Abstract
During Drosophila melanogaster oogenesis, the proper localization of gurken (grk) mRNA and protein is required for the establishment of the dorsal-ventral axis of the egg and future embryo. Squid (Sqd) is an RNA-binding protein that is required for the correct localization and translational regulation of the grk message. We show that Cup and polyA-binding protein (PABP) interact physically with Sqd and with each other in ovaries. We show that cup mutants lay dorsalized eggs, enhance dorsalization of weak sqd alleles, and display defects in grk mRNA localization and Grk protein accumulation. In contrast, pAbp mutants lay ventralized eggs and enhance grk haploinsufficiency. PABP also interacts genetically and biochemically with Encore. These data predict a model in which Cup and Sqd mediate translational repression of unlocalized grk mRNA, and PABP and Enc facilitate translational activation of the message once it is fully localized to the dorsal-anterior region of the oocyte. These data also provide the first evidence of a link between the complex of commonly-used trans-acting factors and Enc, a factor that is required for grk translation.
Keywords: Drosophila, oogenesis, gurken mRNA, translational control, axis specification
INTRODUCTION
Many cells display inherent asymmetries, and polarity is often accompanied by restricting the expression of certain mRNAs to specific regions of the cell by localizing the RNAs and regulating their translation. In Drosophila melanogaster oogenesis, RNA localization followed by localized translation plays an important role in the establishment of the major body axes of the egg and future embryo. Localization of gurken (grk) mRNA during mid-oogenesis establishes the dorsoventral axis, and localization of oskar (osk) and bicoid (bcd) mRNAs result in the formation of the embryonic anterior-posterior axis (reviewed in Huynh and St Johnston, 2004). Similar to localized RNAs in other systems, these RNAs are packaged into large protein complexes that facilitate both their microtubule-dependent transport and their translational control (Chekulaeva et al., 2006; reviewed in Johnstone and Lasko, 2001; and Wilhelm and Smibert, 2005).
Dorsoventral asymmetries in the Drosophila egg are easily observed in the eggshell. The dorsal surface of the egg is marked by two respiratory appendages, and the ventral surface is much more rounded than the dorsal side. The asymmetries seen in the mature egg are initiated during oogenesis as the egg chamber develops (reviewed in Ray and Schupbach, 1996). Dorsal fate is established when the epidermal growth factor receptor (Egfr) is activated by the TGF-α like ligand, Gurken (Grk); (Neuman-Silberberg and Schupbach, 1993; Price et al., 1989; Schupbach, 1987). Egfr is expressed uniformly in the follicle cells overlying the oocyte and nurse cells (Sapir et al., 1998). In contrast, grk mRNA is tightly localized to the future dorsal-anterior region of the oocyte to produce a local supply of ligand. As a result, Egfr is activated in only a subset of follicle cells, and these specific cells adopt dorsal fates (Neuman-Silberberg and Schupbach, 1993; Neuman-Silberberg and Schupbach, 1996; Nilson and Schupbach, 1999).
The distribution of Grk is controlled at the level of both RNA localization as well as translational control, and mutants have been identified that disrupt both processes. In squid (sqd) and fs(1)K10 (K10) mutants, grk mRNA is mislocalized along the entire anterior cortex of the oocyte, and the mislocalized RNA is translated, resulting in ectopic activation of Egfr and dorsalized eggshells (Kelley, 1993; Wieschaus, 1979; Wieschaus et al., 1978). Sqd is a heterogeneous nuclear ribonucleoprotein, or hnRNP, a family of proteins that has been implicated in many processes including RNA processing and transport (and Dreyfuss et al., 2002; reviewed in Dreyfuss et al., 1993) and whose members are often able to shuttle between the nucleus and cytoplasm (Michael et al., 1997; Mili et al., 2001; Pinol-Roma and Dreyfuss, 1992; reviewed in Shyu and Wilkinson, 2000).
Previous studies have shown that Sqd is required for the regulated nuclear export, cytoplasmic localization, and translational control of grk mRNA and have led to a model for Sqd in grk expression (Goodrich et al., 2004; Norvell et al., 1999). In this model, grk mRNA is transcribed and processed in the nucleus, and assembled into a protein complex that contains Sqd and other RNA processing factors, such as Hrb27C/Hrp48. The grk message is exported from the nucleus, where Sqd is required to mediate translational repression of grk until the mRNA is properly localized. This repression is reversible, however, and upon localization to the dorsal-anterior region of the oocyte, grk mRNA is translated and the protein is available to activate Egfr in the overlying follicle cells.
In contrast to sqd and K10 mutants, in which both grk RNA localization and translational repression are disrupted, mutations in encore (enc) result in a partial mislocalization of grk mRNA, yet the RNA is maintained in a state of translational repression (Hawkins et al., 1997). Enc protein co-localizes with grk mRNA (Van Buskirk et al., 2000), and taken together, these data suggest that Enc assists in mediating the translational activation of grk mRNA once it is fully localized to the dorsal-anterior region of the oocyte. However, a specific mechanism for Enc's function in grk translational activation has not been described.
Precise spatial expression of many proteins is accomplished by physical localization of the RNA as well as tight translational control of the message (Dreyfuss et al. 2002). Unique expression patterns can be generated by the integration of input from RNA-specific and more general translation control factors. For instance, Smg and Glo bind specifically to the nanos translational control element, however, Smg exerts its effect, at least in part, through interactions with the eIF4E-interacting protein Cup (Kalifa et al., 2006; Nelson et al., 2004). Furthermore, Sqd and Hrb27C/Hrp48 are involved in the proper expression of both grk and osk transcripts, though the requirement for Sqd seems to be much more important for grk (Goodrich et al., 2004; Huynh et al., 2004; Kelley, 1993; Norvell et al., 2005; Norvell et al., 1999; Steinhauer and Kalderon, 2005; Yano et al., 2004). To identify additional factors that mediate the translational control of grk RNA, we have taken a direct approach using mass spectrometry to identify proteins that interact with Sqd protein in ovaries. Cup and PABP (CG5119 at 55B, hereafter referred to as PABP55B to distinguish this factor from the two other PABP-like proteins in Drosophila) were found to interact with Sqd and we have obtained several lines of evidence showing that both factors are involved in the regulation of grk translation. Cup encodes a novel protein, and recent studies have shown that Cup is important for mediating the translational repression of osk and nanos (nos) mRNAs by preventing translation initiation (Chekulaeva et al., 2006; reviewed in Wilhelm and Smibert, 2005). In contrast, PABP is thought to mediate translational activation by interacting with the translational initiation machinery (reviewed in Mendez and Richter, 2001; and Richter and Sonenberg, 2005), but a specific role for PABP in Drosophila oogenesis has not been previously described.
Here we report the isolation of Cup and PABP through biochemical interactions with Sqd. We show that these interactions are sensitive to RNase A digestion and are therefore likely bridged by an RNA molecule. We confirm the functional significance of these biochemical data by demonstrating that cup mutants lay dorsalized eggs, and pAbp55B mutants lay ventralized eggs. These data are consistent with a severe perturbation of grk signaling. In addition, we observed genetic interactions between pAbp55B and grk as well as between cup and sqd. cup mutants also display improper Grk protein accumulation and compromised grk mRNA localization in the cytoplasm. Finally, we have demonstrated a genetic interaction between pAbp55B and enc. Taken together, these data support a model in which the regulation of grk expression occurs in the context of multi-subunit, dynamic RNA-protein complexes. In this model, Cup functions with Sqd and Hrb27C/Hrp48 in a protein complex that mediates the translational repression of grk mRNA before it is properly localized. We hypothesize that once the RNA has reached the future dorsal-anterior region of the oocyte, PABP55B and Enc facilitate the translational activation of grk mRNA. Significantly, the PABP55B-Enc interaction is the first direct biochemical link between a factor that is specifically required for the regulation of grk RNA and the complex containing factors (Cup, PABP55B, and Sqd) that function more generally in the regulation of multiple RNAs during oogenesis.
MATERIALS AND METHODS
Fly stocks
Akira Nakamura provided the cupΔ212 allele, which was generated by mobilizing the P element in cup4506 (Keyes and Spradling, 1997) and resulted in a deletion of the N-terminal third of the Cup protein product, including most of the eIF4E-binding domain (Nakamura et al., 2004). cup5 and cup20 are EMS-generated alleles (Schupbach and Wieschaus, 1991). pAbpk10109 is a lethal P-element insertion residing in the 5’ UTR of pAbp55B, 102 base pairs upstream of the start codon, and precise excision of the P{lacW} element restores viability and reverts the morphological phenotypes of pAbpk10109 heterozygotes to wild type (Sigrist et al., 2000). pAbpEY11561 is a viable P-element insertion in the 5’ UTR of the RA, RF, and RH transcripts of pAbp55B, 762 base pairs upstream of the start codon.
Germline clones of pAbpk10109 were generated using the FLP-DFS (yeast flipase recombination target-site specific recombinase-dominant female sterile) system previously described (Chou and Perrimon, 1992; Chou and Perrimon, 1996). Progeny from yw hsFLP; ovoD FRTG13 / CyO x FRTG13-pAbpk10109 / CyO were heat shocked at 37°C for 2 hours a day for 4 days during the second and third larval instar. sqd1 is a P-element insertion that specifically disrupts germline expression during oogenesis (Kelley, 1993; Matunis et al., 1994; Norvell et al., 1999). grkHF48 (Thio et al., 2000) is an EMS-generated null allele. encQ4 and encUU3 are strong, EMS-generated hypomorphic alleles and encr17 was generated by P-element excision (Hawkins et al., 1997).
Eggs were collected and eggshell morphology was determined as previously described (Schupbach and Wieschaus, 1991). Eggshell characterization represents multiple independent collections scored by two investigators.
Antibodies
The following antibodies were used: monoclonal α-Grk serum (ID12; 1:10 dilution for immunohistochemistry) (Queenan et al., 1999), monoclonal α-Sqd serum (8F3; 1:7 dilution for IP, 1:200 for western blot) (Goodrich et al., 2004), monoclonal α-Dorsal serum (7F12; 1:7 dilution for IP) (provided by Ruth Steward), monoclonal α-pAbp55B ascites (6E2; 1:140 dilution for IP, 1:1000 for western blot) (Matunis et al., 1992), polyclonal α-Cup (#219; rat against amino acids 1−225; 1:70 dilution for IP, 1:10,000 for western blot) (Nelson et al., 2004), polyclonal α-Cup (rabbit against amino acids 597−975; 1:28 dilution of affinity purified antibody for IP, 1:1000 dilution of non-affinity purified serum for western blot) (Nakamura et al., 2004), polyclonal α-Cup (#27; rabbit; 1:70 dilution for IP, 1:500 for western blot) (Verrotti and Wharton, 2000), polyclonal α-Enc (rabbit and rat against amino acids 133−340; 1:28 dilution of rabbit for IP, 1:1000 dilution of rat for western blot) (Van Buskirk et al., 2000), polyclonal α-Hrb27C/Hrp48 (rabbit; 1:20 dilution for IP)(Siebel et al., 1994), monoclonal α-CycB (F2; 1:20 dilution for IP, 1:30 for western blot)(Knoblich and Lehner, 1993), polyclonal α-slbo (rat; 1:500 for immunohistochemistry), (Jekely et al., 2005), HRP-conjugated goat α-mouse (1:10,000; Jackson ImmunoResearch), HRP-conjugated goat α-rabbit (1:7500, Pierce), and HRP-conjugated goat α-rat (1:10,000, Jackson ImmunoResearch).
Immunoprecipitations
Immunoprecipitations were performed as described (Van Buskirk et al., 2000) with the following modifications: instead of adding protease inhibitors individually to the lysis buffer, a complete mini protease inhibitor cocktail tablet was used (Roche); ovarian lysates were not pre-cleared with pre-immune serum, but were sometimes pre-cleared with Protein A/G agarose beads (Santa Cruz Biotechnology) for 30 minutes at 4°C; and ovarian lysates were first incubated with antibodies for 1−2 hours at 4°C and then beads were added to the mixture for an additional 1−2 hours at 4°C. Beads were rinsed 4× in cold lysis buffer before electrophoresis. When indicated, RNase A was included in the lysis and wash buffers at a final concentration 1 mg/mL.
For immunoprecipitations performed for mass spectrometry analysis, the gels were fixed in 7% acetic acid + 10% methanol for 30 minutes, incubated in Sypro Ruby Protein Gel Stain (Molecular Probes) overnight, and destained in fix solution for 1 hour prior to UV visualization. Gel slices containing Sqd interacting proteins were analyzed by the Princeton SynSeq facility.
For immunoprecipitations performed for western blotting, the samples were transferred to nitrocellulose (Amersham), blocked in TBST (Tris-buffered saline + 0.1% Tween-20) + 5% milk + 1% BSA overnight at 4°C, and incubated in primary antibody for 1−2 hours at room temperature. Blots were washed in TBST for 2−3 hours and incubated in secondary antibody for one hour at room temperature, washed in TBST for 2−3 hours, and detected by ECL (Amersham or Pierce).
Immunohistochemistry
Ovaries were dissected in ice cold 1× PBS and fixed for 20 minutes at room temperature in 4% paraformaldehyde in PBST (PBS + 0.3% Triton X-100) and heptane. Ovaries were rinsed three times with PBST and blocked in PBS + 1% Triton X-100 + 2.5% BSA for 1 hour at room temperature. Ovaries were incubated for one hour at room temperature in 1:500 α-slbo and/or 1:10 α-Grk diluted in block buffer and washed overnight at 4°C in PBST. Ovaries were incubated for one hour at room temperature in block buffer containing AlexaFluor 488-conjugated goat α–rat and/or AlexaFluor 568-conjugated goat α-mouse secondary (Molecular Probes) diluted 1:1000 + Hoechst diluted 1:10,000 + Oregon Green or AlexaFluor 633-conjugated phalloidin (Molecular Probes) diluted 1:1000. Ovaries were washed for 2−3 hours at room temperature in PBST and mounted in 1:1 PBS-glycerol or Aqua Poly/Mount (Polysciences, Inc).
In situ hybridization
Ovaries were dissected in ice cold 1× PBS and fixed for 20 minutes at room temperature in 4% paraformaldehyde in PBS + heptane + DMSO. Subsequent steps were performed as previously described (Tautz and Pfeifle, 1989) using a grk RNA probe.
RESULTS
Sqd co-purifies with Cup and PABP55B in ovarian extracts
In order to elucidate the role of Sqd in the regulation of grk expression, we prepared ovarian extracts and performed immunoprecipitations using either an α-Sqd antibody or a negative control antibody. Eleven bands were pulled down using the α-Sqd antibody but not by the negative control antibody (Figure 1A, arrows). The identities of these bands were determined by mass spectrometry. One band of 150 kDa was identified as the translational repressor Cup, and a 65 kDa protein was identified as the translational activator polyA binding protein (PABP55B, encoded by CG5519). The negative control antibody, α-Dorsal (α-Dl) was used because it is a monoclonal antibody that was generated in the same facility as α-Sqd, but Dl is not expressed during oogenesis and therefore should not specifically pull down any ovarian factors.
Figure 1. Cup and PABP55B interact with Sqd in ovarian extracts.
(A) Immunoprecipitations were performed out of ovarian lysates using either α-Sqd or a negative control antibody, α-Dorsal. Specific Sqd interactors (marked by arrows) were excised from the gel and sequenced by mass spectrometry. Positively identified bands are labeled. (B-E) Immunoprecipitations were performed in the presence or absence of RNase (if indicated) using α-Sqd, α-Cup, α-PABP55B, or α-Dorsal. The lysate lane represents 5% of the sample probed for interactions. Western blots were probed with α-PABP55B (B and E) or α-Cup (C-E). These experiments demonstrate a strong interaction between Sqd, Cup, and PABP55B. (E) Additional immunoprecipitations were performed including α-Hrb27C/Hrp48 and probed with α-Cup and α-PABP55B. In addition to the regular exposure of the Cup western, an extended exposure is shown to clearly demonstrate the somewhat weaker interaction of Hrb27C/Hrp48 with Cup.
To verify the identity of the bands sequenced by mass spectrometry, we performed co-immunoprecipitations using antibodies that recognize Sqd (Goodrich et al., 2004), Cup (Keyes and Spradling, 1997; Nakamura et al., 2004; Nelson et al., 2004; Verrotti and Wharton, 2000), or PABP55B (Matunis et al., 1992). In these assays, α-Sqd, but not α-Dl, immunoprecipitates Cup and PABP55B in an RNA-dependent manner (Figure 1B and C), confirming the results of the mass spectrometry analysis. We further characterized the approximate stoichiometry of the Sqd/Cup interaction and found that sqd is complexed with approximately 5−7% of the Cup in ovarian lysates (supplemental Figure 1). Given that Cup is known to regulate other RNAs, this level of interaction is significant. We also wanted to determine whether the interaction between Sqd and Cup or PABP55B is specific for one of the Sqd isoforms (Kelley, 1993), so we used an antibody that recognizes the HA epitope to perform immunoprecipitations out of ovarian extracts expressing either HA-SqdA or HA-SqdS. In these experiments, Cup and PABP55B were able to interact with both Sqd isoforms, indicating that these interactions are not isoform-specific (data not shown). Given that the hnRNP Hrb27C/Hrp48 interacts with Squid (Goodrich et al., 2004), we tested whether it also interacts with Cup and PABP55B. Indeed we were able to immunoprecipitate both Cup and PABP55B with antibodies against Hrb27C/Hrp48 (Fig. 1E).
cup and PABP55B females lay eggs with dorsoventral patterning defects
Cup has been shown to interact with other proteins in oogenesis, and cup mutations have multiple phenotypes, indicating that Cup regulates a number of targets in oogenesis (Keyes and Spradling, 1997; Nakamura et al., 2004; (Wilhelm et al., 2003; Zappavigna et al., 2004). To determine whether the physical interactions between Cup, Squid and PABP were relevant to grk function, we examined the eggs laid by pAbp55B or cup mutant females and observed that these eggs display dorsoventral patterning defects. We obtained two P-element insertion alleles in the 5’ UTR of pAbp55B (pAbpk10109 and pAbpEY11561). The pAbpk10109 insertion is homozygous and hemizygous lethal whereas the pAbpEY11561 insertion is homozygous viable. We examined the eggs that were laid by pAbpk10109/pAbpEY11561 trans-heterozygous and pAbpEY11561 homozygous females. Significantly, these females lay 3.9 ±0.7% (n=355) and 8.4 ±1.2% (n=286) ventralized eggs respectively. These eggs are characterized by partial or complete fusion of the normally distinct dorsal appendages (Figure 2A compared to 2B). This phenotype is consistent with a reduction in Grk protein accumulation at the dorsal-anterior of the oocyte and was consistently observed. While this phenotype is not direct measure of Grk translation, the biochemical and genetic tests described below further demonstrate that the phenotype is a clear indication of a ventralization of the egg chambers.
Figure 2. cup and pAbp55B females lay eggs with dorsoventral patterning defects.
Most pAbpEY11561 / pAbpk10109 and pAbpEY11561 eggs display wild-type morphology (A), but approximately 4% of eggs laid by pAbpEY11561 / pAbpk10109 females and 8% of eggs laid by pAbpEY11561 homozygous females (B) exhibit partially or completely fused dorsal appendages. A small percentage of cup5 eggs (n=135) have wild-type morphology (A), but approximately 95% of eggs laid by cup5 / cup5 females have thick, fused dorsal appendages (C), a crown of dorsal appendage material (D), or open chorions (E).
Since the strong pAbpk10109 allele is lethal, we produced germline clones (Chou and Perrimon, 1996) to determine whether Grk protein expression is reduced in egg chambers with a germline homozygous for pAbpk10109. Unfortunately, these germline clones were cell-lethal and did not permit the growth of egg chambers past stage 3 (data not shown). This demonstrates that there is an essential requirement for pAbp55B early in oogenesis. It is not likely that the pAbpEY11561 homozygous egg chambers would demonstrate a detectable reduction in Grk levels given the low penetrance of the eggshell ventralization phenotype that we observed with this much weaker allele of pAbp55B. Unfortunately this prevented the further analysis of Grk protein or grk mRNA in mid-oogenesis in germ cells homozygous for pAbp55B mutations.
In contrast, homozygous cup5 females lay 94.5 ± 7.8% (n=135) dorsalized eggs, characterized by either one broad dorsal appendage or a crown of dorsal appendage material encircling the anterior of the egg (Figure 2C-E). cupΔ212 seems to be a weaker allele, as homozygous females lay 95 ± 4.9% (n=398) eggs with broad, fused appendages, a weaker phenotype than a crown of appendage material (data not shown). Both of these phenotypes are reminiscent of mutations in sqd and Hrb27C/Hrp48 (Norvell et al., 1999), and are consistent with ectopic expression of Grk protein in the oocyte. However, the phenotype also suggests that peak levels of grk signaling are not achieved in the mutant, since the dorsal midline fate is often missing.
Grk protein is inefficiently localized in cup ovaries
To test whether the eggshell phenotypes seen in cup mutants were due to defective grk expression, we monitored Grk protein by indirect immunofluorescence. Consistent with the dorsalized eggshells, Grk protein accumulates inefficiently in cup mutants (Figure 3). cup mutants exhibit a range of oogenesis defects, such as abnormally small oocytes and nurse cell chromosomes that fail to properly disperse (Keyes and Spradling, 1997), therefore, defining the stage of these egg chambers was extremely difficult. For this reason, the egg chambers were not staged according to traditional definitions in these experiments, but instead Grk localization was analyzed and scored in every egg chamber in which the oocyte nucleus had achieved an asymmetric localization within the oocyte, indicating that the egg chamber had at least reached stage 8 of oogenesis. In the wild-type controls, only stage 8 and 9 egg chambers were counted, whereas stage 10 and older egg chambers were disregarded because they were overrepresented in the wild-type samples relative to stage 10 egg chambers in cup alleles. Omitting stage 10 egg chambers from wild-type scoring increases the percentage of egg chambers displaying unlocalized Grk in the wild-type control, thus increasing the stringency of this control.
Figure 3. Grk protein is not properly localized in cup mutants.
OregonR, cup5, and cupΔ212 egg chambers were stained for Grk (red), F-actin (green), and DNA (blue). Grk protein distribution was categorized as either dorsal-anterior only (A), dispersed throughout the oocyte with a dorsal-anterior bias (B), or evenly dispersed throughout the ooplasm (C). +/− indicates standard deviation of multiple microscopy sessions. Binomial probabilities for the frequency of each classification in cup mutants were calculated relative to OregonR. *, p < 0.05; ** p < 0.01.
In scoring, Grk localization was categorized as either localized properly to the future dorsal-anterior of the oocyte (Figure 3A), dispersed throughout the oocyte, but somewhat accumulated at the future dorsal-anterior (Figure 3B), or evenly dispersed throughout the oocyte (Figure 3C). Grk protein was localized to the future dorsal-anterior of about 86% (n=548) of wild-type egg chambers, in contrast to 31% (n=196) of cup5 and 61% (n=163) of cupΔ212 egg chambers. The degree of severity of this defect was greater for cup5 mutants than for cupΔ212 mutants, which is also consistent with the degree of severity of the eggshell phenotypes (Figure 3).
We also further confirmed this phenotype by staining egg chambers homozygous for cup5 with the antibody to Grk as well as an antibody to the Slbo protein (Jekely et al., 2005). By assessing the location of the border cells, which migrate through the nurse cell cluster during stage 9 of oogenesis, we were able to stage the cup5 egg chambers by this independent criterion. The squamous morphology of the follicle cells associated with the nurse cells was also used to indicate whether the egg chambers were in early or late stage 9. However, very few egg chambers ever reached late stage 9 in the cup5 homozygous mutant. Nevertheless, a mislocalization of Gurken protein could be seen in these egg chambers, whereas in the wildytpe control, Grk protein was always dorsally localized (data not shown).
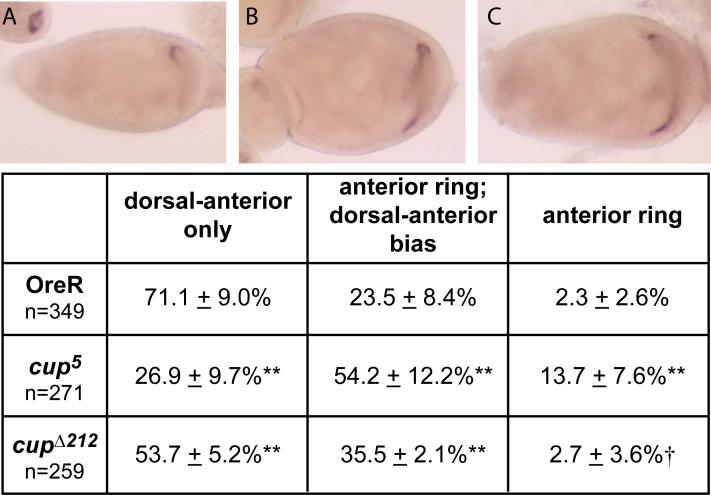
grk mRNA is localized less efficiently in cup mutants
In order to determine whether the ectopic Grk protein expression is a result of unrestrained translation alone, we analyzed the localization of grk mRNA in cup mutants (Figure 4). In contrast to wild-type egg chambers, in which about 71% (n=349) of stage 8−9 egg chambers demonstrate grk mRNA localized to the future dorsal-anterior of the oocyte, only 27% (n=271) of cup5 and 54% (n=259) of cupΔ212 egg chambers display this localization. Consistent with the eggshell phenotypes and Grk protein localization data, grk mRNA localization was less severely disrupted in cupα212 than in cup5 (Figure 4). This might suggest that Cup is required for grk mRNA localization, however, this effect of cup mutants may also be indirect. Perhaps removing Cup from the localization/repression complex built upon grk mRNA compromises the stable architecture of the complex, resulting in less efficient localization.
Figure 4. grk mRNA is localized less efficiently in cup mutants.
In situ hybridization using a grk RNA probe was performed on OregonR, cup5, and cupΔ212 egg chambers. grk mRNA distribution was categorized as either dorsal-anterior only (A), an anterior ring with a dorsal-anterior bias (B), or an anterior ring (C). +/−indicates standard deviation of multiple microscopy sessions. Binomial probabilities for the frequency of each classification in cup mutants were calculated relative to wild type. †, p = 0.16; **, p < 0.01.
Importantly, the grk mRNA localization data show that Cup is required for the translational repression of unlocalized grk mRNA. This effect is also seen in sqd mutants (Norvell et al., 1999), and is in contrast to many ventralizing mutants in which grk mRNA is mislocalized but remains in a translationally repressed state, such as encore or the spindle class genes (Ghabrial et al., 1998; Gonzalez-Reyes et al., 1997; Hawkins et al., 1997).
Genetic and biochemical interactions with cup and PABP55B
Females transheterozygous for a weak allelic combination of sqd (sqd1/sqdk12) lay only 13% wild-type eggs (n=355), and the remaining eggs display mild to strong dorsalized phenotypes. Females homozygous for the weak allele sqdk12 lay only 5% weakly dorsalized eggs (n=160), but the frequency and severity of dorsalization in both sqd allelic combinations was dramatically enhanced by heterozygosity for cup20 (Figure 5), a strong cup allele (Keyes and Spradling, 1997). This effect has been observed over multiple independent experiments, and representative data are shown (Table 1). This synergistic genetic interaction presents further evidence that the observed Cup-Sqd biochemical interaction is functionally relevant for grk translational repression.
Figure 5. sqd interacts genetically with cup, and grk interacts genetically with pAbp.
Heterozygosity for cup20 enhances the moderately dorsalized phenotype of sqdk12 / sqd1 transheterozygotes and of sqdk12 / sqdk12 at 29°C (A). Eggs were characterized as either wild-type like, weakly dorsalized (single, broad fused appendage), moderately dorsalized (widely-spaced appendages), or strongly dorsalized (crown of appendage material). In addition, grkHF48 / pAbpk10109 transheterozygotes lay an increased percentage of and more severely ventralized eggs at 25°C than does either heterozygote alone (B). Eggs were characterized as either wild-type like, weakly ventralized (appendages fused at the base), moderately ventralized (single, slender fused appendage), or strongly ventralized (no appendage material). +/− indicates standard deviation of multiple egg collections scored by independent investigators. Binomial probabilities for the frequency of each eggshell classification were calculated for cup20/+ ; sqdk12 / sqd1 relative to sqdk12 / sqd1, cup20/+ ; sqdk12 / sqdk12 relative to sqdk12 / sqdk12, and grkHF48 / pAbpk10109 relative to grkHF48 / +. All p-values were less than 0.01.
Females heterozygous for a null allele of grk, grkHF48, lay 3% weakly ventralized eggs (n=1146), and reducing pAbp55B by one copy was able to greatly enhance this ventralization (n=1212, Figure 5). This synergistic genetic interaction is consistent with PABP55B functioning positively in grk translation.
Immunoprecipitation experiments show that α-PABP55B is able to specifically pull down Cup protein (Figure 1D and E) and α-Cup pulls down PABP55B (Figure 1E). Considering that Cup and PABP55B interact biochemically with each other and with Sqd, cup and pAbp55B females lay eggs with opposite phenotypes, and cup-sqd and pAbp55B-grk interact genetically, we propose that Cup and PABP55B work antagonistically to regulate grk mRNA expression.
PABP55B interacts with Encore, a potential cytoplasmic scaffolding protein
Encore (Enc) is a large, novel, cytoplasmic protein that co-localizes with grk mRNA (Van Buskirk et al., 2000) and is required for proper grk mRNA localization (Hawkins et al., 1997). In contrast to sqd and K10 mutants, in which unlocalized grk mRNA is translated and dorsalized eggs are laid (Kelley, 1993; Norvell et al., 1999; Wieschaus et al., 1978), unlocalized grk mRNA in enc mutants is maintained in a translationally repressed state and ventralized eggs are laid (Hawkins et al., 1997). These data are consistent with a role for Enc in the translational activation of grk mRNA.
Enc has previously been shown to be a large cytoplasmic protein that interacts with subunits of the proteasome in early oogenesis (Ohlmeyer and Schupbach, 2003; Van Buskirk et al., 2000). Considering its function early in oogenesis, and given its cortical localization in the oocyte during mid-oogenesis, Enc may act as a scaffolding protein mediating the transition from translational repression to activation of grk mRNA. To test this hypothesis, we performed immunoprecipitations and showed that α-Enc specifically immunoprecipitates the translational activator PABP55B. Furthermore, the data suggest that the interaction may be direct and not bridged by an RNA molecule, as α-Enc is able to immunoprecipitate PABP55B even when RNase A is added to the extract (Figure 6A).
Figure 6. pAbp55B interacts with Enc biochemically and genetically.
(A) Immunoprecipitations were performed out of ovarian lysates in the presence or absence of RNase using α-Enc or α-Dorsal. The lysate lane represents 5% of the sample probed for interactions. Western blots were probed with α-PABP55B. (B) Heterozygosity for pAbpk10109 enhances the weakly ventralized phenotype of encQ4 / encQ4 and of encUU3/encUU3 homozygotes at 25°C, increases the percentage of collapsed eggs, and decreases the overall number of eggs laid. Eggs were characterized as either wild-type like, weakly ventralized (appendages fused at the base), moderately ventralized (single, slender fused appendage), strongly ventralized (no appendage material), or collapsed. Binomial probabilities for the frequency of each eggshell morphology classification were calculated for encQ4 / encQ4 ; pAbpk10109 / + relative to encQ4 / encQ4 mutants and encUU3 / encUU3 ; pAbpk10109/+ relative to encUU3 / encUU3 mutants. †, p > 0.05; *, p < 0.05; **, p < 0.01.
In addition to the biochemical interaction, pAbp55B and enc also interact genetically. enc mutants are cold-sensitive for ventralization (Hawkins et al., 1997) and display much weaker ventralization at 25°C than at 18°C. However, heterozygosity for pAbpk10109 enhances the both the frequency and severity of ventralized eggs laid by enc homozygotes at 25°C (Figure 6B). Although this enhancement is a subtle phenotype, the effect has been observed reproducibly over several independent experiments using two different enc alleles (encQ4 and encUU3). This genetic interaction suggests that the PABP55B-Enc biochemical interaction is functionally relevant in grk expression.
In addition to the enhanced ventralization, heterozygosity for pAbpk10109 dramatically reduces the number of eggs that are laid by enc homozygotes and increases the percentage of collapsed eggs for which the eggshell phenotype cannot be determined. In fact, heterozygosity for pAbpk10109 causes encr17 and encR1 homozygous females to lay no eggs. Taken together, the genetic interaction and RNA-independent physical association between Enc and PABP55B suggest that these proteins may function together to activate translation of grk mRNA.
DISCUSSION
Sqd binds to RNA localization and translation factors in ovarian extracts
We have taken a direct approach to identify proteins that interact with Sqd protein in ovaries. Using a Sqd antibody, we performed immunoprecipitations out of ovarian extracts, isolated proteins that specifically interacted with Sqd, and identified those proteins by mass spectrometry. Four of the Sqd-interacting proteins were positively identified in the mass spectrometry analysis: Cup, PABP55B, Imp, and Hrb27C/Hrp48. The remaining bands were not identified with certainty. Imp and Hrb27C/Hrp48 are two factors that have previously been shown to be involved in RNA localization (Geng and Macdonald, 2006; Goodrich et al., 2004; Munro et al., 2006), and both Hrb27C/Hrp48 and Imp bind to grk mRNA (Geng and Macdonald, 2006; Goodrich et al., 2004). The identification of these two factors confirmed that the immunoprecipitation method could successfully identify functional Sqd interactors.
Cup and translational repression
One of the Sqd interactors identified in the mass spectrometry analysis was the novel 150 kDa protein Cup. cup mutants display egg chambers with nurse cell nuclear morphology defects and eggs with open chorions (Keyes and Spradling, 1997). Cup interacts with several factors known to be required for osk localization and translation, such as Exu, Yps, eIF4E, Me31B, and Bruno (Nakamura et al., 2004; Wilhelm et al., 2003; Zappavigna et al., 2004), and independent studies have shown that osk mRNA is prematurely translated in cup mutants (Chekulaeva et al., 2006; Nakamura et al., 2004; Wilhelm et al., 2003). Cup co-localizes with the cap-binding protein, eIF4E, and eIF4E is not properly localized to the oocyte posterior pole in cup mutants (Wilhelm et al., 2003; Zappavigna et al., 2004). Cup competes away eIF4G, another translation initation factor, for binding to eIF4E (Nelson et al., 2004; Zappavigna et al., 2004), thereby repressing translation. Together, these data are consistent with the following model for Cup-mediated translational repression; Cup represses the translation of RNAs containing BREs through interactions with Bruno. In this complex Cup binds directly to eIF4E and interferes with eIF4G binding to eIF4E. Because eIF4G binding to eIF4E is a prerequisite for translation initiation, Cup represses translation by blocking this interaction (reviewed in Richter and Sonenberg, 2005). Direct biochemical data supporting this model have recently been obtained by Checkulaeva et al. (2006). We propose that Cup represses grk translation by a similar mechanism prior to its localization to the dorsal anterior of the oocyte.
Cup activity is used by several transcript-specific factors to mediate translational repression of that RNA in a developmentally appropriate context. For instance Cup is required to mediate the translational repression of the nanos (nos) transcript. Cup has been shown to interact with Nos protein, and co-localizes with Nos in the germarium. cup and nos also interact genetically, as heterozygosity for cup suppresses nos-induced phenotypes in early oogenesis (Verrotti and Wharton, 2000). Later in development, Cup binds to Smaug, a factor that specifically binds to nos RNA and is required for its translational repression in embryos (Crucs et al., 2000; Dahanukar et al., 1999; Smibert et al., 1996). In this example, Cup is required for Smaug to interact with eIF4E and mediate nos repression. Consistent with this biochemical model, Smaug-mediated translational repression is less efficient in cup mutants (Nelson et al., 2004).
Here, we have shown that Cup is also required for grk translational repression. This contrasts with previous reports that grk expression is normal in cup mutants (Nakamura et al., 2004; Wilhelm et al., 2003), but these earlier reports used relatively weak cup alleles and monitored Grk levels by immunofluorescence. In contrast, we were able to use alleles that allowed us to assess the eggshell phenotype in cup mutants, which provides the most sensitive assay for defects in Grk levels. Our own analyses showed that the different cup alleles vary greatly in phenotypic strength and range of phenotypes (data not shown).
Using two different alleles of cup from two distinct genetic backgrounds, we have shown that cup mutants lay dorsalized eggs, display defects in Grk protein accumulation, and display less efficient grk mRNA localization. Furthermore, Cup interacts biochemically with Sqd and Hrb27C/Hrp48 in ovarian extracts. Finally, heterozygosity for cup is able to enhance the moderate dorsalization observed in weak allelic combinations of sqd. Together, these data strongly support a model in which Cup functions with Sqd and Hrb27C/Hrp48 to mediate the translational repression of the grk message (Figure 7).
Figure 7. Model for the role of Sqd, Cup, PABP55B, and Enc in grk expression.

Before grk mRNA is fully localized to the dorsal-anterior region of the oocyte, translational repression is mediated by Sqd, Hrb27C/Hrp48, Otu and Cup. Once the RNA is localized, PABP55B mediates translational activation of the localized message. Enc, bound to PABP55B, could function in association with a cytoplasmic anchor to mediate the transition from translational repression to activation of grk mRNA.
PABP55B and translational activation
Once grk mRNA is properly localized to the future dorsal/anterior of the oocyte, translational control must be switched from repressive to promoting. In many cellular situations, this activation is accomplished by binding of PABP to polyA tails of transcripts. In fact, PABP55B contains four RNA-recognition motifs (RRMs) that directly bind to polyA tails (Preiss and Hentze, 1999). PABP55B also has a C-terminal polyA domain that is used for oligomerization of PABP55B on polyA tails (Kuhn and Pieler, 1996). Once PABP55B is bound to RNA, it binds to eIF4G, and this interaction helps to increase the affinity of eIF4G for eIF4E. With this increased affinity, eIF4G is able to effectively compete with Cup for binding to eIF4E, and translation is able to begin (reviewed in Richter and Sonenberg, 2005; Tarun and Sachs, 1996; Tarun et al., 1997).
There are at least three polyA-binding proteins in the Drosophila genome (CG5119 at 55B, CG4612 at 60D, and CG2163 at 44B) which are predicted to function as general translation factors, so it is conceivable that PABP55B could regulate a subset of RNAs. CG2163 has also been designated as PABP2, and has been shown to have essential roles in germ line development and in early embryogenesis (Benoit et al., 2005). Here we have shown that PABP55B mediates the translational activation of fully localized grk mRNA. Specifically, heterozygous pAbp55B mutants lay ventralized eggs in certain genetic combinations, and heterozygosity for pAbp55B also enhances the weakly ventralized phenotype of grk heterozygotes, consistent with a role in translational activation of grk.
PABP55B functions with Enc to mediate translational activation of grk
We have also shown that PABP55B binds to Enc in ovarian extracts, and that this interaction may be direct and not bridged by an RNA molecule. Furthermore, heterozygosity for pAbp55B is able to enhance the weakly ventralized phenotype of enc mutants raised at 25°C. Taken together, the biochemical and genetic interactions suggest that PABP55B and Enc function together to mediate the translational activation of grk mRNA once it is localized to the dorsal anterior of the oocyte.
Previously, Enc has been shown to be required for activation of grk translation in mid-oogenesis (Hawkins et al., 1997). An effect on osk mRNA localization has also been previously observed in enc mutants, but it is unclear at what level this process is affected, or whether this effect is direct (Van Buskirk et al., 2000). In addition, Enc has been shown to interact with subunits of the proteasome early in oogenesis (Ohlmeyer and Schupbach, 2003). Because of its large size and its ability to interact with several different proteins, Enc may play multiple roles during oogenesis. Considering the function of Enc in grk translational activation (Hawkins et al., 1997) and its localization to the dorsal-anterior region of the oocyte (Van Buskirk et al., 2000), we hypothesize that Enc could function as a scaffolding protein that helps to mediate the transition from translational repression to activation of grk mRNA.
Sqd, Cup, PABP, and Enc mediate translational control of grk RNA
We have shown that Cup functions with Sqd in a protein complex that mediates the translational repression of grk mRNA before it is properly localized. It is clear from the analysis of mutants such as spn-F and encore, in which mislocalized grk mRNA is translationally silent, that these two steps can be uncoupled (Abdu et al., 2006; Hawkins et al., 1997). We propose that once the RNA has reached the future dorsal-anterior region of the oocyte, PABP, Sqd, and Enc facilitate the translational activation of grk mRNA (Figure 7). In Figure 7 PABP is shown associating with the complex once it is fully localized, however it is possible that PABP associates with the grk transport complex in an inactive form that is remodeled following its anchorage at the dorsal-anterior of the oocyte.
Previous studies have shown that Bruno (Bru) binds directly to Cup protein (Chekulaeva et al., 2006; Nakamura et al., 2004; Wilhelm et al., 2003) and is required for the translational repression of osk. Bru binds to specific sequence elements in the osk 3’ UTR called Bruno Response Elements (BREs), and mutations in these BREs have been shown to reduce Bru binding and result in ectopic Osk accumulation in the oocyte (Kim-Ha et al., 1995; Webster et al., 1997). Similarly, Bru has also been shown to bind to grk mRNA and to Sqd protein. Overexpression of bru cDNA leads to ventralization of the eggshell, consistent with reduced Grk protein expression in the oocyte. (Filardo and Ephrussi, 2003; Kim-Ha et al., 1995; Norvell et al., 1999). Furthermore, disrupting bru expression in certain genetic contexts has been shown to result in excess Grk protein in the oocyte, consistent with Bru being required to mediate grk translational repression (Yan and Macdonald, 2004). In light of the results presented here, we propose that this phenotype is the result of Bru-mediated repression of grk translation by Cup.
The mechanism of grk translation and the trans-acting factors required for translational control largely parallel the mechanism employed by osk RNA, so an important question to be answered is how these two different RNAs are differentially transported and translationally regulated in distinct parts of the oocyte at the appropriate stage in oogenesis. Since the same group of trans-acting factors is involved in the expression of both RNAs, the specificity could be provided by cis-acting sequences within the RNA molecules themselves that affect the activity of common trans-acting factors. Alternatively, RNA-specificity could be generated by as-yet unidentified trans-acting factors. Given that Enc functions in grk translational activation, but is not required for osk translational activation (Van Buskirk et al., 2000), it is possible that Enc is providing some degree of specificity to the commonly-used machinery that mediates translational control of multiple, unrelated transcripts. Currently, Enc is the only factor known to function uniquely in the translational activation of grk mRNA, and our results provide the first evidence of a link between this factor and the general translational control machinery that is used by multiple RNAs in oogenesis.
ACKNOWLEDGMENTS
The authors would like to thank Akira Nakamura, Craig Smibert, Allan Spradling, Robin Wharton, Pernille Rorth, Don Rio, Ruth Steward and Gideon Dreyfuss for providing antibodies and fly stocks; Joe Goodhouse for help with confocal microscopy and Saw Kyin for mass spectrometry analysis. We are very grateful to Amanda Norvell and Roshan Jain and Stas Shvartsman for critical comments on the manuscript and to members of the Schüpbach and Wieschaus laboratories for helpful discussions and suggestions. This work was supported by the Howard Hughes Medical Institute and US Public Health Service Grant PO1 CA41086 and RO1 GM077620; as well as a New Jersey Cancer predoctoral fellowship #03−2011-CCR-EO and the DOD fellowship DAMD17−03−1−0393 to KNC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Antibodies were used to immunoprecipitate Sqd or Cup from ovarian lysates. Cyclin B was used as a second negative control for immunoprecipitation in addition to the α-Dorsal antibody used in figures 1 and 6. 5% of the lysate probed for interactions was loaded in the input lane. Cup, Sqd, and CycB were detected by western blotting. Images were acquired by CCD imaging and pixel intensities in the bands were integrated using AlphaEaseFC software (Alpha Innotech). All signals were within the dynamic range of the camera. 16% (S.D. 6.7%) of the Sqd in the lysate was recovered in the IP. After correcting for the efficiency of the Sqd IP, 6% (S.D. 1.9%) of the Cup in ovarian lysates was found to be associated with Sqd. Data are the result of three independent experiments. No Sqd was detected in Cup or PABP immunoprecipitates (PABP data not shown). These data may indicate that the Sqd:Cup stoichiometry on grk transcripts is biased towards multiple Cup molecules for each Sqd molecule. Alternatively the Cup antibody may recognize an epitope that is required for interactions with Bru or other trans-acting factors necessary for the association of Cup with grk, and by association Sqd. In the case of PABP, which binds most poly-adenylated messages, the population of grk mRNA that comprises a PABP immunoprecipitate is expected to be quite small. Immunoprecipitation with α-Sqd antibodies, however, pulls down a more selective pool of RNA of which grk, and by association PABP and Cup, are a larger component.
REFERENCES
- Abdu U, Bar D, Schupbach T. spn-F encodes a novel protein that affects oocyte patterning and bristle morphology in Drosophila. Development. 2006;133:1477–84. doi: 10.1242/dev.02319. [DOI] [PubMed] [Google Scholar]
- Benoit B, Mitou G, Chartier A, Temme C, Zaessinger S, Wahle E, Busseau I, Simonelig M. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Dev Cell. 2005;9:511–22. doi: 10.1016/j.devcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–33. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics. 1992;131:643–53. doi: 10.1093/genetics/131.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–9. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crucs S, Chatterjee S, Gavis ER. Overlapping but distinct RNA elements control repression and activation of nanos translation. Mol Cell. 2000;5:457–67. doi: 10.1016/s1097-2765(00)80440-2. [DOI] [PubMed] [Google Scholar]
- Dahanukar A, Walker JA, Wharton RP. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol Cell. 1999;4:209–18. doi: 10.1016/s1097-2765(00)80368-8. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Filardo P, Ephrussi A. Bruno regulates gurken during Drosophila oogenesis. Mech Dev. 2003;120:289–97. doi: 10.1016/s0925-4773(02)00454-9. [DOI] [PubMed] [Google Scholar]
- Geng C, Macdonald PM. Imp associates with squid and Hrp48 and contributes to localized expression of gurken in the oocyte. Mol Cell Biol. 2006;26:9508–16. doi: 10.1128/MCB.01136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial A, Ray RP, Schupbach T. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 1998;12:2711–23. doi: 10.1101/gad.12.17.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, Elliott H, St Johnston D. Oocyte determination and the origin of polarity in Drosophila: the role of the spindle genes. Development. 1997;124:4927–37. doi: 10.1242/dev.124.24.4927. [DOI] [PubMed] [Google Scholar]
- Goodrich JS, Clouse KN, Schupbach T. Hrb27C, Sqd and Otu cooperatively regulate gurken RNA localization and mediate nurse cell chromosome dispersion in Drosophila oogenesis. Development. 2004;131:1949–58. doi: 10.1242/dev.01078. [DOI] [PubMed] [Google Scholar]
- Hawkins NC, Van Buskirk C, Grossniklaus U, Schupbach T. Post-transcriptional regulation of gurken by encore is required for axis determination in Drosophila. Development. 1997;124:4801–10. doi: 10.1242/dev.124.23.4801. [DOI] [PubMed] [Google Scholar]
- Huynh JR, Munro TP, Smith-Litiere K, Lepesant JA, St Johnston D. The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Dev Cell. 2004;6:625–35. doi: 10.1016/s1534-5807(04)00130-3. [DOI] [PubMed] [Google Scholar]
- Huynh JR, St Johnston D. The origin of asymmetry: early polarisation of the Drosophila germline cyst and oocyte. Curr Biol. 2004;14:R438–49. doi: 10.1016/j.cub.2004.05.040. [DOI] [PubMed] [Google Scholar]
- Jekely G, Sung HH, Luque CM, Rorth P. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev Cell. 2005;9:197–207. doi: 10.1016/j.devcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- Kalifa Y, Huang T, Rosen LN, Chatterjee S, Gavis ER. Glorund, a Drosophila hnRNP F/H homolog, is an ovarian repressor of nanos translation. Dev Cell. 2006;10:291–301. doi: 10.1016/j.devcel.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Kelley RL. Initial organization of the Drosophila dorsoventral axis depends on an RNA-binding protein encoded by the squid gene. Genes Dev. 1993;7:948–60. doi: 10.1101/gad.7.6.948. [DOI] [PubMed] [Google Scholar]
- Keyes LN, Spradling AC. The Drosophila gene fs(2)cup interacts with otu to define a cytoplasmic pathway required for the structure and function of germ-line chromosomes. Development. 1997;124:1419–31. doi: 10.1242/dev.124.7.1419. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–12. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Lehner CF. Synergistic action of Drosophila cyclins A and B during the G2-M transition. Embo J. 1993;12:65–74. doi: 10.1002/j.1460-2075.1993.tb05632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- Matunis EL, Kelley R, Dreyfuss G. Essential role for a heterogeneous nuclear ribonucleoprotein (hnRNP) in oogenesis: hrp40 is absent from the germ line in the dorsoventral mutant squid. Proc Natl Acad Sci U S A. 1994;91:2781–4. doi: 10.1073/pnas.91.7.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Matunis EL, Dreyfuss G. Isolation of hnRNP complexes from Drosophila melanogaster. J Cell Biol. 1992;116:245–55. doi: 10.1083/jcb.116.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol. 2001;2:521–9. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Michael WM, Eder PS, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. Embo J. 1997;16:3587–98. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Shu HJ, Zhao Y, Pinol-Roma S. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol Cell Biol. 2001;21:7307–19. doi: 10.1128/MCB.21.21.7307-7319.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro TP, Kwon S, Schnapp BJ, St Johnston D. A repeated IMP-binding motif controls oskar mRNA translation and anchoring independently of Drosophila melanogaster IMP. J Cell Biol. 2006;172:577–88. doi: 10.1083/jcb.200510044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Leidal AM, Smibert CA. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. Embo J. 2004;23:150–9. doi: 10.1038/sj.emboj.7600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schupbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell. 1993;75:165–74. [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schupbach T. The Drosophila TGF-alpha-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech Dev. 1996;59:105–13. doi: 10.1016/0925-4773(96)00567-9. [DOI] [PubMed] [Google Scholar]
- Nilson LA, Schupbach T. EGF receptor signaling in Drosophila oogenesis. Curr Top Dev Biol. 1999;44:203–43. doi: 10.1016/s0070-2153(08)60471-8. [DOI] [PubMed] [Google Scholar]
- Norvell A, Debec A, Finch D, Gibson L, Thoma B. Squid is required for efficient posterior localization of oskar mRNA during Drosophila oogenesis. Dev Genes Evol. 2005;215:340–9. doi: 10.1007/s00427-005-0480-2. [DOI] [PubMed] [Google Scholar]
- Norvell A, Kelley RL, Wehr K, Schupbach T. Specific isoforms of squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev. 1999;13:864–76. doi: 10.1101/gad.13.7.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmeyer JT, Schupbach T. Encore facilitates SCF-Ubiquitin-proteasome-dependent proteolysis during Drosophila oogenesis. Development. 2003;130:6339–49. doi: 10.1242/dev.00855. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–2. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Preiss T, Hentze MW. From factors to mechanisms: translation and translational control in eukaryotes. Curr Opin Genet Dev. 1999;9:515–21. doi: 10.1016/s0959-437x(99)00005-2. [DOI] [PubMed] [Google Scholar]
- Price JV, Clifford RJ, Schupbach T. The maternal ventralizing locus torpedo is allelic to faint little ball, an embryonic lethal, and encodes the Drosophila EGF receptor homolog. Cell. 1989;56:1085–92. doi: 10.1016/0092-8674(89)90641-7. [DOI] [PubMed] [Google Scholar]
- Queenan AM, Barcelo G, Van Buskirk C, Schupbach T. The transmembrane region of Gurken is not required for biological activity, but is necessary for transport to the oocyte membrane in Drosophila. Mech Dev. 1999;89:35–42. doi: 10.1016/s0925-4773(99)00196-3. [DOI] [PubMed] [Google Scholar]
- Ray RP, Schupbach T. Intercellular signaling and the polarization of body axes during Drosophila oogenesis. Genes Dev. 1996;10:1711–23. doi: 10.1101/gad.10.14.1711. [DOI] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–80. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Sapir A, Schweitzer R, Shilo BZ. Sequential activation of the EGF receptor pathway during Drosophila oogenesis establishes the dorsoventral axis. Development. 1998;125:191–200. doi: 10.1242/dev.125.2.191. [DOI] [PubMed] [Google Scholar]
- Schupbach T. Germ line and soma cooperate during oogenesis to establish the dorsoventral pattern of egg shell and embryo in Drosophila melanogaster. Cell. 1987;49:699–707. doi: 10.1016/0092-8674(87)90546-0. [DOI] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–36. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF. The double lives of shuttling mRNA binding proteins. Cell. 2000;102:135–8. doi: 10.1016/s0092-8674(00)00018-0. [DOI] [PubMed] [Google Scholar]
- Siebel CW, Kanaar R, Rio DC. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes Dev. 1994;8:1713–25. doi: 10.1101/gad.8.14.1713. [DOI] [PubMed] [Google Scholar]
- Sigrist SJ, Thiel PR, Reiff DF, Lachance PE, Lasko P, Schuster CM. Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature. 2000;405:1062–5. doi: 10.1038/35016598. [DOI] [PubMed] [Google Scholar]
- Smibert CA, Wilson JE, Kerr K, Macdonald PM. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 1996;10:2600–9. doi: 10.1101/gad.10.20.2600. [DOI] [PubMed] [Google Scholar]
- Steinhauer J, Kalderon D. The RNA-binding protein Squid is required for the establishment of anteroposterior polarity in the Drosophila oocyte. Development. 2005;132:5515–25. doi: 10.1242/dev.02159. [DOI] [PubMed] [Google Scholar]
- Tarun SZ, Jr., Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. Embo J. 1996;15:7168–77. [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Jr., Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci U S A. 1997;94:9046–51. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–5. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Thio GL, Ray RP, Barcelo G, Schupbach T. Localization of gurken RNA in Drosophila oogenesis requires elements in the 5' and 3' regions of the transcript. Dev Biol. 2000;221:435–46. doi: 10.1006/dbio.2000.9690. [DOI] [PubMed] [Google Scholar]
- Van Buskirk C, Hawkins NC, Schupbach T. Encore is a member of a novel family of proteins and affects multiple processes in Drosophila oogenesis. Development. 2000;127:4753–62. doi: 10.1242/dev.127.22.4753. [DOI] [PubMed] [Google Scholar]
- Verrotti AC, Wharton RP. Nanos interacts with cup in the female germline of Drosophila. Development. 2000;127:5225–32. doi: 10.1242/dev.127.23.5225. [DOI] [PubMed] [Google Scholar]
- Webster PJ, Liang L, Berg CA, Lasko P, Macdonald PM. Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev. 1997;11:2510–21. doi: 10.1101/gad.11.19.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschaus E. fs(1)K10, a female sterile mutation altering the pattern of both the egg covering and the resultant embryos in Drosophila. In: Le Douarin N, editor. “Cell Lineage, Stem Cells, and Cell Determination”. Vol. 10. Elsevier; Amsterdam: 1979. pp. 291–302. Vol. Inserm Symposium No. [Google Scholar]
- Wieschaus E, Marsh JL, Gehring W. fs(1)K10, a germline-dependent female sterile mutation causing abnormal chorion morphology in Drosophila Melanogaster. Roux's Arch. Dev. Biol. 1978;184:75–82. doi: 10.1007/BF00848670. [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol. 2003;163:1197–204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Smibert CA. Mechanisms of translational regulation in Drosophila. Biol Cell. 2005;97:235–52. doi: 10.1042/BC20040097. [DOI] [PubMed] [Google Scholar]
- Yan N, Macdonald PM. Genetic interactions of Drosophila melanogaster arrest reveal roles for translational repressor Bruno in accumulation of Gurken and activity of Delta. Genetics. 2004;168:1433–42. doi: 10.1534/genetics.104.033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Lopez de Quinto S, Matsui Y, Shevchenko A, Shevchenko A, Ephrussi A. Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev Cell. 2004;6:637–48. doi: 10.1016/s1534-5807(04)00132-7. [DOI] [PubMed] [Google Scholar]
- Zappavigna V, Piccioni F, Villaescusa JC, Verrotti AC. Cup is a nucleocytoplasmic shuttling protein that interacts with the eukaryotic translation initiation factor 4E to modulate Drosophila ovary development. Proc Natl Acad Sci U S A. 2004;101:14800–5. doi: 10.1073/pnas.0406451101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Antibodies were used to immunoprecipitate Sqd or Cup from ovarian lysates. Cyclin B was used as a second negative control for immunoprecipitation in addition to the α-Dorsal antibody used in figures 1 and 6. 5% of the lysate probed for interactions was loaded in the input lane. Cup, Sqd, and CycB were detected by western blotting. Images were acquired by CCD imaging and pixel intensities in the bands were integrated using AlphaEaseFC software (Alpha Innotech). All signals were within the dynamic range of the camera. 16% (S.D. 6.7%) of the Sqd in the lysate was recovered in the IP. After correcting for the efficiency of the Sqd IP, 6% (S.D. 1.9%) of the Cup in ovarian lysates was found to be associated with Sqd. Data are the result of three independent experiments. No Sqd was detected in Cup or PABP immunoprecipitates (PABP data not shown). These data may indicate that the Sqd:Cup stoichiometry on grk transcripts is biased towards multiple Cup molecules for each Sqd molecule. Alternatively the Cup antibody may recognize an epitope that is required for interactions with Bru or other trans-acting factors necessary for the association of Cup with grk, and by association Sqd. In the case of PABP, which binds most poly-adenylated messages, the population of grk mRNA that comprises a PABP immunoprecipitate is expected to be quite small. Immunoprecipitation with α-Sqd antibodies, however, pulls down a more selective pool of RNA of which grk, and by association PABP and Cup, are a larger component.