Summary
During vertebrate eye development, the transcription factor MITF plays central roles in neuroepithelial domain specification and differentiation of the retinal pigment epithelium. MITF, however, is not a single protein but represents a family of isoforms that are generated from a common gene by alternative promoter/exon use. To address the question of the role and regulation of these isoforms, we first determined their expression patterns in developing mouse eyes and analyzed the role of some of them in genetic models. We found that two isoforms, A- and J-Mitf, are present throughout development in both retina and pigment epithelium while H-Mitf is detected preferentially and D-Mitf exclusively in the pigment epithelium. We further found that a genomic deletion encompassing the promoter/exon regions of H-, D- and B-Mitf leads to novel mRNA isoforms and proteins translated from internal start sites. These novel proteins lack the normal, isoform-specific aminoterminal sequences and are unable to support the development of the pigment epithelium but capable of inducing pigmentation in the ciliary margin and the iris. Moreover, in mutants of the retinal Mitf regulator Chx10, reduced cell proliferation and abnormal pigmentation of the retina are associated with a preferential upregulation of H- and D-Mitf. This retinal phenotype is corrected when H- and D-Mitf are missing in double Mitf/Chx10 mutants. The results suggest that Mitf regulation in the developing eye is isoform-selective both temporally and spatially, and that some isoforms, including H- and D-Mitf, are more critical than others in effecting normal retina and pigment epithelium development.
Keywords: retina, retinal pigment epithelium, Mitf-red-eyed-white, Mitf-black-eyed-white, Chx10-ocular retardation, internal start codons
Introduction
Alternative promoter use is a common principle by which genes generate transcript diversity and contribute to an organism’s phenotypic complexity. Often, promoter choice does not affect the sequence of the corresponding proteins because translation is invariably initiated from a single downstream start site, but the untranslated 5′ regions can influence mRNA stability and translational efficiency and so refine gene regulation beyond the spatial and temporal control afforded at the transcriptional level. With some genes, however, variant 5′ exons encode unique amino acid sequences and hence provide an additional level of regulation as protein isoforms with distinct amino termini can differ in their function (Carninci et al., 2006; Sandelin et al., 2007). Nevertheless, despite a wealth of sequence information for many species, relatively little is known about the role and regulation of alternative promoter use in vivo, both during development and in adulthood.
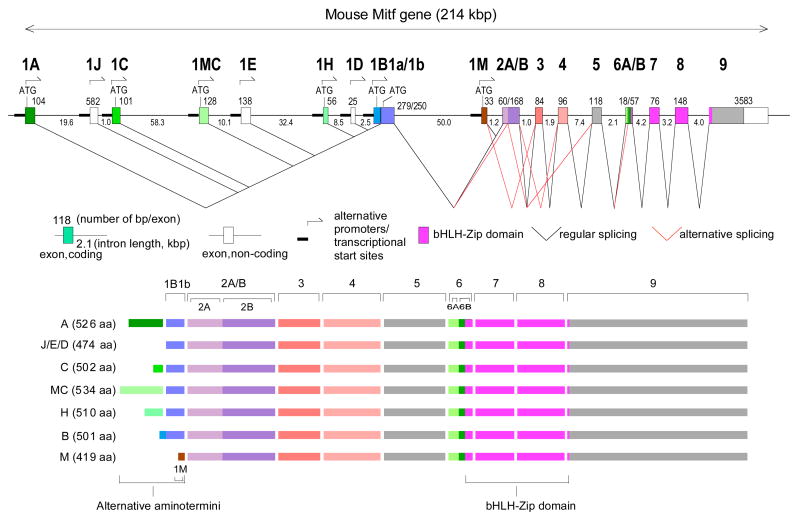
A prime example of a gene with multiple promoters is Mitf (microphthalmia-associated transcription factor) which encodes a basic helix-loop-helix-leucine zipper protein that is critical for mammalian eye development (Hodgkinson et al., 1993; Nakayama et al., 1998; Nguyen and Arnheiter, 2000). In mice and humans, the gene contains nine distinct promoters, six of which are linked to different coding exons and three to non-coding exons (Fig. 1) (Hallsson et al., 2000; Steingrimsson et al., 2004; Hershey and Fisher, 2005; Arnheiter et al., 2006; Hallsson et al., 2007). Using probes that do not distinguish among the corresponding isoforms, it has been shown that Mitf is initially expressed in mice throughout the budding optic vesicle and retained in the future retinal pigment epithelium (RPE), the ciliary body, and the iris but downregulated in the future retina and optic stalk (Bora et al., 1998; Nguyen and Arnheiter, 2000). In Mitf mutants in which all isoforms are equally affected, the RPE can develop as a laminated second retina (Müller, 1950; Bumsted and Barnstable, 2000; Nguyen and Arnheiter, 2000), and in mutants in which Mitf is abnormally upregulated in the retina, the retina can develop as a single-layered RPE (Rowan et al., 2004; Horsford et al., 2005; Bharti et al., 2006). Based on these results it is unclear, however, which isoforms are normally expressed during RPE and retina formation and whether any of these isoforms have selective roles or are entirely interchangeable.
Fig. 1.
Top: Schematic diagram of the mouse Mitf gene with its multiple promoters and regular and alternative splice choices. Bottom: Schematic diagram of the seven protein isoforms differing in their aminoterminal sequences due to different promoter choice.
The question of isoform-specific function and expression also touches on the problem of whether distinct isoforms are co-regulated or regulated separately. In fact, two lines of evidence argue in favor of co-regulation. One is the observation, made in mice, that the combined genetic lack of the eye transcription factors PAX2 and PAX6 is associated with a total absence of all MITF protein. This finding is supported by the notion that the human A-MITF promoter, which like the mouse A-Mitf promoter is located at the 5′ end of the gene, is stimulated by PAX2/PAX6 in vitro, and hence may serve as a control region for the entire Mitf locus in the eye in vivo (Bäumer et al., 2003). The other is the recent observation that the paired-like homeodomain transcription factor CHX10, which is involved in the retinal downregulation of Mitf (Liu et al., 1994; Burmeister et al., 1996; Rowan et al., 2004; Horsford et al., 2005), may stimulate a cluster of genes encoding microRNAs that serve to suppress Mitf mRNAs by recognizing their common 3′ UTRs (Xu et al., 2007). Without more detailed information about the developmental expression pattern of each Mitf isoform, however, an isoform-selective regulation cannot be excluded a priori.
Here, we use a combination of tissue dissection, reverse transcriptase-PCR, and isoform-specific antibodies to show that some Mitf isoforms are ubiquitously expressed throughout development in both ocular and extraocular tissues, that others are more restricted to the optic neuroepithelium, and still others are retained exclusively in the RPE. We then analyze two different mouse mutations associated with over- or under-expression of Mitf to assess the functional relevance of some of these isoforms in both retina and RPE. The results indicate that contrary to previous assumptions, the regulation of Mitf during eye development is not simply global, affecting all transcriptional isoforms indiscriminately, but in fact is isoform-selective. They also suggest that some isoforms are more critical than others for early cell fate decisions in the developing eye, and hence for the formation of a functional adult eye.
Materials and methods
Animals
C57BL/6J control mice were from Jackson Laboratories (Bar Harbor, Maine) and albino Swiss Webster (SW-Tyrc) mice were from Taconic (Rockville, Maryland). The following mutant mice were bred locally and kept according to NINDS-approved animal study proposals: C57BL/6J-Mitfmi-rw, mixed [C57BL/6J;C3H/HeJ]-Mitfmi-bw, C57BL/6J•C3H/HeJ-Mitfmi-vga9, used as N7F2, and C57BL/6J•129S1/Sv-Chx10orJ, used as N10F12-F16.
Antibodies, immunohistochemistry, immunofluorescence, and in situ hybridization
A list of antibodies and their preparation and purification is given in Supplemental Table S1. Immunolabeling was done on 14-micrometer cryostat sections as described previously (Nakayama et al., 1998) except for the exon-specific anti-1B1b antibody which required antigen retrieval by boiling the sections for 30 minutes. In situ hybridization was done using a pan-specific Mitf probe as described (Nakayama et al., 1998), or a PCR-generated exon 1B1b-specific probe (primers see Supplemental Table S2).
Reverse transcriptase reactions and real time PCR
For RNA extractions using RNeasy mini kit (Qiagen), E9.5 and 10.5 eye primordia were manually dissected along with the surrounding tissue, and from E11.5-P0, retinal tissue was separated manually from the surrounding RPE/mesenchyme except where indicated. Complementary DNA was prepared with Superscript RT-PCR kit (Invitrogen), using one microgram of total RNA, and cDNA corresponding to 25 ng of total template RNA was used for each PCR reaction. In some experiments, RNA was isolated with Picopure RNA extraction kit (Arcturus) from 100 cells that were microscopically selected after trypsinization, using micromanipulator-controlled glass microinjection pipettes. In these cases, the amount of cDNA used for amplification with two rounds of PCR using nested primer pairs was estimated to correspond to approximately 1–10 pg of template RNA. Real time PCR was performed using an ABI Prism 7000 real time PCR machine (Applied Biosystems) and is described in detail in Supplemental Materials. All RT-PCR products were sequenced at least once to confirm primer specificity. In the eye, sequencing gave no evidence for alternative splicing events linking exon 1A with exon 1J1b and exon 1J1b with exon 1C1b as reported for cultured cell lines (Hershey and Fisher, 2005). Therefore, exons 1J and 1C are here considered as single exons. All primer sets used for RT-PCR and real time PCR analysis are listed in Supplemental Table S2.
Molecular characterization of the Mitfmi-rw mutation, constructs, in vitro mutagenesis, and reporter assays
The Mitfmi-rw mutation was characterized by PCR and Southern blots as shown in Supplemental Fig. S1. The various Mitf isoforms were cloned and their activities assayed in vitro as detailed in Supplemental Fig. S2. The Supplemental Materials also describe in vitro mutagenesis and the analysis of the mutated constructs.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays using anti-CHX10 antibodies and amplification of Mitf promoter DNA are described in Supplemental Fig. S3 and S4 and Supplemental Table S1 and S2.
Results
Mitf mRNA and protein expression in the developing mouse eye
Previous studies using pan-specific in situ hybridizations probes and antibodies have shown that Mitf expression in the developing eye begins at around embryonic day (E) 9.5 and lasts until postnatal day (P) 0 (Nakayama et al., 1998; Nguyen and Arnheiter, 2000). To analyze the isoform composition of this pan-specific signal, we first used an in situ hybridization probe specific for exon 1B1b which is common to all isoforms except M-Mitf (Fig. 1), a major component of neural crest-derived choroidal and iris melanocytes. As shown in Fig. 2A, exon 1B1b is expressed in the optic vesicle at E9.5, is downregulated in the distal optic vesicle at E10.0, and then is concentrated in the developing RPE as previously reported for a single time point (Amae et al., 1998). As expected, no labeling is seen in choroidal and iris melanocytes which start to accumulate at around E11.5 on the proximal side of the RPE and normally are labeled with pan-specific probes (Nakayama et al., 1998). Also, no labeling is seen on sections from Mitfmi-rw (Mitf red-eyed white) embryos (Fig. 2A, right-most panel) which carry a genomic deletion of exon 1B (Hallsson et al., 2000) but express Mitf as detectable with pan-specific probes (see later in this study, Fig. 5F).
Fig. 2.
Expression of Mitf RNAs and proteins containing exon 1B1b. (A) In situ hybridization with an exon 1B1b-specific probe on frontal sections of the developing eye at the indicated time points. Arrowheads at E9.5 indicate expression throughout the optic vesicle, and arrowhead at E10.5 indicates reduced expression in the distal optic vesicle. In a control section of an Mitfmi-rw/mi-rw embryo, which lacks exon 1B1b, there is no labeling. (B) Antibody characterization and Mitf expression in RPE/choroid and retina. A-, H-, D-, and M-Mitf cDNAs were expressed in NIH3T3 cells and extracts blotted with polyclonal anti-1B1b MITF antibodies (top panel) or monoclonal anti-MITF carboxyl terminal-domain antibodies (anti-CTD, middle panel). The bottom left panel shows a combined immunoprecipitation (IP)/immunoblot (IB) of NIH-3T3 cells transfected with A-Mitf. The bottom right panel shows a similar IP/IB of extracts from RPE/choroidal and retinal fractions from 100 wild type eye primordia (E12.5). Arrowheads point to MITF isoforms with distinct electrophoretic mobilities. (C) Immunohistochemistry of frontal cryosections of embryos of the indicated genotypes and ages using anti-1B1b MITF and anti-pan MITF antibodies. Arrows in panel labeled wt-E15.5 point to M-MITF expressing neural crest-derived melanocytes that are only seen with pan-MITF-specific antibodies. Note that “wt” refers to Tyrc (albino) embryos carrying a wild type Mitf allele. RPE, retinal pigment epithelium; ret: retina; CMZ: ciliary margin zone.
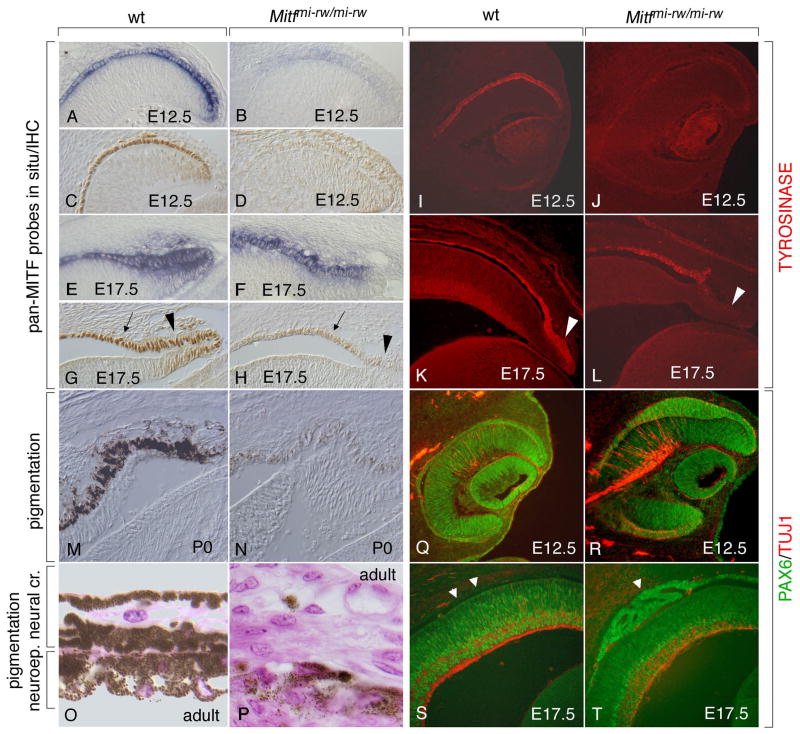
Fig. 5.
The Mitfmi-rw allele allows for the expression of Mitf and its target gene tyrosinase in the anterior RPE as well as for residual pigmentation in the iris but leads to an abnormal RPE dorsally. Sections of wild type and Mitfmi-rw/mi-rw mutant eyes of the indicated ages were stained with a pan-Mitf in situ probe (A,B,E,F), a pan-MITF antiserum (C,D,G,H), antibodies against TYROSINASE (I–L), or against PAX6 (green) and TUJ1 (red) (Q–T). Note that the mutant eye expresses Mitf RNA (B,F) and low levels of MITF protein, along with TYROSINASE, particularly at later stages (compare G and H, arrows; and K and L). Also note that in the mutant at E17.5, the distal ciliary margin, though positive for Mitf RNA (F), is relatively free of MITF and TYROSINASE (arrowhead in H and L, compare with arrowhead in G and K). (M,N) Strong pigmentation at P0 in wild type (M) and weak pigmentation in rw (N). (O,P) Pigmentation in adult wild type iris (O) and rw iris (P). (Q,R) Dorsal thickening of the E12.5 RPE in rw (R) compared to wild type (Q). (S,T) RPE abnormalities in rw at E17.5. Arrowheads in (S) mark the location of the wt RPE which now is free of PAX6 staining. In contrast, mutant RPE retains PAX6 staining and in this eye showed epihelial folds rather than homogeneous thickening (arrowhead in T).
To determine whether exon 1B1b-containing mRNAs are translated into corresponding proteins, we then performed a series of immunoblot, immunoprecipitation, and immunohistochemical analyses. The specificity of an antibody for exon 1B1b-containing MITF (reactivity with A-, J- and D-Mitf but not M-Mitf) is shown in Fig. 2B, along with the pan-specific reactivity of a mouse monoclonal antibody to the MITF carboxyl terminal domain (CTD) and the absence of reactivity of either antibody with normal fibroblast extracts. Using these antibodies, we then performed a combined immunoprecipitation/immunoblot assay of extracts of RPE/choroids and retinas that were manually dissected and separately pooled from approximately 100 eyes of wild type E12.5 embryos. The results revealed at least three bands in RPE/choroid and one band of lower electrophoretic mobility in retina (Fig. 2B, bottom right panel, marked by arrowheads). This finding indicates that RPE/choroid and retina express distinct exon 1B1b-containing isoforms, but based on electrophoretic mobility alone, their precise identification is not feasible because post-translational modifications may potentially modify their apparent molecular weights.
In immunohistochemical assays (Fig. 2C), both exon 1B1b- and pan-specific rabbit antibodies gave overlapping expression patterns in the neuroepithelial parts of the developing eye, starting from E9.5, when MITF is expressed in the entire optic vesicle, through E17.5 and beyond (not shown), when it is expressed predominantly in the RPE. Neural crest-derived choroidal melanocytes, however, were only labeled with the pan-specific antibody (arrows in Fig. 2C, panel labeled wt E15.5). Both exon 1B1b- and pan-specific antibodies gave a low-level labeling in the retina and surrounding mesenchyme when compared to equally treated sections from Mitfmi-rw/mi-rw embryos, which lack exon 1B (Hallsson et al., 2000), or Mitfmi-vga-9/mi-vga-9 embryos, which lack MITF entirely (Hodgkinson et al., 1993; Nakayama et al., 1998) (Fig. 2C, panels labeled Mitfmi-rw/mi-rw or Mitfmi-vga-9/mi-vga-9). This result is consistent with the observation presented in Fig. 2B showing that E12.5 retinas continue to express at least some isoform(s) of MITF.
Expression of promoter-specific Mitf isoforms
To determine the expression profile of individual isoforms, we exclusively used RT-PCR, primarily because it provides the sensitivity to detect low abundance isoforms. For the early time points (E9.5-E10.5), RNA was obtained from whole eye primordia including the surrounding tissue, and for the later time points (E11.5-P0), separately from RPE/choroidal and retinal fractions. Complementary DNAs were amplified using either the respective 5′ exon-specific forward primers in combination with a reverse primer in exon 1B1b, or, in the case of M-Mitf, a reverse primer in exon 2A. Pan-specific primers corresponded to exons 2A and 4. The number of PCR cycles was adapted for each primer pair to give amplification in the linear range. As shown in Fig. 3A, the first signal was seen at E9.5 with pan-specific primers and weakly with A- and J-Mitf-specific primers while H- and D-Mitf-specific amplification gave the first signals at E10.5. No other isoforms were detected at these early stages.
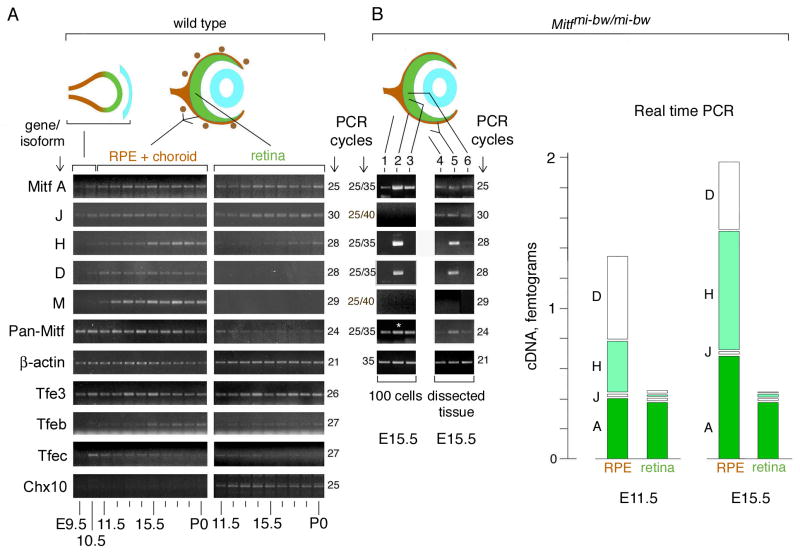
Fig. 3.
Differential expression of Mitf isoforms during mouse eye development. (A) RT-PCR analysis on RNA isolated from wild type whole optic vesicles (E9.5–10.5) or separate RPE+choroidal and retinal fractions (E11.5-P0). For primer choice and number of PCR cycles, see text and Supplemental Table S2. (B) RT-PCR and real-time PCR for RNA isolated from Mitfmi-bw/mi-bw eyes which lack neural crest-derived melanocytes. Left panel: E15.5 eyes were dissected as in (A) and 100 individual cells were microscopically selected as described in materials and methods. Lane 1, mesenchymal cells; Lane 2, RPE cells; Lane 3, retinal cells. The asterisk in lane 2 indicates that the corresponding cDNA was diluted 1:10 for the reaction with pan-specific primers. Middle panel: Pooled fractions from dissected E15.5 eyes as indicated. Lane 4, mesenchymal fraction; Lane 5, RPE/mesenchymal fraction; Lane 6, retinal fraction. Right panel: Real time PCR. RNA was prepared from separately pooled RPE/mesenchymal and retinal fractions from E11.5 and E15.5 eyes. Results are expressed as mean of absolute amounts and standard deviations (vertical lines in each column) of the respective cDNAs and are calculated taking into account that RPE cells represent 7% of the cells in the RPE/mesenchymal fraction (for details, see Supplemental Materials). Significance of the difference between RPE and retina (p values, Student’s t test, pools of 20 RPE/mesenchymal and retinal fractions each for E11.5; and 12 RPE/mesenchymal and retinal fractions each for E15.5, four separate assays in triplicates per pool): E11.5: A-Mitf, >0.1; J-Mitf, <0.1; H-Mitf, <0.001; D-Mitf, <0.00001. E15.5: A-Mitf, <0.01; J-Mitf, <0.01; H-Mitf, <0.0001; D-Mitf, <0.001.
During the subsequent developmental time points, the different isoforms can be grouped into four distinct expression patterns (Fig. 3A). First, throughout the period from E11.5 to P0, A- and J-Mitf showed similar expression profiles both in the retina and the RPE/choroid. Second, H-Mitf was preferentially found in the RPE/choroid but was present at low levels in the retina as well, particularly after E15.5. Third, D- and M-Mitf were found only in the RPE/choroid and not the retina. Fourth, C-, MC-, E-, and B-Mitf were undetectable, or only barely detectable, throughout development in either retina or RPE/choroid (not shown). In support of these results, from E11.5-P0, pan-specific amplification showed higher expression levels in RPE/choroid compared to retina. Additional assays indicated that the Mitf-related genes Tfe3, Tfeb and Tfec (Hallsson et al., 2007) were also expressed in the developing eye, and amplification for Chx10, which in the eye is retina-specific (Liu et al., 1994), gave a signal only in the retinal fraction. This latter finding, together with the fact that D- and M-Mitf were only seen in the RPE/choroidal fraction, indicates that efficient tissue separation had been achieved.
The above analysis established a temporal expression profile for each isoform but did not address the question of which isoform is expressed specifically in the RPE because the RPE/choroidal fraction contains other cells, including choroidal melanocytes, that may contribute exon 1B1b-containing isoforms detectable by PCR. Therefore, we repeated the above analysis by specifically selecting retinal, mesenchymal and RPE cells. For these experiments, we used eyes from Mitfmi-bw (Mitf black-eyed white) embryos whose only pigmented cells are RPE cells as they lack choroidal melanocytes (Yajima et al., 1999). In postnatal eyes of such mice, M-Mitf mRNAs are missing, full-length exon 1B1b-containing mRNAs are reduced, and skipping of common downstream exons is increased (Yajima et al., 1999). At E11.5 through E17.5, however, we find no differences between Mitfmi-bw and wild type eyes in Mitf expression except that M-Mitf is lacking in the mutant (not shown). Hence, the use of separately trypsinized RPE/mesenchymal and retinal fractions of E15.5 Mitfmi-bw/mi-bw allowed us to select microscopically small populations (100 cells) of pure mesenchymal, RPE, and retinal cells (for details see Materials and methods). The respective cDNAs were amplified in two rounds of PCR, using nested primer sets as indicated in Supplemental Table S2. This analysis confirmed that A-Mitf is ubiquitous and showed that H- and D-Mitf are present in the RPE, and in fact enriched in the RPE compared to retina and mesenchyme. No signals were found for the low-abundance J-Mitf and, as expected, for M-Mitf (Fig. 3B, lanes 1–3). The results are consistent with those obtained from eye tissues dissected in standard ways (Fig. 3B, lanes 4–6) where A- and low levels of J-Mitf are found in all fractions (lanes 4–6), H- and D-Mitf are enriched in the fraction that contains RPE and mesenchyme (lane 5), and M-Mitf is absent in all fractions.
The above experiments showed which isoform was expressed in which cell type but did not address the question of their expression levels. In fact, because different primer pairs amplify cDNAs with distinct efficiencies, differences in band intensities cannot be used for comparisons between isoforms. We, therefore, subjected cDNAs generated from separately pooled RPE/mesenchymal and retinal RNA fractions to real time PCR using isoform-specific standard curves as described in detail in Supplemental Materials. Fig. 3B, right panel, shows the mean amounts of cDNAs obtained from four repeat assays each for E11.5 and E15.5. Consistent with the results above, A- and J-Mitf accumulate to similar levels in RPE and retina, and H- and D-Mitf reach higher levels in RPE compared to retina. We conclude, therefore, that both the future retina and the future RPE continue to express A- and J-Mitf and that the major isoforms in RPE are D-Mitf, and, increasingly with developmental time, H-Mitf.
Genomic deletion of Mitf exons 1H, 1D, and 1B leads to increased expression of novel mRNA and protein isoforms
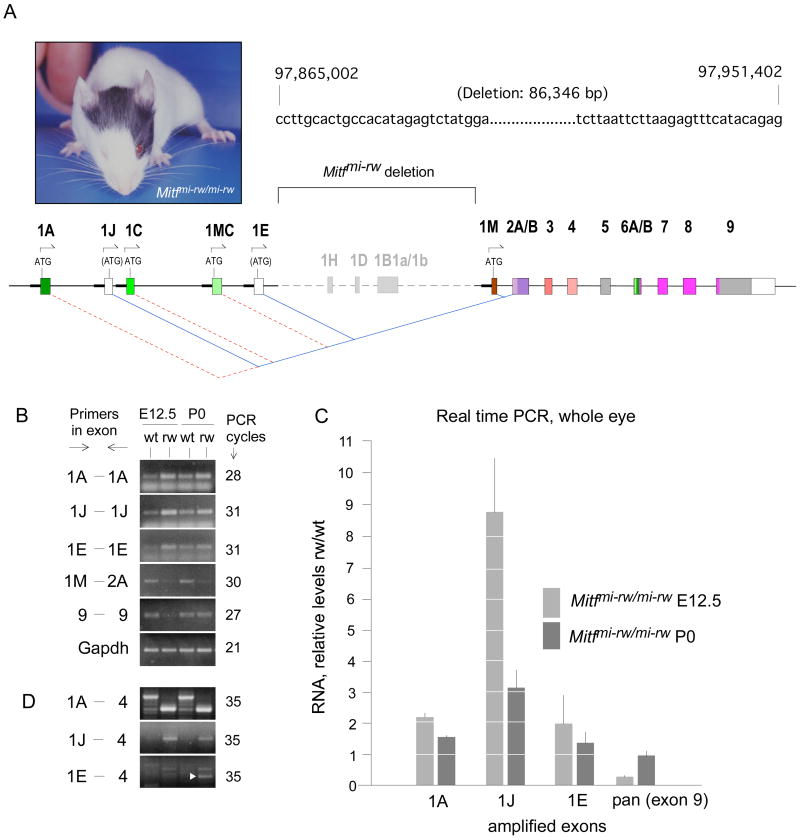
To address the question of whether the major RPE isoforms, H- and D-Mitf, are functionally relevant, we then analyzed mice homozygous for the Mitfmi-rw (Mitf red-eyed white, for short, rw) allele which is characterized by a genomic deletion that encompasses the 1H-, 1D- and 1B- exons (Steingrimsson et al., 1994; Hallsson et al., 2000). Such mice retain a patch of pigmented coat on the head and occasionally on the belly and have eyes whose sizes can vary both between individuals and between the left and right side of the same individual (example shown in Fig. 4A, left side). We first determined the exact boundaries of the deletion and found that it comprises a stretch of 86,346 bp starting downstream of exon 1E and ending upstream of exon 1M (Fig. 4A and Supplemental Fig. S1). The deletion has the consequence that the upstream exons, whether coding or not, splice into the next available downstream exon, exon 2A, rather than the normal exon 1B1b (Fig. 4A). Consequently, if translation initiates at the normal start codons in exon 1A, 1C and 1MC, no open reading frames are maintained beyond exon 2 (marked by red dotted lines in Fig. 4A). Novel open reading frames, however, are generated within the normally non-coding exons 1J and 1E (blue lines in Fig. 4A) but the potential AUG start codons do not conform to Kozak rules for efficient translation initiation (Kozak, 1984; Kozak, 1986).
Fig. 4.
Mitfmi-rw/mi-rw mutant mice carry a genomic deletion in Mitf encompassing the exons 1H, 1D and 1B. (A) Phenotype of an Mitfmi-rw/mi-rw mutant mouse. Generally, coat pigment patches and eye size are not correlated, as eye size is determined by the development of the RPE and coat patches by neural crest-derived melanocytes. The sequence on the right marks the base pairs flanking the genomic deletion, with the numbers of the first and last base of the depicted sequence referring to the positions on chromosome 6 according to assembly NCBIM36. The schematic diagram below shows the extent of the deletion and highlights the novel splice junctions that are generated between the upstream exons 1A, 1J, 1C, 1MC and 1E and the downstream exon 2A. For details, see text. (B) RT-PCR using RNA from E12.5 wild type and Mitfmi-rw (rw) mutant embryos and the corresponding P0 newborns. Note increased band intensity in rw with exons 1A, 1J and 1E but decreased band intensity at E12.5 with pan-specific primers (exon 9). (C) Real time PCR from RNA of whole eyes harvested at the indicated ages, using primers as in (B). The results are expressed as RNA levels in rw mutants relative to those in corresponding wild type embryos (groups of 14 eyes each, three measurements each in triplicates). The increases in rw over wild type in 1A and 1J as well as the decrease in exon 9 at E12.5 are statistically significant (p<0.01, Student’s t test). P values for the increase in 1E are <0.02 at E12.5 and =0.13 at P0. No significant difference was found for exon 9 at P0 (p=0.27). (D) RT-PCR using primers spanning four exons. Note different-size products in wt and rw for isoform 1A. For isoform 1J, the expected larger product in wt is not seen because its relative level is low (see Fig. 2), but the smaller-size product in rw is visible. In 1E-4, white arrowhead points to the correct E-Mitf band.
To analyze the transcriptional profile of Mitf in eyes of rw embryos, we again used RT-PCR as described above, except that whole eye tissue served as source of RNA because dissection into RPE/choroid and retina was not feasible with the mutant eyes. Moreover, we used primers covering only the respective single exons to allow for equal product sizes in wt and rw. The results (Fig. 4B) indicate that the expression levels of the upstream exons 1A, 1J and 1E were increased while those of the downstream exon 1M and the pan-specific exon 9 were decreased. These findings were confirmed by real time PCR (Fig. 4C). The results show that for instance at E12.5, an approximately two-fold increase in A-Mitf and a nine-fold increase in the minor J-Mitf cannot compensate for the lack of the major isoforms, H- and D-Mitf, and the reduction of M-Mitf which is detectable in these whole eye extracts (Fig. 4B). At P0, however, isoform-specific increases and decreases canceled each other out so that pan-specific amplification of exon 9 gave a rw/wt ratio of approximately 1.0. Using primers in the specific upstream exons and exon 4, we further confirmed that the over-expressed upstream exons were indeed part of spliced RNAs (Fig. 4D). Note, however, that even though the exon 1E-specific forward and reverse primers gave a visible band in wild type (Fig. 4B), no correct-size band was seen in wild type with primers in exon 1E and 4 (Fig. 4D). This confirms the finding mentioned above that spliced transcripts containing exon 1E are below threshold in the developing eye.
The lack of normal open reading frames in the internally deleted mRNAs suggested that Mitfmi-rw is a functional null allele. Intriguingly, however, when we tested corresponding expression constructs in transfection or in vitro transcription/translation assays, we found that MITF proteins were generated that reacted with the antibody to the MITF carboxyl terminal domain. Further analyses showed that the translation of these proteins is initiated predominantly from an internal AUG codon corresponding to methionine-62 in exon 2B and that the aminoterminally truncated proteins accumulate in the cell nucleus and are transcriptionally active on target gene promoters, including the promoter of the melanin biosynthetic enzyme gene tyrosinase (Supplemental Fig. S2). These results suggest that rw mice may retain Mitf activity associated with the novel protein isoforms despite the lack of H-and D-Mitf.
Deletion of Mitf exons 1H, 1D and 1B interferes with normal eye development but allows for partial pigmentation
To determine the in vivo consequence of the rw deletion, we then analyzed the development of rw eyes in detail. As shown in Fig. 5, in situ hybridization with a pan-Mitf-specific probe indicated that Mitf mRNAs are indeed expressed at the expected locations in the developing rw eyes. At E12.5, however, their levels were reduced compared to wild type but at E17.5, they were close to normal (compare Fig. 5A and B; and E and F). Immunohistochemistry using pan-specific antibodies indicated a reduced signal in rw at both ages, likely reflecting a reduced translational efficiency of the rw mRNAs compared to wild type RNAs (compare Fig. 5C and D; and G and H). Despite the low MITF protein levels, however, a clear signal for tyrosinase was found in rw RPE, particularly at E17.5 (Fig. 5I–L), in contrast to developing eyes from Mitf null mutants which lack tyrosinase expression (Nakayama et al., 1998). Consistent with this result, pigmentation, although at low levels, could be found in the rw peripheral RPE and iris (Fig. 5M,N) as well as in the neuroepithelial part of the adult iris (Fig. 5O,P). Nevertheless, RPE development was perturbed, either because the overall MITF protein levels were too low or because full-length proteins containing the normal amino-terminal sequences were missing. Much as in developing eyes of other Mitf mutants (Müller, 1950; Bumsted and Barnstable, 2000; Nguyen and Arnheiter, 2000), the dorsal RPE became thick (Fig. 5Q,R), retained high level PAX6 expression (Fig. 5Q-T), and eventually transdifferentiated into a laminated second retina (not shown). In some eyes, however, the RPE abnormalities were characterized not by a general thickening but by foldings (Fig. 5S,T), a much milder abnormality than seen in other rw eyes. The results suggested that the lack of H- and D-MITF and exon 1B leads to RPE abnormalities of variable severity but that the novel MITF protein isoforms are still capable of inducing pigmentation of the ciliary margin and the iris.
Lack of CHX10, a negative regulator of Mitf, leads to an increase in H- and D-Mitf in the retina
The downregulation of Mitf during retinal development involves a pathway that is initiated by extracellular signals emanating from the surface ectoderm, notably FGF1/2, and includes the paired-like homeodomain transcription factor CHX10 that negatively regulates Mitf expression in the retina (schematically indicated in Fig. 6A). As previously shown, in Chx10orJ/orJ mutant embryos, which lack functional copies of Chx10, the future retina is hypoplastic (Konyukhov and Sazhina, 1966; Burmeister et al., 1996; Green et al., 2003) and Mitf mRNA and protein are increased (Rowan et al., 2004; Horsford et al., 2005). To confirm that the mutant retina hypoproliferates, we used phosphohistone H3 labeling to mark the cells at the G2/M transition. As expected, the thinner Chx10or-J/or-J retina shows much fewer labeled cells (Fig. 6B,C). As shown in Fig. 6D,E, this phenotype is correlated with the retinal retention of Mitf RNAs containing exon 1B1b. To dissect the isoform composition of the upregulated RNA, we then used limiting cycle RT-PCR of RNA extracted from wild type and Chx10 mutant eyes at E13.5. The results, shown in Fig. 6F, show a substantial increase in H- and D-Mitf, but a barely perceptible increase in A- and J-Mitf. Theoretically, a specific retinal upregulation of A- and J-Mitf may have been partially masked by the ubiquitous expression of these isoforms outside the retina. This is unlikely, though, because retinal changes in the expression of CyclinD1, which is also expressed outside the retina, can easily be detected in the whole-eye extracts, and A- and J-Mitf continue to be expressed throughout development in the Chx10-positive wild type retina (see Fig. 2).
Fig. 6.
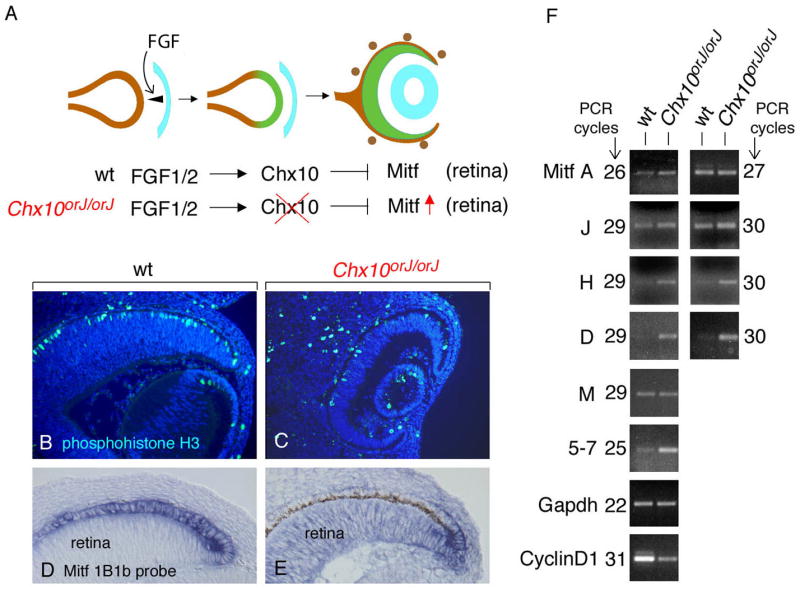
Lack of downregulation of Mitf in Chx10orJ/orJ retinas predominantly affects H- and D-Mitf. (A) Genetic pathway showing the downregulation of Mitf in the future retina by FGFs emanating from the surface ectoderm (light blue) and Chx10 operating in the distal optic neurepithelium. In Chx10 mutant animals, Mitf is not downregulated in the future retina and the retina hypoproliferates. (B–E) Cryostat sections from E13.5 wild type eyes (B,D) and Chx10orJ/orJ eyes (C,E) labeled for phosphohistone H3 (B,C) or by in situ hybridization using an Mitf exon 1B1b probe (D,E). Note upregulation of exon-1B1b containing RNA in mutant retina (E) compared to wild type retina (D). Also note that the section in (E) comes from an embryo with a pigmented RPE (brown stain) while the one in (D) comes from an albino embryo. (F) Limited cycle RT-PCR analysis performed on RNA isolated from wild type and Chx10orJ/orJ whole eyes at E13.5. Primers for A-, J-, H-, D- and M-Mitf are as those used for Fig. 3A. For pan-specific amplification, primers in exon 5 and 7 were used. Note that for A- and J-Mitf, at the lower number of PCR cycles, the intensities of the bands are slightly increased in mutant compared to wild type eyes, but at the higher number of cycles, the difference is no longer visible. No differences are seen for M-Mitf. H- and D-Mitf, however, show a clear difference between wild type and mutant at both 29 and 30 cycles of amplification. The use of primers specific for CyclinD1 indicates a reduction in mutant, consistent with the corresponding retinal hypoproliferation.
To determine whether CHX10 interacts in vivo with Mitf promoters, we performed chromatin immunoprecipitation (ChIP) assays using extracts of eye primordia at E10.5-E11.0, corresponding to the period during which Mitf is downregulated in the retina. The results, described in detail in Supplemental Figs. S3 and S4, indicate a weak interaction of CHX10 with a binding site in the A-Mitf promoter, a strong interaction with a site in the H-Mitf promoter, and an equally strong interaction with at least two sites in the D-Mitf promoter. Taken together, the results suggest that H- and D-Mitf are the major, and likely direct, targets of the Chx10-mediated Mitf downregulation in the retina while A- and J-Mitf are not much affected by Chx10.
Deletion of Mitf exons 1H, 1D and 1B corrects the retinal hypoproliferation phenotype in Chx10 mutant embryos
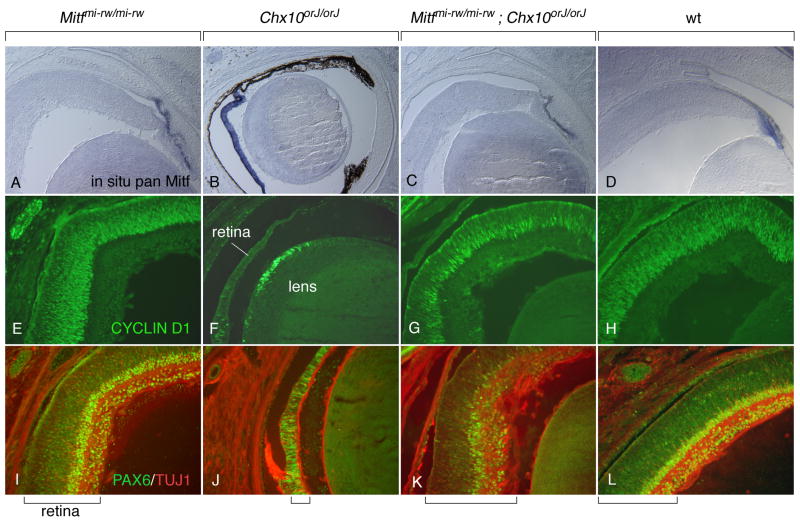
The above results prompted us to finally ask whether the lack of H- and D-MITF and exon 1B, as seen in rw embryos, might be sufficient to correct the Chx10-mediated retinal hypoproliferation phenotype. Such a correction has been described so far only with dominant-negative Mitf mutations that equally affect all isoforms (Konyukhov and Sazhina, 1966; Horsford et al., 2005; Bharti et al., 2006). Therefore, we intercrossed Chx10orJ/orJ mice with Mitfmi-rw/mi-rw mice to generate double homozygotes. Using CYCLIN D1 staining in conjunction with PAX6 staining and staining for the neuronal marker TUJ1 indicated that, as expected, eyes from Mitfmi-rw newborns (Fig. 7A,E,I) had a retinal thickness and lamination similar to wild type (Fig. 7D,H,L) while eyes from Chx10orJ newborns showed the severe retinal hypoproliferation and retina-to-RPE transdifferentiation described previously (Burmeister et al., 1996; Rowan et al., 2004; Horsford et al., 2005) (Fig. 7B,F,J). In the double Mitfmi-rw;Chx10orJ homozygotes, however, CYCLIN D1 staining and retinal thickness were nearly normal although retinal lamination was still abnormal (Fig. 7C,G,K). This result indicated that in Chx10 mutants, Mitf upregulation, once deprived of the contribution of H- and D-Mitf and exon 1B, no longer leads to retinal hypoproliferation. This result is consistent with earlier observations showing that dominant-negative Mitf mutations partially correct the Chx10 mutant phenotype (Konyukhov and Sazhina, 1966; Horsford et al., 2005).
Fig. 7.
The Mitfmi-rw allele partially rescues the Chx10orJ mutant phenotype. Eyes from newborns of the indicated genotypes were sectioned and processed for in situ hybridization with a pan-Mitf probe (A–D), CYCLIND1 immunofluorescence (E–H), or PAX6/TUJ1 double immunofluorescence (I–L). Compared with wild type, Mitfmi-rw/mi-rw retinas appear normal both in thickness and in staining. In contrast, Chx10orJ/orJ retinas retain Mitf expression and are severly hypoplastic, with a pigmented monolayer replacing the retina particularly in the distal part (B). Moreover, they show fewer CYCLIND1-positive and PAX6-positive cells (F,J). Eyes from Mitfmi-rw/mi-rw;Chx10orJ/orJ double mutants, however, have retinas of relatively normal appearance and thickness even though their PAX6 staining and lamination is still abnormal (C,G,K). Brackets at the bottom mark the thickness of the retina.
Discussion
The dissection of the expression patterns of Mitf isoforms has shown that some isoforms, such as H- and D-Mitf, are present in a temporally and spatially restricted manner while others, such as A- and J-Mitf, are found ubiquitously both within and outside the optic neuroepithelium. The notion that H- and D-Mitf are more important for eye development than other isoforms rests on several lines of reasoning. First, H- and D- Mitf consistently accumulate to higher levels in the RPE than other isoforms. Second, retinal hypoproliferation in Chx10 mutants is associated with abnormally high levels of H-and D-Mitf but not A- and J-Mitf or other isoforms. Third, the lack of H- and D-Mitf in Mitf red-eyed white mutants, although confounded by complex changes in mRNAs and protein isoforms, is clearly associated with developmental eye defects. Fourth, when H- and D-Mitf are missing in Chx10 mutants, retinal development is restored. It appears, therefore, that H- and D-Mitf need to be precisely regulated during eye development while the absence of other isoforms, or their presence in the retina, is more readily tolerated.
The fact that upstream Mitf regulatory regions contain distinct transcription factor binding motifs that are conserved across mammalian and avian species (Hallsson et al., 2007) argues strongly for a transcriptional control of the different Mitf isoforms. For instance, the ubiquitously active A-Mitf promoter has potential binding sites for transcription factors with widespread expression patterns such as MYC/MAX, STAT, and SP1 (Hallsson et al., 2007). In contrast, the H- and D-Mitf promoters have potential binding sites for the retina-specific transcription factor CHX10 (Hallsson et al., 2007; and this paper), precisely the factor involved in the retinal downregulation of these two promoters. In fact, ChIP assays clearly indicate that at least some of these sites are occupied by CHX10 in vivo, suggesting a direct regulation of the respective Mitf promoters by this transcription factor.
A separate control of each promoter does not exclude, however, the presence of additional mechanisms by which distinct Mitf mRNAs are co-regulated. A transcriptional co-regulatory mechanism has been suggested by the observation, made by immunofluorescence, that Pax2/Pax6 double knock-out mice lack MITF protein expression in the optic neuroepithelium and that the human A-MITF promoter contains binding sites for PAX2/PAX6, suggesting that it acts as a control region for all ocular Mitf expression (Bäumer et al., 2003). These binding sites, however, are not well conserved across mammalian and avian species. Hence, it is quite possible that even in Pax2/Pax6 double knock-out mice, the ubiquitously expressed A- and J-MITF, which are undetectable by immunofluorescence on histological sections (data not shown), may continue to be present while only the major and immunofluorescently detectable H- and D-MITF are absent. A different, post-transcriptional co-regulatory mechanism is suggested by the conservation, in the 3′UTR, of potential recognition sites for microRNAs, some of which are thought to be controlled by Chx10 (Hallsson et al., 2007; Xu et al., 2007). In fact, microRNAs are excellent candidates to contribute to the retinal downregulation of all Mitf mRNAs that share the same 3′UTR, but the contribution of these microRNAs is likely small and may not exceed the minor effects that Chx10 has, for instance, on the levels of A- and J-Mitf.
The fact that each of the nine promoters is linked to a unique exon also leads to the important question of whether multiple promoters exist solely to provide the correct levels of MITF protein at each developmental time point and location, or whether they exist to generate mRNAs or proteins with isoform-specific functions. Indeed, some evidence suggests that sequence differences at the MITF aminoterminus can influence the proteins’ activities. For instance, ectopic expression of ascidian Mitf, whose aminoterminal sequence resembles A-MITF, induces additional pigment cells in ascidian larvae, and so does mouse A-MITF, but not mouse H- or M-MITF (Yajima et al., 2003). Furthermore, A-, MC- and E-MITF regulate distinct sets of target genes in Mitf null mutant mast cells (Shahlaee et al., 2007). There is also evidence, however, that expression levels are more important than specific sequences to determine MITF activities. For instance, regardless of whether A-MITF or M-MITF was expressed in quail neuroepithelial cells, pigmented colonies arose that consisted of RPE-like, epithelial cells, in which MITF was low, and of neural crest-like, dendritic cells, in which MITF was high (Planque et al., 2004). Our findings with the natural Mitfmi-rw deletion suggest, in fact, that low levels of aminoterminally truncated MITF proteins are capable of inducing pigmentation in the ciliary margin and the neuroepithelial portions of the iris, while in the complete absence of Mitf, no such pigmentation is ever seen (Hodgkinson et al., 1998; Nakayama et al., 1998). This finding is consistent with the fact that aminoterminally truncated MITFs retain activity on promoters of pigmentation genes, including the promoter of tyrosinase which encodes the rate-limiting enzyme for melanin synthesis (see Supplemental Fig. S2). On the other hand, the dorsal RPE is still abnormal in Mitfmi-rw embryos. It is conceivable, therefore, that full RPE development, in contrast to mere pigmentation, depends on the presence of the upstream coding exons, in particular perhaps exon 1B1b. This exon contains a glutamine-rich domain, thought to be involved in protein-protein interactions, that is conserved in vertebrates and flies (Hallsson et al., 2007) and is reminiscent of similar domains in other transcription factors such as CREB or SP1. That cell differentiation does not require this domain, however, is evident from the fact that M-MITF is sufficient to induce pigmentation upon experimental expression in cultured cells (Bejar et al., 2003), and that Mitfmi-rw mice show RPE and iris pigmentation as well as some pigmented spots in the coat. In any event, a definitive answer to the question of why there are distinct promoters must await results from targeted promoter/exon knockouts and isoform-specific rescue experiments.
In addition to highlighting the importance of alternative promoter use, our study of the Mitfmi-rw deletion also draws attention to alternative translation initiation. Alternative translation start sites have been observed in several genes and, much as alternative promoter use, can contribute to functional diversity in wild type and alter disease in mutants (Sandelin et al., 2007; Scheper et al., 2007; Zhang et al., 2007). The fact that in the rw mutant mice, translation can be initiated from downstream start codons suggests that these codons may be utilized for initiation even in the context of a wild type sequence, although at low efficiency. Whether this is biologically relevant, however, remains to be explored.
Finally, the fact that the Mitfmi-rw deletion is associated with random bilateral asymmetries in eye development merits special consideration. The fact that genetically identical cells, even if exposed to the same environment, can show substantial variations in phenotypic manifestations is thought to result from “stochastic” variations in gene expression, likely due to variant epigenetic chromatin states (for review, see Kaern et al., 2005). It is reasonable to assume that high levels of gene expression will generally buffer cells against random fluctuations in their molecular composition and so assure a normal embryonic development. If, however, only threshold levels of gene expression are achieved, as is the case for Mitf in rw mice with their abnormal Mitf transcripts, stochastic fluctuations in epigenetic states may lead to major phenotypic differences between individual cells. Given that epigenetic states can be passed on through cell divisions, cellular variations may lead to tissue variations and so to differences between organs that are normally bilaterally symmetrical. Hence, our study of Mitf isoforms not only touches on the complexities by which a single gene controls organogenesis but also on the intriguing question of how bilateral symmetries are being generated and maintained.
Supplementary Material
Acknowledgments
We thank Dr. Vincent Hearing for antibodies to tyrosinase, and the NINDS Sequencing Facility and the Animal Health and Care Section for excellent services. We also thank Drs. Dubois-Dalcq and Brian Howell and the members for the Mammalian Development Section for helpful comments on the manuscript. This work was supported in part by grants from the Italian Telethon Foundation to S.B. and by the Intramural Research Program of the NIH, NINDS.
References
- Amae S, Fuse N, Yasumoto K, Sato S, Yajima I, Yamamoto H, Udono T, Durlu YK, Tamai M, Takahashi K, et al. Identification of a novel isoform of microphthalmia-associated transcription factor that is enriched in retinal pigment epithelium. Biochem Biophys Res Commun. 1998;247:710–715. doi: 10.1006/bbrc.1998.8838. [DOI] [PubMed] [Google Scholar]
- Arnheiter H, Hou L, Nguyen MTT, Bismuth K, Csermely T, Murakami H, Skuntz S, Liu W, Bharti K. Mitf—A matter of life and death for the developing melanocyte. In: Hearing V, Leong SPL, editors. From Melanocytes to Melanoma: The progression to malignancy. Totowa, NJ: Humana Press; 2006. [Google Scholar]
- Bäumer N, Marquardt T, Stoykova A, Spieler D, Treichel D, Ashery-Padan R, Gruss P. Retinal pigmented epithelium determination requires the redundant activities of Pax2 and Pax6. Development. 2003;130:2903–2915. doi: 10.1242/dev.00450. [DOI] [PubMed] [Google Scholar]
- Bejar J, Hong Y, Schartl M. Mitf expression is sufficient to direct differentiation of medaka blastula derived stem cells to melanocytes. Development. 2003;130:6545–6553. doi: 10.1242/dev.00872. [DOI] [PubMed] [Google Scholar]
- Bharti K, Nguyen MT, Skuntz S, Bertuzzi S, Arnheiter H. The other pigment cell: specification and development of the pigmented epithelium of the vertebrate eye. Pigment Cell Research. 2006;19:380–394. doi: 10.1111/j.1600-0749.2006.00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bora N, Conway SJ, Liang H, Smith SB. Transient overexpression of the Microphthalmia gene in the eyes of Microphthalmia vitiligo mutant mice. Dev Dyn. 1998;213:283–292. doi: 10.1002/(SICI)1097-0177(199811)213:3<283::AID-AJA5>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Bumsted KM, Barnstable CJ. Dorsal retinal pigment epithelium differentiates as neural retina in the microphthalmia (mi/mi) mouse. Invest Ophthalmol Vis Sci. 2000;41:903–908. [PubMed] [Google Scholar]
- Burmeister M, Novak J, Liang MY, Basu S, Ploder L, Hawes NL, Vidgen D, Hoover F, Goldman D, Kalnins VI, et al. Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nature Genetics. 1996;12:376–384. doi: 10.1038/ng0496-376. [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nature Genetics. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Green ES, Stubbs JL, Levine EM. Genetic rescue of cell number in a mouse model of microphthalmia: interactions between Chx10 and G1-phase cell cycle regulators. Development. 2003;130:539–552. doi: 10.1242/dev.00275. [DOI] [PubMed] [Google Scholar]
- Hallsson JH, Favor J, Hodgkinson C, Glaser T, Lamoreux ML, Magnusdottir R, Gunnarsson GJ, Sweet HO, Copeland NG, Jenkins NA, et al. Genomic, transcriptional and mutational analysis of the mouse microphthalmia locus. Genetics. 2000;155:291–300. doi: 10.1093/genetics/155.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallsson JH, Haflidadottir BS, Schepsky A, Arnheiter H, Steingrimsson E. Evolutionary sequence comparison of the Mitf gene reveals novel conserved domains. Pigment Cell Research. 2007;20:185–200. doi: 10.1111/j.1600-0749.2007.00373.x. [DOI] [PubMed] [Google Scholar]
- Hershey CL, Fisher DE. Genomic analysis of the Microphthalmia locus and identification of the MITF-J/Mitf-J isoform. Gene. 2005;347:73–82. doi: 10.1016/j.gene.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Nakayama A, Li H, Swenson LB, Opdecamp K, Asher JH, Jr, Arnheiter H, Glaser T. Mutation at the anophthalmic white locus in Syrian hamsters: haploinsufficiency in the Mitf gene mimics human Waardenburg syndrome type 2. Hum Mol Genet. 1998;7:703–708. doi: 10.1093/hmg/7.4.703. [DOI] [PubMed] [Google Scholar]
- Horsford DJ, Nguyen MT, Sellar GC, Kothary R, Arnheiter H, McInnes RR. Chx10 repression of Mitf is required for the maintenance of mammalian neuroretinal identity. Development. 2005;132:177–187. doi: 10.1242/dev.01571. [DOI] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nature Reviews Genetics. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- Konyukhov BV, Sazhina MV. Interaction of the genes of ocular retardation and microphthalmia in mice. Folia Biol (Praha) 1966;12:116–123. [PubMed] [Google Scholar]
- Kozak M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature. 1984;308:241–246. doi: 10.1038/308241a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Liu IS, Chen JD, Ploder L, Vidgen D, van der Kooy D, Kalnins VI, McInnes RR. Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron. 1994;13:377–393. doi: 10.1016/0896-6273(94)90354-9. [DOI] [PubMed] [Google Scholar]
- Müller G. Eine entwicklungsgeschichtliche Untersuchung über das erbliche Kolobom mit Mikrophthalmus bei der Hausmaus. Z Mikrosk Anat Forsch. 1950;56:520–558. [Google Scholar]
- Nakayama A, Nguyen MT, Chen CC, Opdecamp K, Hodgkinson CA, Arnheiter H. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech Dev. 1998;70:155–166. doi: 10.1016/s0925-4773(97)00188-3. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Arnheiter H. Signaling and transcriptional regulation in early mammalian eye development: a link between FGF and MITF. Development. 2000;127:3581–3591. doi: 10.1242/dev.127.16.3581. [DOI] [PubMed] [Google Scholar]
- Planque N, Raposo G, Leconte L, Anezo O, Martin P, Saule S. Microphthalmia transcription factor induces both retinal pigmented epithelium and neural crest melanocytes from neuroretina cells. J Biol Chem. 2004;279:41911–41917. doi: 10.1074/jbc.M404964200. [DOI] [PubMed] [Google Scholar]
- Rowan S, Chen CM, Young TL, Fisher DE, Cepko CL. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development. 2004;131:5139–5152. doi: 10.1242/dev.01300. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nature Reviews Genetics. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Scheper GC, van der Knaap MS, Proud CG. Translation matters: protein synthesis defects in inherited disease. Nature Reviews Genetics. 2007;8:711–723. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- Shahlaee AH, Brandal S, Lee YN, Jie C, Takemoto CM. Distinct and shared transcriptomes are regulated by microphthalmia-associated transcription factor isoforms in mast cells. J Immunol. 2007;178:378–388. doi: 10.4049/jimmunol.178.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D’Amare AR, Burley SK, Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, Jenkins NA. Melanocytes and the microphthalmia transcription factor network. Annual Review of Genetics. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA transcriptome of mouse retina and identification of a sensory organ specific miRNA cluster. J Biol Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- Yajima I, Sato S, Kimura T, Yasumoto K, Shibahara S, Goding CR, Yamamoto H. An L1 element intronic insertion in the black-eyed white (Mitf[mi-bw]) gene: the loss of a single Mitf isoform responsible for the pigmentary defect and inner ear deafness. Hum Mol Genet. 1999;8:1431–1441. doi: 10.1093/hmg/8.8.1431. [DOI] [PubMed] [Google Scholar]
- Yajima I, Endo K, Sato S, Toyoda R, Wada H, Shibahara S, Numakunai T, Ikeo K, Gojobori T, Goding CR, et al. Cloning and functional analysis of ascidian Mitf in vivo: insights into the origin of vertebrate pigment cells. Mech Dev. 2003;120:1489–1504. doi: 10.1016/j.mod.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Zhang J, Cai J, Li Y. A genome-wide survey of alternative translational initiation events in Homo sapiens. Science in China. 2007;50:423–428. doi: 10.1007/s11427-007-0040-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.