO EBs. The calcium concentration for KO EBs (patterned bars) was significantly lower than the calcium concentration for wild-type EBs (filled bars). LRAP partially rescued the reduced calcium secretion phenotype seen in KO ES cells. WT = wild-type ES cells. KO = amelogenin-null ES cells. Basal = basal media, Control = control media, LRAP= LRAP media. Relative calcium concentration was calculated by normalization of the absolute calcium concentration to total protein concentration in each sample. **p<0.05.
Quantification of the calcium accumulation in the matrix was achieved using the QuantiChrom™ calcium assay kit to measure the amount of free calcium. In accordance with the visual record from Alizarin red staining, the calcium concentration in the control group was significantly higher than the basal group. And the calcium accumulation in the LRAP-treated group was significantly higher than the control group in both RW4 and KO ES cells (Fig. 2B). Comparing RW4 and KO ES cells, a significant decreased level of calcium accumulation was observed in the basal, control, and LRAP-treated groups. Noticeably, LRAP could partially rescue the reduced level of calcium deposited in the matrix created by the KO ES cells (Fig. 2B, Supplemental Table S2).
LRAP induces the expression of bone marker genes in osteogenic-induced ES cells
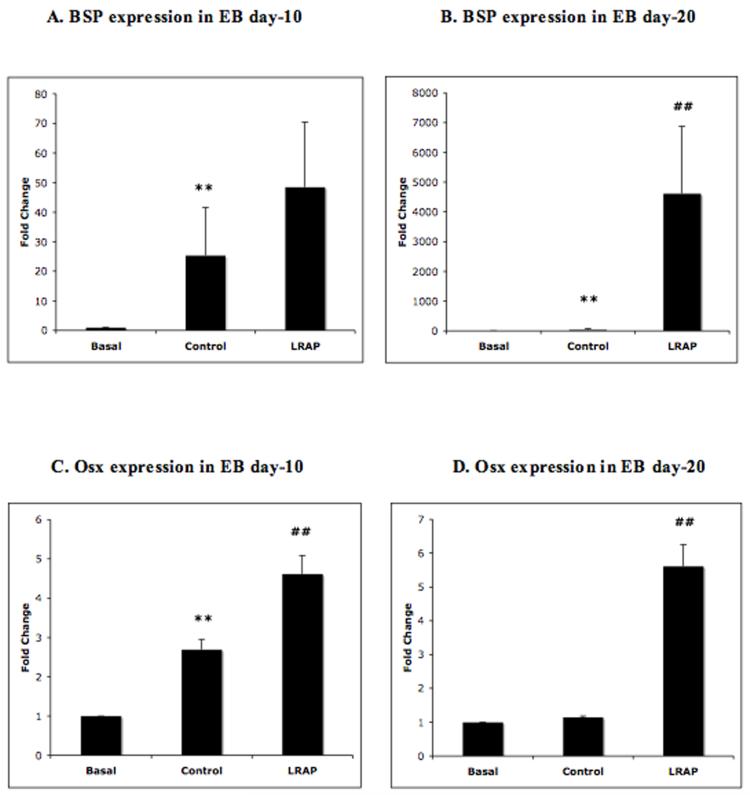
To explore the effect of LRAP on osteogenic gene expression, RNA from osteogenic-induced RW4 EB culture at day-10 and day-20 was collected to represent the EBs at early and late stages of differentiation, respectively. Gene expression was analyzed by quantitative real-time RT-PCR. At day-10, the expression of two osteogenic marker genes, bone sialoprotein (BSP) and osterix (Osx), in the control group was markedly increased. Analyses of the effect of LRAP on gene expression revealed significant enhancement in the expression of BSP (Fig. 3A) and Osx (Fig. 3C). At day-20, the EBs in the control group expressed increased level of BSP, but not Osx. The addition of LRAP in the media resulted in marked increase in both BSP (Fig. 3B) and Osx expression (Fig. 3D).
Figure 3. LRAP induced BSP and Osx expression.
Wild-typed ES cells were induced to osteogenic differentiation. EBs at day-10 and day-20 of culture duration were selected for RT-PCR analysis to represent early and late stages of osteogenesis, respectively. Marked increase in the expression of BSP (3A) and Osx (3C) was observed in LRAP-treated EB as early as at day-10 of culture, when compared to the values from the mineralization control group. A more pronounced increase of BSP expression was observed in LRAP-treated EB at day-20 of culture (3B). Osx expression was significantly increased in LRAP-treated EB day-20, but no change in Osx expression was observed in EB at day-20 of culture in control media. Values on Y-axis represent fold change of real-time PCR CT value compared to GAPDH that was used as an internal control. Basal = basal media, Control = control media, LRAP = LRAP media. **p< 0.05; compared to basal group, ##p<0.05; compared to control group.
DISCUSSION
In the current study, we provide data supporting the function of LRAP as a signaling molecule that enhances osteoblastic cell differentiation in mouse ES cells. This conclusion is supported by our findings that LRAP increases mineral matrix formation and increases calcium accumulation in the matrix from both wild-type ES cells and amelogenin-null ES cells. In addition, we show that the osteogenic effect of LRAP results in increased expression of BSP and Osx at both early- and late-stage of osteogenic differentiation. These results suggest a unique role for LRAP during osteogenesis of mouse embryonic stem cell, and support the previously reported osteogenic functions of LRAP in other cell types [10; 11; 17].
A previous study of the effects of BMP-2 on osteogenic differentiation of mouse ES cells suggested that in embryoid body cultures for 20 days, BMP-2 enhances mineral formation and increases osteocalcin expression by approximately 4-fold as analyzed by semi-quantitative RT-PCR [16]. Here, we demonstrate that LRAP increases as much as 4000-fold for BSP expression, 5-fold for Osx expression, and 6-fold for calcium accumulation in LRAP-treated EBs at the 20th day of culture. This dramatic increase in gene expression and calcium accumulation suggests that LRAP exerts an important role as an osteo-inductive molecule that is equal to or more potent than BMP-2 [18] during osteogenic differentiation of mouse ES cells.
However, other investigators suggest that the ability of LRAP to induce osteogenesis is dependent upon the cell types, the stage of differentiation and the local environment at the site of action [19]. Previous in vitro studies on the effect of LRAP on expression of bone marker genes in several progenitor cell types demonstrated that LRAP enhances osteogenesis. In mouse muscle fibroblast, LRAP treatment caused an immediate upregulation of Runx2, but the expression of Runx2 was diminished after 48 hours [10]. In mouse cementoblasts, LRAP increased expression of osteopontin in 72 hours, but had no effect on Runx2 or BSP expression [11]. Runx2 expression is critical for early osteoblast differentiation and osteoblastic cell lineage commitment [20]. Based on these observations, one may predict that Runx2 expression is more susceptible to modulation in less differentiated cells, such as fibroblasts, when compared to more differentiated cells, such as cementoblasts. However, in the case of pluripotent ES cells, we observed little change in Runx2 expression for embryoid bodies at day-10 and at day-20 of culture. On the other hand, we detected a substantial increase in expression of BSP and Osx, two markers for bone development detected in more fully differentiated osteoblasts [21; 22]. In our study, we selected two time points of EB culture that represent the early (EB day-10) and the late stage of ES cell differentiation (EB day-20). Therefore, it is possible that by day-10 of culture, differentiation has progressed sufficiently, as indicated by the expression of the late markers of differentiation, BSP and Osx, that further changes in Runx2 expression are not required. We hypothesize that Runx2 expression may be upregulated at an earlier time point than the EB day-10.
In the amelogenin-null (KO) ES cell experiment, we explored the ability of LRAP to rescue the reduced osteogenesis observed in the KO ES cells as a consequence of the loss of endogenous amelogenin expression. We have shown that although LRAP significantly increases mineral deposition and calcium accumulation when compared to the control media used in KO ES cells, LRAP can only partially rescue the amelogenin null phenotypes even when the concentration of LRAP is increased to from 10 ng/ml to 100 ng/ml (Fig. 2B). The failure of LRAP to fully rescue osteogenesis suggests that other amelogenin isoforms may be required to work coordinately with LRAP to fully induce the osteogenic gene expression. One of the candidate amelogenin isoforms for such a function is the mouse amelogenin splicing product expressed predominately by mouse ameloblasts called M180. The M180 protein is encoded by exon 2, 3, 5, 6, and 7, consisting of 180 amino acids (Figure S1), and is believed to be involved principally in controlling enamel crystal habit [15; 23]. A recent study in cementoblasts suggested that LRAP and p172, the porcine ortholog of mouse M180, work together to promote cell proliferation and migration in cementoblasts and in periodontal ligament cells from the amelogenin-knockout mouse [24].
For tissue engineering application, there are limitations to the use of conventional grafting therapies primarily due to immuno-rejection and post-operative complications. In addition, mesenchymal stem cells or lineage-committed cells have been reported to lose their proliferative capacity and identity during ex vivo expansion [25]. Therefore, the result from this study of directed differentiation of ES cells is predicted to provide another option for the potential therapeutic use of LRAP as an alternative pharmacologic agent in bone tissue engineering and for regenerative application in the repair of craniofacial and axial skeletal defects. The concept of moonlighting proteins suggests that one protein can exert two or more unique functions in different tissues [14]. We propose that LRAP, in addition to its original function in enamel formation, also moonlights as a signaling molecule to induce bone formation at other anatomical sites during development. Studies are now underway to identify the signaling pathway(s) responsible for LRAP-mediated osteo-induction, and to extend these studies to the function of LRAP on human stem cells.
The study of LRAP to function as a signaling molecule began only in the past decade [9] [10]. Several in vitro and in vivo studies have identified the role of LRAP as a potential signaling molecule implicated in bone formation in different cell types [10; 11; 17]. In the present study, we chose to use embryonic stem cells in our assays of amelogenin isoform signaling since these cells are pluripotent and display no previous overt differentiation pattern. Thus, their response to exogenous LRAP is unconstrained. We show that during their differentiation as embryoid bodies, amelogenin isoforms are expressed and are available to provide instructive signals in the absence of a preexistent bias to cell commitment that may have hampered prior experimental strategies. We identify that LRAP functions as a signaling molecule in the differentiation of mouse ES cells, based on the ability of LRAP to induce bone marker gene expression and induce mineral deposition in these ES cells.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Baruch Frenkel, Charles Shuler, Songtao Shi, and anonymous reviewers for their appraisal and constructive criticisms. This work was supported by NIDCR grants R01DE13045 (MLS), R03DE017362 (YZ), James H. Zumberge Faculty Research & Innovation Fund, University of Southern California (YZ) and Thammasat University School of Dentistry, Thailand (RW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Weiner S, Sagi I, Addadi L. Structural biology. Choosing the crystallization path less traveled. Science. 2005;309:1027–8. doi: 10.1126/science.1114920. [DOI] [PubMed] [Google Scholar]
- [2].Snead ML. Amelogenin protein exhibits a modular design: implications for form and function. Connect Tissue Res. 2003;44(Suppl 1):47–51. [PubMed] [Google Scholar]
- [3].Tarasevich BJ, Howard CJ, Larson JL, Snead ML, Simmer JP, Paine M, Shaw WJ. The nucleation and growth of calcium phosphate by amelogenin. Journal of Crystal Growth. 2007;304:407–415. doi: 10.1016/j.jcrysgro.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Iacob S, Veis A. Identification of temporal and spatial expression patterns of amelogenin isoforms during mouse molar development. Eur J Oral Sci. 2006;114(Suppl 1):194–200. doi: 10.1111/j.1600-0722.2006.00287.x. discussion 201-2, 381. [DOI] [PubMed] [Google Scholar]
- [5].Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–5. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- [6].Fincham AG, Belcourt AB, Termine JD, Butler WT, Cothran WC. Amelogenins. Sequence homologies in enamel-matrix proteins from three mammalian species. Biochem J. 1983;211:149–54. doi: 10.1042/bj2110149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen E, Yuan ZA, Wright JT, Hong SP, Li Y, Collier PM, Hall B, D’Angelo M, Decker S, Piddington R, Abrams WR, Kulkarni AB, Gibson CW. The small bovine amelogenin LRAP fails to rescue the amelogenin null phenotype. Calcif Tissue Int. 2003;73:487–95. doi: 10.1007/s00223-002-0036-7. [DOI] [PubMed] [Google Scholar]
- [8].Jiang J, Goodarzi G, He J, Li H, Safavi KE, Spangberg LS, Zhu Q. Emdogain-gel stimulates proliferation of odontoblasts and osteoblasts. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102:698–702. doi: 10.1016/j.tripleo.2006.02.011. [DOI] [PubMed] [Google Scholar]
- [9].Hammarstrom L. The role of enamel matrix proteins in the development of cementum and periodontal tissues. Ciba Found Symp. 1997;205:246–55. doi: 10.1002/9780470515303.ch17. discussion 255-60. [DOI] [PubMed] [Google Scholar]
- [10].Veis A, Tompkins K, Alvares K, Wei K, Wang L, Wang XS, Brownell AG, Jengh SM, Healy KE. Specific amelogenin gene splice products have signaling effects on cells in culture and in implants in vivo. J Biol Chem. 2000;275:41263–72. doi: 10.1074/jbc.M002308200. [DOI] [PubMed] [Google Scholar]
- [11].Boabaid F, Gibson CW, Kuehl MA, Berry JE, Snead ML, Nociti FH, Jr., Katchburian E, Somerman MJ. Leucine-rich amelogenin peptide: a candidate signaling molecule during cementogenesis. J Periodontol. 2004;75:1126–36. doi: 10.1902/jop.2004.75.8.1126. [DOI] [PubMed] [Google Scholar]
- [12].Lacerda-Pinheiro S, Jegat N, Septier D, Priam F, Bonnefoix M, Bitard J, Kellermann O, Tompkins K, Veis A, Goldberg M, Poliard A. Early in vivo and in vitro effects of amelogenin gene splice products on pulp cells. Eur J Oral Sci. 2006;114(Suppl 1):232–8. doi: 10.1111/j.1600-0722.2006.00320.x. discussion 254-6, 381-2. [DOI] [PubMed] [Google Scholar]
- [13].Li Y, Yuan ZA, Aragon MA, Kulkarni AB, Gibson CW. Comparison of body weight and gene expression in amelogenin null and wild-type mice. Eur J Oral Sci. 2006;114(Suppl 1):190–3. doi: 10.1111/j.1600-0722.2006.00286.x. discussion 201-2, 381. [DOI] [PubMed] [Google Scholar]
- [14].Jeffery CJ. Moonlighting proteins: old proteins learning new tricks. Trends Genet. 2003;19:415–7. doi: 10.1016/S0168-9525(03)00167-7. [DOI] [PubMed] [Google Scholar]
- [15].Zhu D, Paine ML, Luo W, Bringas P, Jr., Snead ML. Altering biomineralization by protein design. J Biol Chem. 2006;281:21173–82. doi: 10.1074/jbc.M510757200. [DOI] [PubMed] [Google Scholar]
- [16].Phillips BW, Belmonte N, Vernochet C, Ailhaud G, Dani C. Compactin enhances osteogenesis in murine embryonic stem cells. Biochem Biophys Res Commun. 2001;284:478–84. doi: 10.1006/bbrc.2001.4987. [DOI] [PubMed] [Google Scholar]
- [17].Nebgen DR, Inoue H, Sabsay B, Wei K, Ho CS, Veis A. Identification of the chondrogenic-inducing activity from bovine dentin (bCIA) as a low-molecular-mass amelogenin polypeptide. J Dent Res. 1999;78:1484–94. doi: 10.1177/00220345990780090201. [DOI] [PubMed] [Google Scholar]
- [18].Cheng H, Jiang W, Phillips FM, Haydon RC, Peng Y, Zhou L, Luu HH, An N, Breyer B, Vanichakarn P, Szatkowski JP, Park JY, He TC. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs) J Bone Joint Surg Am. 2003;85-A:1544–52. doi: 10.2106/00004623-200308000-00017. [DOI] [PubMed] [Google Scholar]
- [19].Lacerda-Pinheiro S, Septier D, Tompkins K, Veis A, Goldberg M, Chardin H. Amelogenin gene splice products A+4 and A-4 implanted in soft tissue determine the reorientation of CD45-positive cells to an osteo-chondrogenic lineage. J Biomed Mater Res A. 2006;79:1015–22. doi: 10.1002/jbm.a.30912. [DOI] [PubMed] [Google Scholar]
- [20].Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–71. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- [21].Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- [22].Ganss B, Kim RH, Sodek J. Bone sialoprotein. Crit Rev Oral Biol Med. 1999;10:79–98. doi: 10.1177/10454411990100010401. [DOI] [PubMed] [Google Scholar]
- [23].Fincham AG, Moradian-Oldak J. Recent advances in amelogenin biochemistry. Connect Tissue Res. 1995;32:119–24. doi: 10.3109/03008209509013713. [DOI] [PubMed] [Google Scholar]
- [24].Hatakeyama J, Philp D, Hatakeyama Y, Haruyama N, Shum L, Aragon MA, Yuan Z, Gibson CW, Sreenath T, Kleinman HK, Kulkarni AB. Amelogenin-mediated regulation of osteoclastogenesis, and periodontal cell proliferation and migration. J Dent Res. 2006;85:144–9. doi: 10.1177/154405910608500206. [DOI] [PubMed] [Google Scholar]
- [25].Mauney JR, Kaplan DL, Volloch V. Matrix-mediated retention of osteogenic differentiation potential by human adult bone marrow stromal cells during ex vivo expansion. Biomaterials. 2004;25:3233–43. doi: 10.1016/j.biomaterials.2003.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.