Abstract
The homeodomain transcription factor Crx is required for expression of many photoreceptor genes in the mammalian retina. The mechanism by which Crx activates transcription remains to be determined. Using protein–protein interaction assays, Crx was found to interact with three co-activator proteins (complexes): STAGA, Cbp and p300, all of which possess histone acetyl-transferase (HAT) activity. To determine the role of Crx–HAT interactions in target gene chromatin modification and transcriptional activation, quantitative RT–PCR and chromatin immunoprecipitation were performed on Crx target genes, rod and cone opsins, in developing mouse retina. Although cone opsins are transcribed earlier than rhodopsin during development, the transcription of each gene is preceded by the same sequence of events in their promoter and enhancer regions: (i) binding of Crx, followed by (ii) binding of HATs, (iii) the acetylation of histone H3, then (iv) binding of other photoreceptor transcription factors (Nrl and Nr2e3) and RNA polymerase II. In Crx knockout mice (Crx−/−), the association of HATs and AcH3 with target promoter/enhancer regions was significantly decreased, which correlates with aberrant opsin transcription and photoreceptor dysfunction in these mice. Similar changes to the opsin chromatin were seen in Y79 retinoblastoma cells, where opsin genes are barely transcribed. These defects in Y79 cells can be reversed by expressing a recombinant Crx or applying histone deacetylase inhibitors. Altogether, these results suggest that one mechanism for Crx-mediated transcriptional activation is to recruit HATs to photoreceptor gene chromatin for histone acetylation, thereby inducing and maintaining appropriate chromatin configurations for transcription.
INTRODUCTION
Rods and cones are the two types of photoreceptor neurons in the vertebrate retina that are specialized for carrying out phototransduction, the process of converting light into a neuronal signal. These neurons preferentially express a set of genes, the so-called photoreceptor genes, which encode proteins that are required for photoreceptor function. Of these genes, rhodopsin and cone opsins have been studied extensively. Rhodopsin is specifically expressed by the rod photoreceptors that are highly sensitive to dim light and thus responsible for night vision. M/L- and S-cone opsins are expressed by the corresponding cone photoreceptors that are responsible for color vision in bright light. Appropriate expression of opsin genes, along with other photoreceptor genes, is required for normal photoreceptor development and maintenance, as over-expression or under-expression of these genes can lead to developmental defects or photoreceptor degeneration (1,2).
Development of various neurons in the mammalian retina follows a sophisticated choreographic process involving coordinated progenitor cell proliferation and differentiation. All major cell types in the retina are generated from common multi-potent progenitor cells (3,4) in an highly conserved order and overlapping phases (5): ganglion cells, horizontal cells and cone cells are born in early phases, whereas rods, bipolar cells and Muller glial cells are born in late phases (6,7). Despite a wide range of birth dates, developing photoreceptor cells express opsins, the terminal differentiation markers, much later during development (8). As an example, rhodopsin transcription in mouse was detected around postnatal day 3 (P3) and peaks at about P14 (9), which is much later than the birth dates (~P0) of the majority of rods. The mechanism for this ‘lagging’ period between birth dates and opsin expression is largely unknown. However, this phenomenon might be important for generating appropriate ratios of various photoreceptor subtypes.
Increasing evidence suggests that cell type specification and differentiation of the retinal progenitor cells is controlled by intrinsic factors and extrinsic signals (10,11). A series of homeodomain and basic helix–loop–helix proteins have been reported to serve as intrinsic factors (12,13). For rod and cone photoreceptor cells, interplays among members of homeodomain, basic leucine zipper and nuclear receptor families appear to play an important role in their cell fate specification and patterning (14). For example, rod development requires the action of homeodomain factors Otx2 (15) and Crx (16–18), the neuroretina leucine-zipper protein Nrl (19) and the photoreceptor-specific nuclear receptor Nr2e3 (20). M/L cone development requires thyroid hormone receptor beta 2 isoform (Trβ2) (21) in addition to Otx2/Crx. In contrast, S-cone specification appears to involve combinatorial actions of many factors, including all the factors mentioned above and several other members of the nuclear receptor family, such as the retinoid-related orphan receptor beta (Rorβ) (22), the retinoid X receptor gamma (Rxrγ) (23), the orphan nuclear receptor Nr2e1 (Tlx) (24) and possibly Nr1d1 (25) that interacts with Nr2e3. Despite recent progress in identifying photoreceptor transcription factors and their role in photoreceptor subtype specification, little is known about their mechanism of action. Both Crx and Nrl have been shown to act as transcription activators (16,26), whereas Nr2e3 is a dual regulator that activates the expression of some rod genes but represses cone genes (25,27–29).
Increasing evidence indicates that chromatin remodeling is an important mechanism for transcription regulation. Two major types of chromatin remodeling are known: (i) ATP-dependent nucleosome positioning, which alters the accessibility of nucleosomal DNA to regulatory factors (reviewed in 30); (ii) covalent modifications of histone tails, including acetylation, phosphorylation and methylation (reviewed in 31), which also alters the configuration of chromatin for interaction with transcription factors and the basal transcriptional machinery. Among these modifications, histone acetylation has been studied extensively. Enhanced transcription is often associated with increased histone acetylation, particularly lysine acetylation of histone H3 (at K9/K14 positions) (32), whereas silencing is commonly correlated with histone hypoacetylation (33–35). Histone acetylation is a dynamic process controlled by histone acetyl-transferases (HATs) and histone deacetylases (HDACs) (36,37). Several co-activator proteins or complexes are known to possess intrinsic HAT activity, including Gcn5/Pcaf (38,39), which catalyze K9/K14 acetylation of histone H3, and Cbp/p300 (40), which catalyze acetylation of both histone H3 and H4 (33). These HAT containing co-activators can be recruited to selected regulatory DNA regions (promoters/enhancers) by specific transcription factors to catalyze histone H3/H4 acetylation for transcriptional activation (reviewed in 41). Histone deacetylase inhibitors (HDACi) that block the reverse direction of this pathway can also enhance histone acetylation, thereby altering transcription (42). HDACi therapy is being developed to treat certain types of cancers that are characterized by transcription dysregulation (42,43).
It was observed a long time ago that mature rod and cone photoreceptors have distinct nuclear architecture and heterochromatin patterns (44). The mature rod nuclei possess a large and highly condensed heterochromatin territory surrounded by a thin and less condensed euchromatin layer. This charateristic chromatin pattern arises during rod maturation when rod-specific gene expression reaches a peak (45). Highly transcribed rod genes such as rhodopsin and transcription factors that regulate their expression appear to co-localize in the euchromatin area, excluded from the condensed heterochromatin territory (46). Interestingly, two regulatory proteins, ataxin-7 and pRb, have been reported to be important for the development or maintenance of this rod phenotype (46,47). Both are known to be involved in chromatin remodeling and transcription regulation. Ataxin-7 is a disease protein that causes spinocerebellar ataxia type 7 (SCA7) with photoreceptor degeneration, when its polyglutamine (poly-Q) tract is expanded (48). Ataxin-7 is a component of STAGA/TFTC chromatin remodeling complexes containing Gcn5 HAT (49,50). Ectopic expression of a poly-Q expanded ataxin-7 in rod photoreceptor cells resulted in dysregulation of photoreceptor genes and degeneration of rods, accompanied by altered histone H3 acetylation and loss of the heterochromatin pattern of affected cells (46). However, it is unclear which change is the cause and which is the consequence of other alterations, and more fundamentally, if any of the epigenetic patterns is directly linked to photoreceptor gene expression.
The retinoblastoma protein pRb is another chromatin modulator that is involved in rod differentiation. pRb is essential for retinal progenitor cell proliferation and differentiation of rod photoreceptors as demonstrated by conditional Rb1 knockout mice (47). Rb1-deficient immature photoreceptor cells exhibit a large nucleus volume, fail to undergo chromatin condensation and are unable to express rhodopsin and other rod markers (45,51). Consistent with this, the Rb1-deficient Y79 retinoblastoma cell line does not express opsins, the terminal differentiation markers for photoreceptors. pRb and its family of pocket proteins (p107 and p130) are known to regulate transcription by interacting with a number of transcription factors and cofactors/enzymes with histone modification and chromatin remodeling capability (52). pRb binds to E2F and converts it from a transcriptional activator to a repressor for genes involved in cell cycle progression through a mechanism that involves the recruitment of HDACs (53–55). Using neonatal mouse retinal explant cultures, HDAC activity was recently reported to be important for appropriate gene expression and differentiation of rods (56). pRb is also known to interact with histone methyltransferases to establish heterochromatin (57,58). Furthermore, pRb influences the accessibility of chromatin through interactions with ATP-dependent helicases, Brg1 and Brm, the catalytic subunits of the SNF/SWI chromatin remodeling complex (59,60). Thus, pRb could promote rod differentiation by activating rod gene expression via modulating chromatin organization and configuration. However, the exact molecular mechanism by which pRb regulates rod differentiation and its role in photoreceptor gene expression remain to be determined.
The cone-rod homeobox protein Crx is a photoreceptor-specific transactivator (16) required for differentiation and maintenance of both cone and rod photoreceptors. Crx contains a homeodomain near the N-terminus (Fig. 1B) that is required for DNA recognition and nuclear localization. It also contains two activation domains, AD-1 and AD-2 in the C-terminal portion (Fig. 1B), that are essential for transcriptional activation (61). Crx mutations cause several forms of retinal degeneration, including cone-rod dystrophy, Leber’s congenital amaurosis (LCA) and retinitis pigmentosa (62–67). Photoreceptors of homozygous Crx knockout (Crx−/−) mice do not develop outer segments and therefore are non-functional and undergo slow degeneration (18). Expression levels of many photoreceptor genes are altered, although not abolished, in Crx−/− mice (18,68,69), suggesting that these genes are either direct or indirect targets of Crx. Using chromatin immunoprecipitation (ChIP) assays, we showed previously that the opsin genes are direct targets of Crx (70,71). The mechanism by which Crx activates transcription is unknown. It has been reported to interact with several transcription regulators, including Nrl (72), Nr2e3 (29), Sp4/Sp1 (73) and Qrx (74). More recently, Crx was found to interact with ataxin-7 in the STAGA complex (75). This interaction is mediated by the poly-Q-rich regions of Crx and ataxin-7 (70). Since Gcn5 is a key component of STAGA, this finding implies that Crx can recruit Gcn5 HAT by interacting with ataxin-7 (50,70). Crx was also reported to interact with Cbp and p300, two other HAT-containing co-activators (76). Thus, we hypothesize that Crx activates transcription by recruiting HAT-harboring co-activators to target gene promoters and promoting histone acetylation. In this paper, we describe experimental evidence supporting this hypothesis. We show that two HATs, Gcn5 and Cbp, and acetylated histone H3 (AcH3) are associated with opsin gene promoters in a Crx-dependent manner in developing and mature mouse retina, as well as in Y79 retinoblastoma cells. This association correlates with opsin transcription. Furthermore, HDACi treatments can enhance K9/K14 acetylation of histone H3 and transcription of endogenous opsin genes in Crx-deficient Y79 cells.
Figure 1.
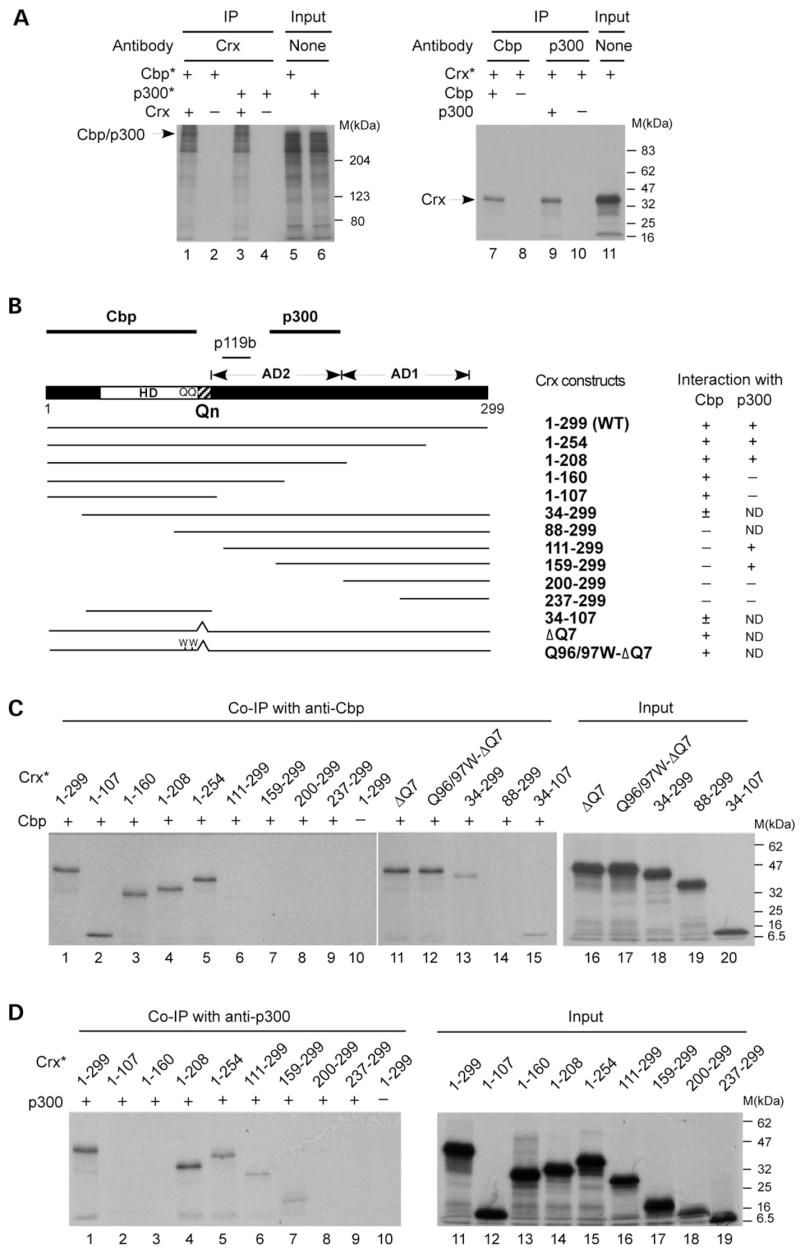
Crx interacts with Cbp and p300 in vitro via distinct domains. Co-ip assays were performed using the indicated IP antibodies and recombinant Cbp, p300 and various deleted/mutated forms of Crx made by in vitro transcription/translation. The results are presented as autoradiograms of SDS–PAGE (8–12%) analysis of co-immunoprecipitated proteins in (A), (C) and (D) and summarized in (B). (A) Co-ip with full-length WT proteins. Left panel: co-ip using anti-Crx (p119b) antibody and 35S-labeled Cbp or p300 (labeled by asterisk) in the presence of Crx (lanes 1 and 3) or the empty pcDNA3.1/HisC vector (lanes 2 and 4). Right panel: co-ip using anti-Cbp (lanes 7 and 8) or anti-p300 (lanes 9 and 10) and 35S-labeled Crx (Crx*) in the presence or absence (pcDNA3.1/HisC empty vector) of Cbp (lane 7 versus 8) or p300 (lane 9 versus 10). Lanes 5, 6 and 11 are input control without immunoprecipitation. (B) Schematic representation of the Crx coding region, showing the location of the homeodomain (HD), glutamine-rich region (Qn) and activation domains 1 and 2 (AD1, AD2). The N-terminus is towards the left, the C-terminus towards the right and the numbers below the schematic represent the amino acids at either end of the molecule. The regions that interact with Cbp, p300 and the p119b antibody are indicated by lines above the schematic. Expression constructs encoding various deleted and mutated forms of Crx are represented by lines below the diagram, and the results of co-ip assays are summarized in the table at the right. (C) 35S-labeled Crx (Crx*) and its deleted or mutated forms were incubated with Cbp (lanes 1–9, 11–15) or with the product of the empty vector (‘–’, lane 10) and immunoprecipitated by an anti-Cbp antibody. Lanes 16–20 are input controls from the mutated constructs without immunoprecipitation. (D) 35S-labeled Crx (Crx*) and its deleted forms were incubated with p300 (lanes 1–9) or with the empty vector (lane 10) and immunoprecipitated by an anti-p300 antibody. Lanes 11–19 are input controls showing the in vitro-translated Crx proteins from the deletion constructs.
RESULTS
Crx interacts with Cbp and p300 via distinct domains, independent of the Qn motif required for Gcn5 recruitment
Crx was previously reported to interact with Cbp and p300 in a mammalian two-hybrid assay (76), although whether this interaction is direct or indirect via other proteins is not known. To determine whether Crx directly interacts with Cbp or p300, co-immunoprecipitation (co-ip) assays with in vitro-translated proteins were carried out. Figure 1A shows that an anti-Crx antibody can co-immunoprecipitate 35S-labeled Cbp (lane 1 versus 2) or p300 (lane 3 versus 4) in a Crx-dependent manner. Reciprocally, an anti-Cbp or anti-p300 antibody can also co-immunoprecipitate 35S-labeled Crx in the presence of each respective antigen (lane 7 versus 8 and lane 9 versus 10). These results suggest that Crx can directly interact with Cbp or p300 in vitro.
To determine which domain(s) of Crx is responsible for interacting with Cbp or p300, in vitro co-ip assays were carried out using a series of deleted and mutated forms of Crx (Fig. 1B). The Crx N-terminal region spanning amino acids 1–107, including the homeodomain, appears to be responsible for interacting with Cbp (Fig. 1B), since deleting this region completely abolished the ability of Crx to interact with Cbp (Fig. 1C, lanes 6–9), whereas a peptide harboring this region alone is sufficient to mediate Crx’s interaction with Cbp (Fig. 1C, lane 2). Within this 1–107 region, amino acids 1–33 N-terminal to the homeodomain appear to contribute to Crx’s ability to interact with Cbp, since deleting these amino acids reduced Crx–Cbp interaction strength (scored as ± in Fig. 1B). This is based on a comparison between the intensity of immunoprecipited Crx from constructs with and without amino acids 1–33 in Figure 1C (lane 1 versus 13 and lane 2 versus 15). This 1–107 region also includes the glutamine-rich (Qn) region and two glutamine residues in the homeodomain C-terminus that are known to be critical for recruiting ataxin-7–STAGA complex containing Gcn5-HAT (70). To test whether Crx–Cbp interaction requires the Qn motif, in vitro co-ip assays were performed using mutant forms of Crx that partially (ΔQ7) or completely (Q96/97WΔQ7) lack the nine Q residues. Both mutants showed a normal ability to interact with Cbp (Fig. 1C, lanes 11 and 12 versus lane 1), suggesting that the Qn motif and C-terminus of the homeodomain are not critical for Crx’s interaction with Cbp. Taken together, these results suggest that the Crx domain responsible for interacting with Cbp maps to amino acids 1–107, independent of the Qn motif, as summarized in Figure 1B. Interestingly, the Crx domain responsible for interacting with p300 maps to a region within Crx transactivation domain 2 (AD-2) (amino acids 160–208) (Fig. 1B), distinct from the Cbp-interaction domain. As shown in Figure 1D, deleting C-terminal amino acids 209–299 (construct 1–208), or N-terminal amino acids 1–158 (construct 159–299), did not abolish Crx’s interaction with p300 (Fig. 1D, lanes 4 and 7), whereas any other deletion mutant missing the 160–208 region showed no detectable interaction with p300. Thus, Crx appears to use distinct domains for interacting with different co-activators, Cbp, p300 and previously reported STAGA-Gcn5 (via ataxin-7), raising the possibility that Crx could recruit all three HAT-containing co-activators simultaneously.
The next question is whether these Crx–co-activator interactions occur in vivo. As an initial step to address this question, co-ip assays were carried out using nuclear extracts made from P14 retinae of wild-type (WT) and Crx knockout (Crx−/−) mice. Figure 2 shows that an anti-Crx antibody (p119b) co-immunoprecipitated Cbp, p300 and Gcn5 with Crx from the nuclear extracts made from WT, but not from Crx−/− mice (lane 3 versus 4). Non-specific IgG failed to pull down the three HATs from WT nuclear extracts (lane 5), further demonstrating the specificity of the co-ip assays. These results suggest that Crx complexes with Cbp, p300 and Gcn5-STAGA in vivo.
Figure 2.
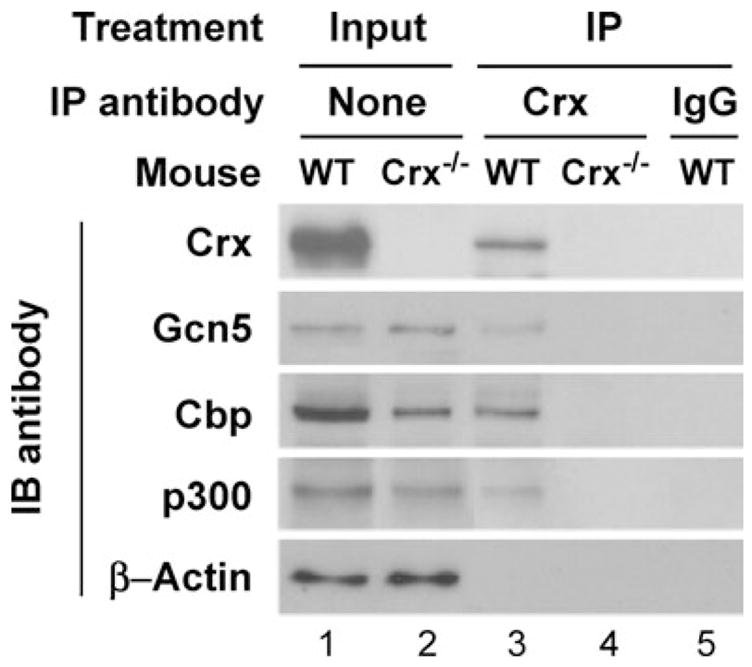
Co-ip of Gcn5, Cbp and p300 with Crx from retinal nuclear extracts. Equivalent amounts of retinal nuclear extracts from WT or Crx−/− mice were incubated with an anti-Crx (p119b) antibody or with normal rabbit IgG as negative control, precipitated by protein A beads, analyzed by SDS–PAGE and immunoblotted (IB) with antibodies against Gcn5 (sc20698), Cbp, p300, Crx or β-actin as a loading control. Lanes 1 and 2 are input controls without immunoprecipitation. Lanes 3–5 are co-immunoprecipitates from the retinas of WT (lanes 3 and 5) or Crx−/− (lane 4) mice using anti-Crx (lanes 3 and 4) or rabbit IgG (lane 5).
In addition, immunoblot analysis of the three co-activators in the nuclear extracts demonstrated that the amount of Cbp protein is significantly reduced in Crx−/− retina when compared with the WT level, although no apparent changes in Gcn5 and p300 protein levels were seen in the Crx−/− retina (Fig. 2, lane 2 versus 1). To confirm these results, quantitative RT–PCR assays were carried out to compare mRNA levels of each co-activator in the retina of Crx−/− versus WT mice. The Crx−/− retina showed lower mRNA levels than the WT retina for Cbp (46.7 ±1.9% WT) and p300 (75.0 ±2.0% WT), but no significant change in Gcn5 mRNA levels (95.6 ±2.1% WT). These results suggest that Crx is important for keeping appropriate expression levels of Cbp (and possibly p300) in the retina.
As a second step to determine whether Crx interacts with the three co-activators in vivo, immunohistochemistry assays were carried out to determine whether they are expressed in the photoreceptor cells where Crx is expressed. Antibodies against each of the four proteins were used to stain paraffin-embedded sections of P28 WT and Crx−/− mouse retina. As expected, Gcn5, Cbp and p300 are each expressed in all three nuclear layers of the mouse retina, whereas Crx is mainly expressed in the outer nuclear layer where photoreceptor nuclei reside (Supplementary Material, Fig. S1). The immunostaining intensity and distribution of each co-activator appear similar in Crx−/− and WT retina. This co-localization of Cbp, p300 and Gcn5 with Crx in photoreceptor nuclei further supports the interaction of Crx with these cofactors in vivo.
Crx recruits HAT-containing co-activators to the promoter/enhancer regions of opsin genes in the retina, triggering histone H3 (K9/K14) acetylation and transcriptional activation
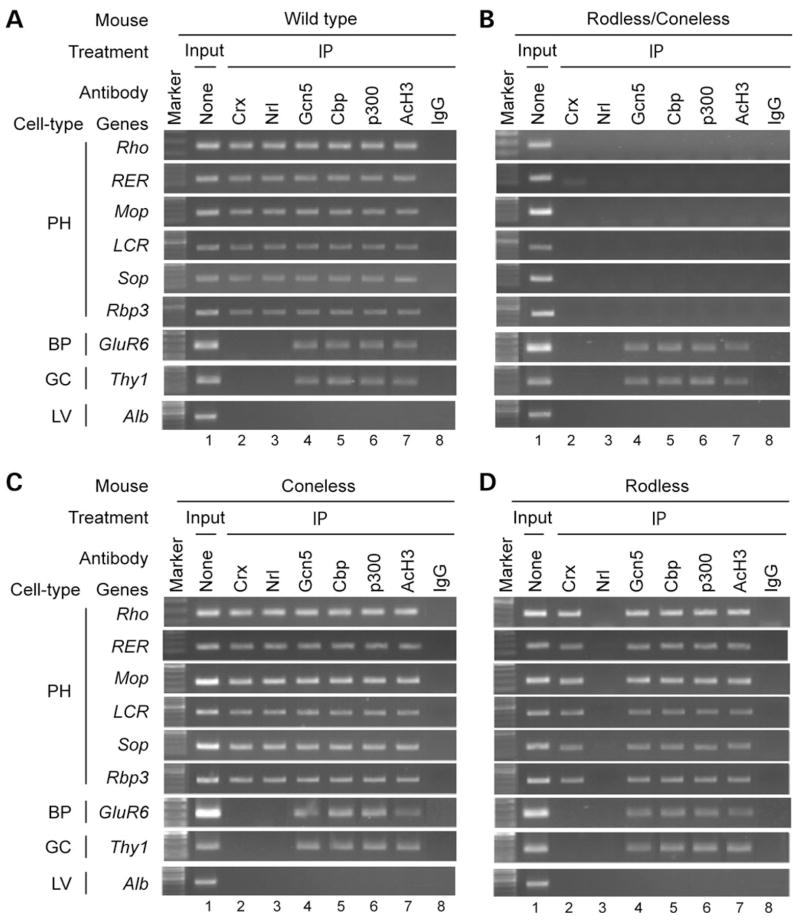
To determine whether HAT-containing co-activators Gcn5, Cbp and p300 contribute to Crx-dependent transcriptional regulation of the opsin genes in vivo, we carried out ChIP assays using mouse retina and antibodies against Crx, each of the three co-activators and one of their products, AcH3 (on K9/K14). The rod-specific transcription factor Nrl was also examined in the ChIP assays. As shown in Figure 3A, two photoreceptor transcription factors, Crx and Nrl, three HATs and AcH3 are all associated with the promoter and enhancer regions of the opsin genes in the retina, but not in the brain or liver (data not shown) where the opsin genes are not expressed. In contrast, in the retina, Crx and Nrl do not bind to the promoter of GluR6, a bipolar-specific gene, or Thy1, a ganglion cell gene, although the HATs and AcH3 are associated with the promoter of GluR6 and Thy1 (Fig. 3A). None of these regulators binds to the liver-specific gene Albumin (Alb) that is not transcribed in the retina (Fig. 3A–D). Furthermore, PCR analysis of IP samples using primers amplifying the 3′ region immediately downstream of each opsin gene did not yield detectable PCR products (data not shown), suggesting that the above transcription regulators bind preferentially to regulatory regions of the opsin genes. To determine whether the association of HATs and AcH3 with the opsin genes occurs in the photoreceptor cells, similar ChIP assays were performed using mouse retinas lacking cones, rods or both by genetic ablation [coneless-Red-DT(A); rodless-Nrl−/− and rodless/coneless-Red-DT(A)/rd1/rd1]. Consistent with photoreceptor-specific opsin expression, this HAT/AcH3-opsin promoter/enhancer association appears to occur only in the photoreceptor cells, as it is absent in rodless/coneless mouse retina where both rods and cones are depleted (Fig. 3B). In contrast, the HAT/AcH3 association with the promoter of bipolar/ganglion cell genes was unaffected in rodless/coneless retina, demonstrating the reliability and cell-type specificity of our ChIP assays. Furthermore, the HAT/AcH3 association with each of the opsin genes appears to occur in either rods or cones, as similar results were obtained using the retina of coneless mice [Red-DT(A)] where cones are genetically ablated (77) (Fig. 3C) or rodless mice (Nrl−/−) where all the photoreceptor cells are cones without detectable expression of rhodopsin or other rod-specific markers due to a null mutation of the rod transcription factor Nrl (19,78). Interestingly, HATs/AcH3 are associated with the rhodopsin promoter/enhancer in the absence of Nrl (Fig. 3D), similar to the findings in the WT and coneless mouse retina, and yet the rhodopsin gene is completely silent in Nrl−/− retina (19). These results suggest that Nrl is not required for mediating HAT/AcH3 association with the rhodopsin promoter/enhancer (discussed subsequently), and the HAT/AcH3 association alone is not sufficient for activating rhodopsin transcription. This implies that a high level of rhodopsin transcription requires the action of multiple transcription factors mediating chromatin remodeling and true transcriptional activation. Another interesting observation is that HATs/AcH3 and the two rod-specific transcription factors Nrl and Nr2e3 are associated with rhodopsin as well as cone opsins in rod photoreceptors (Fig. 3C) (14,71,79), suggesting that additional regulatory mechanisms are involved in silencing cone genes in rods. In summary, the results presented in Figure 3 suggest that, in photoreceptor cells, the three HATs Gcn5, Cbp and p300 are recruited to the promoter/enhancer of opsin genes, which correlates with K9/K14 acetylation of histone H3 and transcriptional activation of these genes.
Figure 3.
HATs and AcH3 are associated with promoter/enhancer regions of opsin genes in photoreceptor cells. (A) ChIP assays using the retinas of adult (P28) WT mice with antibodies to Crx (p119b, lane 2), Nrl (lane 3), Gcn5 (sc20698, lane 4), Cbp (lane 5), p300 (lane 6) or AcH3 (lane 7). Input (chromatin samples without immunoprecipitation) served as the positive control (lane 1) and immunoprecipitation with normal rabbit IgG (lane 8) was the negative control. Immunoprecipitated DNA fragments were analyzed by PCR for promoter/enhancer regions of photoreceptor-specific (PH) genes (Rho, rhodopsin; RER, rhodopsin enhancer region; Mop, M-cone opsin; LCR, cone opsin enhancer/locus control region; Sop, S-cone opsin; Rbp3, inter-photoreceptor binding protein) and non-photoreceptor genes [GluR6, metabotropic glutamate receptor subtype 6 expressed by bipolar cells (BP); Thy1, a marker for ganglion cells (GC) and albumin (Alb), a marker for liver cells (LV)]. The results are shown as combined gel (1% agarose) images. Similar results were obtained for P14 and P56 retinas (data not shown). (B) ChIP assays using the retinas of adult (P90) rodless/coneless [Red-DT(A)/rd1/rd1] mice. (C) ChIP assays using the retinas of adult (P56) coneless [Red-DT(A)] mice. (D) ChIP assays using the retinas of P14 rodless (Nrl−/−) mice.
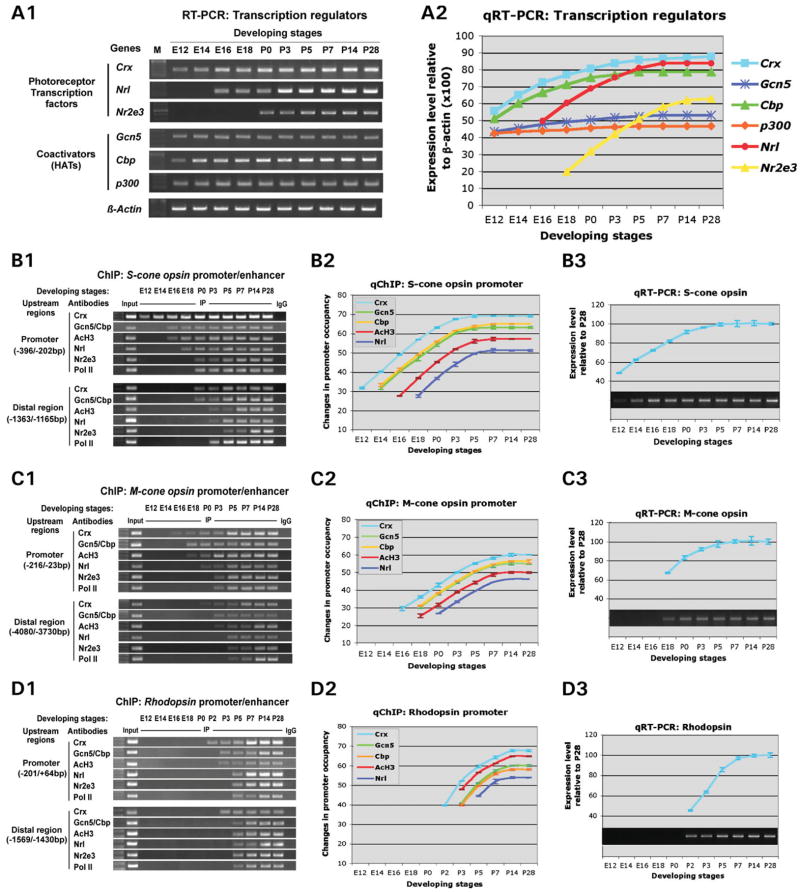
The association of three HATs with Crx in retinal nuclear extracts and on the promoter region of the opsin genes raises a possibility that Crx activates opsin transcription by recruiting these HATs to its targets to promote histone acetylation. This model predicts that, during photoreceptor development, opsin transcription should be preceded by Crx binding to the promoter followed by recruitment of HATs and AcH3. To test this prediction, quantitative RT–PCR and ChIP assays were performed on each opsin gene using developing mouse retinas of various ages from embryonic day 12 (E12) to postnatal day 28 (P28). The results of these assays are shown in Figure 4. First, temporal mRNA expression of known photoreceptor transcription factors, Crx, Nrl and Nr2e3, and three co-activators was measured by RT–PCR (Fig. 4A1 and A2). Crx mRNA was detected at E12, earlier than Nrl (first seen at E16) and Nr2e3 (at P0), substantially increased thereafter and reached a peak at around P7 (Fig. 4A1 and A2). By comparison, Nr2e3 mRNA was initially detected at birth, later than Nrl (E16) and showed kinetics similar to those of Nrl but distinct from those of Crx (Fig. 4A2), consistent with the fact that Nr2e3 requires Nrl for mRNA expression (19). In contrast, the three co-activators are expressed throughout the developmental stages examined without substantial changes in mRNA levels for Gcn5 and p300 (Fig. 4A1 and A2). However, Cbp mRNA also substantially increased during retinal development with kinetics similar to those of Crx (Fig. 4A2), consistent with the observation that Crx regulates the expression of Cbp (discussed earlier).
Figure 4.
Sequence of events on Crx target gene promoters during retinal development. ChIP analysis and qRT–PCR assays were performed on retinas from mice of various ages from embryonic day (E) 12 to postnatal day (P) 28 to determine when each of the various factors bind relative to transcription initiation. (A) RT–PCR analysis of mRNA levels of the indicated photoreceptor transcription factors and co-factors during retinal development with β-actin as a positive control. The results are presented by gel images (A1) and line graphs (A2) of quantified mRNA levels normalized to concurrent β-actin expression. (B) Events leading to S-cone opsin gene transcription. (B1) ChIP assay results are presented as combined gel (1% agarose) images of PCR analysis of the promoter and distal regions of S-cone opsin. (B2) Quantitative ChIP assay results are presented as the change in relative promoter occupancy (changes in ΔCt referenced to P28). (B3) S-cone opsin expression determined by qRT–PCR. The 1% agarose gel image is shown together with a graph of the expression level at each time point relative to the maximal level expressed at P28. (C) Events leading to M-cone opsin gene transcription. (C1) Gel images of ChIP analysis for binding of the transcription factors to the M-cone opsin promoter and distal enhancer (LCR). (C2) qChIP showing parallel kinetics of binding of the various factors to M-opsin promoter. (C3) M-cone opsin expression measured by qRT–PCR. (D) Events leading to rhodopsin gene transcription. (D1) Gel images of ChIP analysis for binding of the indicated factors to rhodopsin promoter and distal enhancer (RER). (D2) qChIP showing kinetics of binding of the various factors to rhodopsin promoter. (D3) rhodopsin expression measured by qRT–PCR.
Next, ChIP analysis using antibodies against the seven transcription regulators was performed on the proximal promoter and distal enhancer region of each opsin gene at various stages of retinal development (Fig. 4B1, B2, C1, C2, D1 and D2). The results demonstrated that binding of the regulators to each opsin gene’s upstream regions follows a sequential order: (i) binding of Crx, (ii) recruitment of Gcn5 and Cbp and appearance of acetylated histone H3, (iii) binding of other transcription factors Nrl and Nr2e3 and lastly (iv) binding of an activated form of RNA polymerase II (CTD Ser5 phosphorylated; Pol II) (Fig. 4B1, C1 and D1). For the first five regulators in this sequential order, the amount of opsin promoter DNA bound by each factor was quantified by real-time PCR (IP versus Input) at each developmental time point shown in Figure 4B1, C1 and D1. Changes in binding at each time point examined were then presented as line graphs in Figure 4B2, C2 and D2. For each opsin promoter, the shape of the lines appears to be similar to that of Crx, the first factor bound to the promoter, although the other lines were shifted to the right (indicating that binding was first detected at later time points). This sequential binding and parallel kinetics of increase correlates with the temporal expression pattern of each corresponding opsin gene measured by quantitative RT–PCR (Fig. 4B3, C3 and D3). Using S-cone opsin as an example, Figure 4B1 and B2 show that Crx was first found on the promoter region weakly at E12, before any other regulators including the HATs Gcn5 and Cbp that are highly expressed at the time (Fig. 4A1 and A2). When Crx’s binding reached a higher level at E14, Gcn5 and Cbp began to bind, followed by appearance of AcH3 at E16, then Nrl at E18 and finally Nr2e3 and Pol II at P0. Consistent with this sequential binding of the regulatory proteins, S-cone opsin transcript was barely detectable at E12, significantly increased at E16 after HAT/AcH3 promoter association was seen and peaked at around P3–P5 when binding of each factor to the promoter reached a plateau (Fig. 4B3). Furthermore, sequential binding of the transcription regulators also occurred in the more distal region (putative enhancer), although the timing shifted to later developmental stages (Fig. 4B1, bottom 6 rows). Thus, S-cone opsin transcription appears to be initiated and regulated by sequential binding of Crx, HATs/AcH3 and other regulators. Similar sequential binding and temporal transcription patterns were seen for the M-cone opsin (Fig. 4C1–C3) and rhodopsin (Fig. 4D1–D3) genes, although their curves were shifted to later ages and the kinetics were also different. For example, the sequential binding events on rhodopsin promoter/enhancer regions were detected after P2 (Fig. 4D1 and D2), much later than the two cone opsins (Fig. 4B1, B2, C1 and C2), consistent with the relatively late phase of rhodopsin transcription (Fig. 4D3) when compared with that of cone opsins (Fig. 4B3 and C3). Furthermore, sequential binding of the factors to the rhodopsin promoter had faster kinetics than the cone opsins, which also correspond to the fast kinetics of rhodopsin mRNA induction during the first 7 days of postnatal mouse life. These results suggest that Crx’s recruiting of HATs to the target promoter/enhancer for histone H3 acetylation is a prerequisite for opsin transcription induction during photoreceptor development.
Crx is required for recruiting Gcn5, Cbp and AcH3 to the opsin promoter/enhancer regions in the mouse retina and Y79 retinoblastoma cells
To further determine the role of Crx in HAT recruitment and histone acetylation, we carried out quantitative ChIP assays to compare the association of three HATs and AcH3 and H4 with the promoter/enhancer regions of three opsin genes in Crx knockout (Crx−/−) and WT mouse retinas. Table 1 shows that, in the absence of Crx, binding of Gcn5 and Cbp, but not p300, to the promoter/enhancer of all three opsins was significantly reduced, ranging from 21 to 58% of the WT levels. Consistent with this, the promoter/enhancer of all three opsin genes had significantly reduced levels of AcH3, ranging from 0.2% (for M-cone opsin locus control region LCR) to 33% (for rhodopsin promoter) of the WT levels. In addition, the promoters of S-opsin and rhodopsin also showed significantly decreased levels of acetylated histone H4 (36 and 64% of WT levels, respectively). These reductions correlate with little (8% for rhodopsin, 4% for S-cone opsin) or no (M-opsin) transcription of the opsin genes in Crx−/− retina reported previously (18). The reduction in Gcn5/Cbp binding cannot be explained fully by possible Gcn5/Cbp deficiency, since (i) western blots and quantitative RT–PCR showed no significant changes in Gcn5 mRNA/protein levels, and the Cbp mRNA/protein levels remain at least 50% in Crx−/− retina (discussed subsequently); (ii) immunohistochemistry showed that Gcn5 and Cbp proteins are present in the outer nuclear layer of Crx−/− retina (Supplementary Material, Fig. S1) and (iii) no significant Gcn5/Cbp binding defects were detected for Rbp3, a Crx-independent gene (18). Thus, these results suggest that Crx is required for recruiting Gcn5 and Cbp, but not p300, to the opsin gene targets as well as for acetylation of histone H3 (and histone H4 on rhodopsin and S-cone opsin promoters). These HAT recruitment and histone modifications are likely important for transcriptional activation of the opsin genes in the retina, since opsin expression levels parallel these decreases in Crx−/− retinas (18).
Table 1.
Relative promoter occupancy (as % of WT) in P14 Crx−/− mouse retina
| Genes/regions | Transcription regulators | ||||||
|---|---|---|---|---|---|---|---|
| Crx | Gcn5 | Cbp | p300 | AcH3 | AcH4 | ||
| Mop | Promoter | 0.0* | 57.7 ±2.7* | 42.3 ±2.4* | 102.1 ±3.8 | 5.5 ±1.7* | 96.3 ±7.7 |
| LCR | 0.0* | 35.5 ±2.2* | 32.8 ±2.7* | 100.0 ±7.7 | 0.2 ±0.9* | 98.7 ±5.6 | |
| Sop | Promoter | 0.0* | 45.7 ±3.3* | 31.9 ±3.9* | 97.7 ±5.9 | 6.8 ±2.1* | 36.4 ±3.7* |
| Rho | Promoter | 0.0* | 35.1 ±4.0* | 46.3 ±2.3* | 96.7 ±5.8 | 33.3 ±4.9* | 64.0 ±4.6* |
| RER | 0.0* | 21.2 ±3.0* | 42.4 ±3.6* | 105.4 ±8.1 | 7.3 ±1.8* | 95.9 ±5.9 | |
| Rbp3 | Promoter | 0.0* | 89.7 ±7.2 | 87.2 ±6.4 | 102.9 ±8.6 | 107.4 ±9.4 | 87.1 ±5.8 |
The promoter or enhancer occupancy was measured by ΔCT (i.e. CTIP − CTInput) for each primer pair. The results from Crx deficient (Crx−/−) retina are presented as % ΔCT of WT and represent mean ± STDEV of three trials.
P < 0.05 based on paired Student’s t-test.
Crx interacts with two rod-specific transcription factors, Nrl and Nr2e3. To determine whether these two factors also contribute to HAT recruitment and histone acetylation of the opsin genes, we carried out quantitative ChIP assays using Nrl knockout mice (Nrl−/−), which lack both Nrl and Nr2e3 (19). Consistent with gel images of ChIP–PCR shown in Figure 3C, no significant alterations in binding of Crx, three HATs and AcH3 to the opsin gene promoters/enhancers were detected (Supplementary Material, Table S1). Thus, unlike Crx, Nrl (or Nr2e3) is not required for recruiting the three HATs and K9/K14 acetylation of histone H3 of the opsin genes. This finding is consistent with the observation that Nrl does not bind to the opsin gene promoters until after Crx and HATs/AcH3 bind during retinal development (Fig. 4).
To further determine whether the above findings from the mouse retina also apply to human photoreceptor genes, we performed similar RT–PCR and ChIP analysis of the opsin genes using a human retinoblastoma cell line Y79 and compared the results with those from a normal human retina sample. Y79 cells are undifferentiated (80) and do not express any opsin (Fig. 5A). These cells do express CRX messenger RNA, but at a much lower level than the human retina (Fig. 5A). As a result, no CRX binding to the promoter of rhodopsin and three cone opsin genes was detected in Y79 cells, although CRX protein bound to the RBP3 promoter was detected in the same immunoprecipitates (Fig. 5B, left panel). In contrast, CRX and AcH3 are associated with the promoter of all four opsin genes in the human retina (Fig. 5B, right panel). Similar to the results obtained in the Crx−/− mouse retina, this lack of CRX target binding in Y79 cells correlates with no detectable binding of GCN5 and CBP, as well as absence of AcH3 on the opsin promoters (Fig. 5B, left panel). In contrast, the HATs and AcH3 are associated with the promoter of the RBP3 gene that is transcribed in Y79 cells. These results further support the finding from mouse studies that opsin transcription requires Crx’s action to recruit HATs for histone acetylation of the chromatin associated with regulatory elements.
Figure 5.
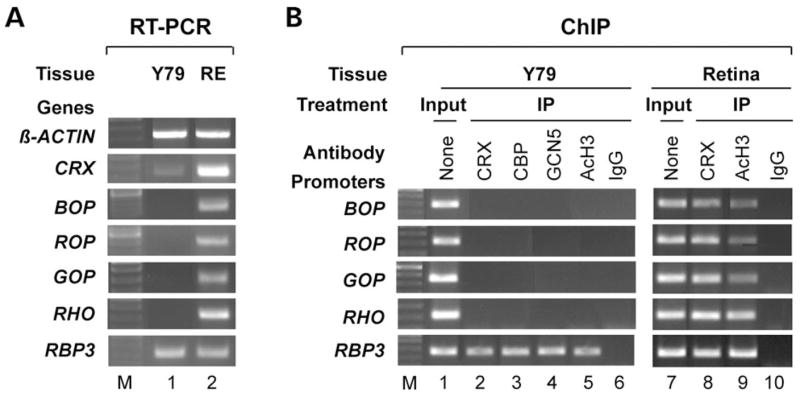
Y79 retinoblastoma cells express low levels of Crx, correlating with defective opsin transcription and HAT/AcH3 association with opsin promoters. (A) RT–PCR analysis of CRX, BOP, ROP, GOP, rhodopsin (RHO) and RBP3 in Y79 cells (lane 1) and human retina (RE, lane 2). β-ACTIN served as a positive control. (B) ChIP assays were performed on opsin promoters from Y79 cells and human retina using antibodies to Crx (lanes 2 and 8), Cbp (lane 3), Gcn5 (lane 4), AcH3 (lanes 5 and 9). Normal rabbit or mouse IgG served as negative controls (lanes 6 and 10), input (chromatin samples without immunoprecipitation) served as positive controls (lanes 1 and 7).
Y79 defects in histone H3 acetylation and opsin transcription can be reversed by expressing a recombinant Crx
To determine whether Crx is sufficient to reverse the Y79 defects in HAT recruitment, histone H3 acetylation and transcription of the opsin genes, a plasmid expressing a recombinant CRX was transiently transfected into Y79 cells. Shown in Figure 6A, transcription of previously silent opsin genes was induced by a recombinant CRX expression vector (WT, lane 3) when compared with untransfected (lane 1) or empty vector-transfected (lane 2) Y79 cells. Quantitative RT–PCR analysis demonstrated that CRX transfection resulted in 4–6-fold induction in transcription of the endogenous opsin genes in Y79 cells (Fig. 6B, WT bars). This CRX-induced opsin transcription correlated with the association of CRX, GCN5, CBP and AcH3 with each opsin promoter as measured by ChIP assays (Fig. 6C, lanes 1–3, 5–7, 9–11 and 13–15). Quantitative ChIP analysis showed an average of 5–7-fold enhancement of HAT/AcH3 association with each opsin promoter (Fig. 6D, solid bars), correlating well with the increase in transcriptional induction. In contrast, CRX expression did not dramatically alter the transcription and HAT/AcH3 association for the RBP3 gene that is expressed in untreated Y79 cells (Fig. 6A–D). To determine whether this induction of opsin transcription by CRX is dependent on its ability to recruit a HAT, we took advantage of a mutated form of CRX, Q96/97WΔQ7 (ΔQ), which lacks all nine Q residues in the end of HD and in the Qn motif, and therefore is unable to recruit STAGA-GCN5 in vitro (70). This mutant CRX, however, can normally interact with CBP (Fig. 1B and C), bind to DNA targets in vitro, and activate transcription of an unintegrated rhodopsin-luciferase reporter in transiently transfected HEK293 cells (70). If CRX-mediated GCN5 recruitment is important for endogenous opsin gene transcription, we would expect this Q96/97WΔQ7 mutant to be less potent than the WT CRX in promoting histone H3 acetylation and opsin transcription. As shown in Figure 6A, the Q96/97WΔQ7 (ΔQ) mutant (lane 4) induces less endogenous opsin transcription than the WT CRX (lane 3) in Y79 cells.
Figure 6.
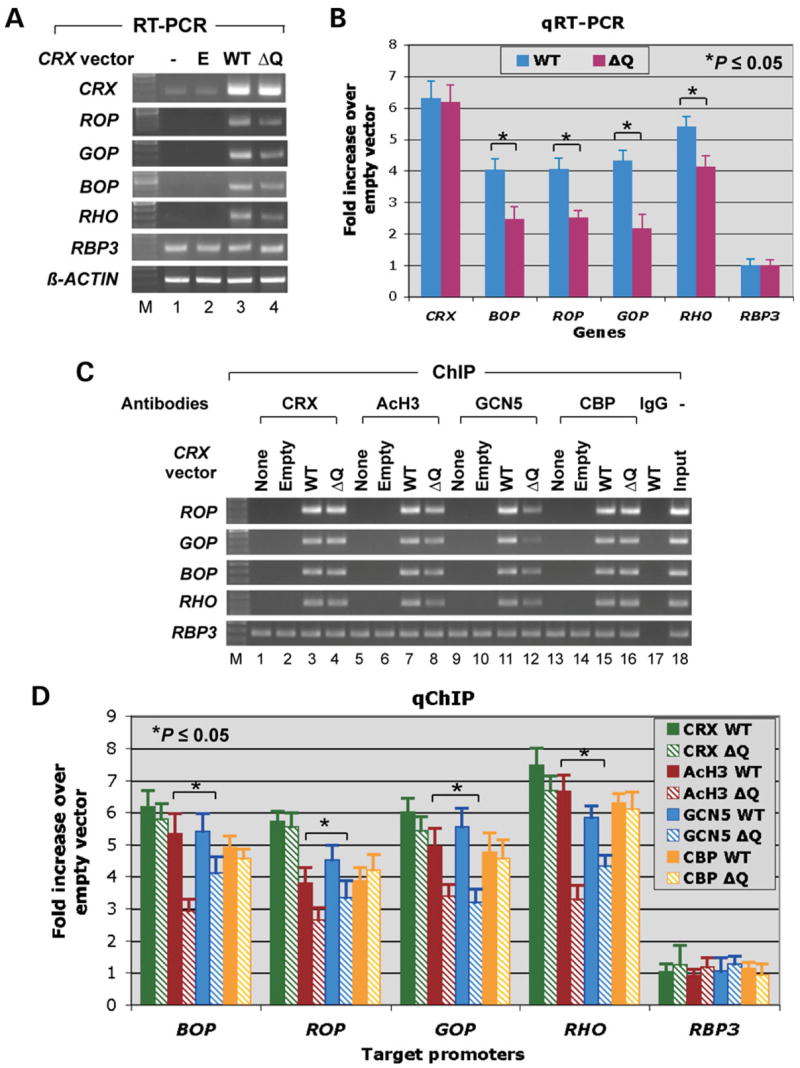
Transient transfection of Y79 cells with CRX induces opsin transcription and the association of GCN5/CBP and AcH3 with the opsin promoters. (A) RT–PCR analysis of the indicated genes comparing untreated Y79 cells (lane 1) and cultures transfected with the empty vector (E, lane 2), vector containing WT CRX (WT, lane 3), or a mutant form of CRX, Q96/97W-ΔQ7 (ΔQ, lane 4). (B) Quantitative RT–PCR results from a parallel experiment confirm that the ΔQ construct is less potent than WT CRX at inducing transcription of the opsin genes, but not CRX itself or RBP3. (C) Gel images of ChIP assays performed on untreated Y79 cells (‘None’) or cells transfected with empty vector (‘Empty’), WT CRX (WT) or CRX-Q96/97W-ΔQ7 (ΔQ). Antibodies to Crx, AcH3, Gcn5 and Cbp were used, and samples were examined for four opsin promoters and RBP3. (D) Quantification of promoter sequences in the ChIP samples, demonstrating that the Qn motif is required for Crx to recruit GCN5 to opsin promoters for histone H3 acetylation in Y79 cells.
The potency for this mutant to activate opsin transcription compared with that of WT CRX was quantified by real-time RT–PCR analysis (Fig. 6B). ΔQ’s mean potency was only 61% [blue cone opsin (BOP)], 62% [red cone opsin (ROP)], 51% [green cone opsin (GOP)] and 76% [rhodopsin (RHO)] of the WT activation levels for the four opsin genes, but no significant difference was seen for induction of CRX and RBP3 transcription. Thus, the ΔQ mutant indeed has a lower potency than the WT CRX to induce transcription of all four opsin genes. Consistent with the above results, quantitative ChIP assays demonstrated that the ΔQ mutant had decreased activities compared with WT CRX to recruit GCN5 and promote AcH3 association to each opsin promoter, although it bound to each opsin promoter and recruited CBP equally well as the WT CRX (Fig. 6C and D). For example, the amount of GCN5 and AcH3 recruited to the GOP promoter was only 57 and 68%, respectively in cells transfected with the ΔQ mutant when compared with the WT CRX. In contrast, no significant difference was detected for the amount of CRX and CBP bound to the GOP promoter (90 and 96%) in cells transfected with ΔQ and WT CRX. These results suggest that the lack of histone H3 K9/K14 acetylation and transcription of the opsin genes in Y79 cells is due to an insufficient level of CRX, which can be reversed by expressing a recombinant CRX. Furthermore, GCN5 appears to be important for CRX-mediated histone H3 acetylation and transcription of the opsin genes.
Y79 defects in histone H3 acetylation and opsin transcription can be reversed by HDACi treatments
If Crx-mediated histone H3 acetylation is critical for opsin transcription, increasing histone acetylation by using HDACi drugs should also enhance opsin transcription in Y79 cells without expressing a recombinant Crx. To test this possibility, we treated cultured Y79 cells with various doses of HDACi drugs. These include three inhibitors for class I/II HDACs (43), sodium butyrate (NaB), trichostatin A (TSA) and suberoylanilide hydroxamic acid (SAHA), as well as one inhibitor for class III NAD-dependent HDACs, sirtinol (81). RT–PCR and ChIP assays were performed 24 h after treatment. As shown in Figure 7, the transcription of the four opsin genes was induced by TSA (Fig. 7A and C), NaB (Fig. 7A and D) and SAHA (Fig. 7A and E) in a dose-dependent manner, but not by Sirtinol (Fig. 7A and F). Quantitative dose–response curves showed that optimal doses of TSA, SAHA and NaB can induce opsin transcription up to 4–7-fold (Fig. 7C–E), comparable to the levels induced by Crx transfection, although the induction capacity varies among different drugs and individual opsin genes. The induction in opsin transcription by the class I/II HDACi correlated with increased AcH3 levels on the opsin gene promoters/enhancers (Fig. 7B). In contrast, sirtinol, the inhibitor for class III HDACs does not exert any effect on AcH3 levels of the opsin gene promoters. Furthermore, the effects of the class I/II HDACi on transcription appear to be specific to Crx-regulated photoreceptor genes, as no significant dose-dependent inductions were observed in transcription or AcH3 promoter association for the Crx-independent photoreceptor gene RBP3 or non-Crx target genes GLUR6 and THY1 that are not expressed in Y79 cells or retinal photoreceptor cells. In addition, none of the HDACi drugs, nor the CRX expression vector, can induce histone H3 acetylation and transcription of the endogenous opsin genes in cultured cells of non-retinal origin, such as HEK293 cells (data not shown), suggesting that additional factors are needed to make such cells responsive to these treatments.
Figure 7.
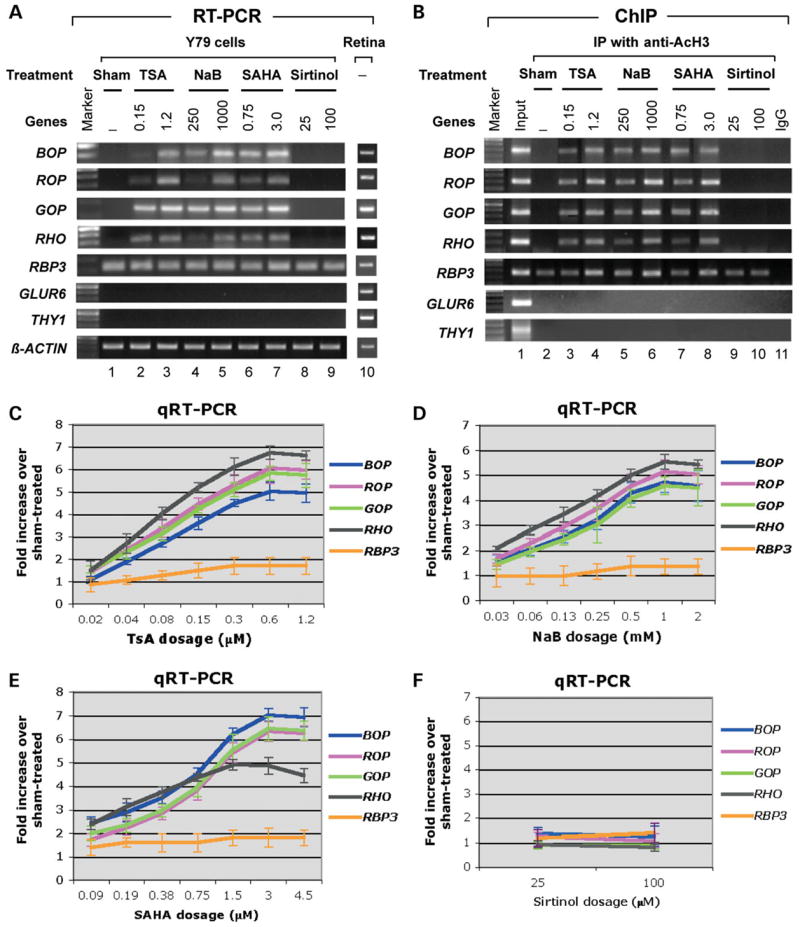
HDAC inhibitors enhance opsin gene transcription in Y79 cells in the absence of exogenous Crx. (A) Gel images of RT–PCR analysis of the four opsin genes, RBP3, GLUR6, THY1 and β-actin from Y79 cell cultures treated with vehicle alone (‘Sham’) or HDAC inhibitors TSA, NaB, SAHA and sirtinol at two doses (μM). RT–PCR results of human retinas served as positive controls. (B) ChIP assays of AcH3 levels on the promoters of the genes examined in (A). Lane 1 is the input control without immunoprecipitation. Lane 2 is the sham-treated control. Lanes 3–8 show that increases in promoter-associated histone H3 acetylation induced by TSA (lanes 3 and 4), NaB (lanes 5 and 6), SAHA (lanes 7 and 8) correlated with the increased opsin transcription seen in (A). However, histones associated with the non-photoreceptor gene promoters GLUR6 and THY1 did not show histone H3 acetylation. The class III HDAC inhibitor sirtinol (lanes 9 and 10) does not exert any effect on AcH3 level of the opsin genes. IP with normal rabbit or mouse IgG served as negative controls (lane 11). (C–F) Dose–response curves showing quantification of expression of the opsin genes and RBP3 in cultures treated with different doses of each of the HDAC inhibitors reveal subtle differences in the sensitivity of each of the promoters to the different drugs. Expression levels are shown as fold increase over expression levels in sham-treated cultures.
DISCUSSION
The role of Crx in HAT recruitment and chromatin modifications
In this paper, we described several pieces of evidence indicating that Crx is involved in inducing chromatin remodeling of target genes by interacting with three co-factors with HAT activity. Crx interacts with Cbp or p300 in vitro in the absence of other mammalian transcription regulators. Crx has also been reported to interact with Gcn5 of the STAGA complex in vitro and in the Y79 cells via a bridge protein, ataxin-7 (50). However, this Crx–HAT interaction was not detected in retinal extracts (46), raising a question whether the interaction actually occurs in vivo. Here we describe several pieces of evidence that strongly support Crx’s interaction with Gcn5, Cbp and p300 in vivo on target genes: (i) all three HATs can be co-immunoprecipitated with Crx from mouse retinal nuclear extracts in a Crx-dependent manner; (ii) Gcn5, Cbp and p300 are associated with Crx targets, not only the opsin genes, but also several other photoreceptor genes such as Arr3, Pde6a and Pde6b (data not shown); (iii) The HAT-target association with Crx occurs at the right time (always preceded by Crx binding to target promoters during development) and in the right place (in photoreceptors); (iv) Gcn5/Cbp association with Crx target genes requires the presence of a WT Crx, as this target association does not happen or is dramatically reduced in Crx knockout mouse retina and in Y79 retinoblastoma cells which express an extremely low level of Crx. Together, our results suggest that one mechanism of Crx action is to recruit HAT-harboring co-activators to the target genes for chromatin remodeling.
It is interesting that different Crx domains interact with each of the three different HATs. Cbp and p300 are two closely related family members, and yet they interact with Crx at the N-terminus (Cbp) and AD-2 region (p300), respectively. These two proteins share 61% homology overall, but are almost 90% identical in several discrete regions (41). A previous study suggested that the C-terminal domain of p300 is important for interacting with Crx (76). It would be interesting to further determine which domain on each protein mediates the interaction with Crx and whether these domains share sequence homology. Cbp and p300 have overlapping but distinct functions in transcription (76). It remains to be determined whether all three HATs could interact with Crx simultaneously, especially on the same target gene.
As demonstrated in Table 1, Cbp and Gcn5 are dependent on Crx for binding to the opsin gene promoters. Although we did not observe apparent changes in Cbp protein distribution in Crx−/− retina (Supplementary Material, Fig. S1A–F), the reduction of Cbp on opsin promoters/enhancers in Crx−/− retina could be partially attributed to the reduction in Cbp mRNA and protein expression levels (Fig. 2). During normal retinal development, Cbp mRNA levels are induced following Crx mRNA induction (Fig. 4). These findings suggest that Crx may regulate Cbp activity via both transcriptional regulation and protein–protein interactions.
It is important to point out that some other regulatory factors in addition to Crx also contribute to HAT recruitment to opsin promoter regions and histone acetylation modifications. First, in the absence of Crx, p300 binds to the opsin promoters/enhancers normally (Table 1). Second, extremely low but persistent binding of Gcn5/Cbp to the opsin promoters/enhancers is detectable in Crx−/− mice. Consistent with these observations, in Crx−/− retina, the transcription of rhodopsin and S-cone opsin remains at 8–12 and 4–9% of the WT levels, respectively (14,18). Third, during retinal development, Gcn5/Cbp binding to the S-cone opsin promoter appears to be delayed for a few days after Crx binds to its targets. The reason for this ‘slow’ recruitment to the cone genes remains to be determined. One possibility is that this delay of HAT/AcH3 binding is attributable to a limited sensitivity of the ChIP assays to detect binding in cones in a rod-dominated retina. The other possibility beyond technical limitations, however, is that HAT/AcH3 recruitment to the cone genes during development requires some other cone transcription activator(s). Nuclear receptor family members such as TRβ2 (21), Rorβ (22) and Rxrγ (23) are known to be important for the development of cone subtypes through interactions with co-activators/co-repressors with chromatin remodeling capability (reviewed in 14,82,83). These nuclear receptors therefore could contribute to Crx-regulated chromatin remodeling and transcription of the cone genes. Fourth, transfecting Crx into HEK293 cells does not promote HAT association with the opsin genes and their transcription even when Crx was co-transfected with Nrl (data not shown). Nrl can synergize with Crx in activating rhodopsin-luciferase reporters in transfected HEK293 cells (16) and was recently reported as a regulatory factor necessary and sufficient for directing rod development (19,79). Nrl, however, is unlikely to be a key player in recruiting HATs and mediating histone H3 K9/K14 acetylation on the opsin promoters, because Nrl does not bind to the opsin targets until after HAT/AcH3 recruitment (Fig. 4B–D), and Nrl deficiency does not affect the levels of HATs/AcH3 bound to the opsin genes (Supplementary Material, Table S1). These observations, however, do not rule out the possibility that Nrl is involved in chromatin modifications other than K9/K14 acetylation of histone H3. In contrast, Otx2, a close family member of Crx, is more likely a key contributor to histone acetylation modifications of the opsin genes. Otx2 contains several domains that are highly homologous to those Crx counterparts responsible for interaction with Cbp and Gcn5 (ataxin-7). Otx2 is required for photoreceptor development by acting upstream of Crx. Importantly, Otx2 is up-regulated in Crx−/− mouse retina (18) and, thus, could possibly compensate for Crx function in recruiting HATs and promoting histone acetylation of target genes in Crx−/− retina. It is also possible that, during development, Otx2 and Crx act together to mediate HAT and AcH3 recruitment.
The role of HAT-containing co-activators (co-activator complexes) in photoreceptor gene expression
The in vivo role of Gcn5, Cbp and p300 in retinal gene transcription, development and maintenance remains to be determined. Cbp/p300 is known to be involved in lens crystallin gene expression and development by interacting with c-Maf, a transcription factor closely related to Nrl, and a homeodomain protein, Prox-1 (84). Cbp and p300 have been shown to interact or complex with numerous transcription factors (85), many of which are clearly involved in retinal development and maintenance, including c-Fos/c-Jun (86–88), Pax6 (89), pRb regulatory pathways (47,90) as well as the nuclear receptor families. In contrast, fewer STAGA-Gcn5 interacting partners have been reported, including estrogen nuclear receptor alpha (91), the tumor suppressor protein BRCA1 (92), E2Fs (93) and c-Myc oncogenes (94). Our immunohistochemistry studies showed that Gcn5, Cbp and p300 are expressed in all the nuclear layers of the adult (Supplementary Material, Fig. S1) and P14 (data not shown) retina. In transient co-transfection assays of HEK293 cells with a rhodopsin-luciferase reporter, over-expression of Cbp, p300 and Gcn5 did not significantly enhance the potency of Crx (or Crx plus Nrl) to activate the rhodopsin promoter reporter (data not shown). However, these negative results do not rule out a role of these co-activators in Crx-mediated transcriptional activation, since transiently transfected reporters do not have the same native chromatin configuration as the corresponding endogenous genes. As an example, the importance of the rhodopsin enhancer (RER) was not detected by transient transfection assays, even though RER is known to be required for rhodopsin transcription in vivo (95). Furthermore, HEK293 cells may already have sufficient amounts of endogenous GCN5, CBP and p300 for Crx-mediated transcriptional activation of the reporter genes so that the presence of additional amounts of each as a result of transfection would not increase the reporter activity. This possibility is supported by a previous report that knockdown of GCN5 using an RNAi strategy significantly reduced the potency of Crx and Nrl to activate a rhodopsin-luciferase reporter (50).
Another important finding shown in the current study is that the defects in GCN5/CBP recruitment, histone H3 acetylation and transcription of the endogenous opsin genes in Y79 cells can be reversed by expressing a WT recombinant CRX, but the ΔQ CRX mutant that is defective in recruiting GCN5 is far less effective. Ectopic expression of CRX also induced the expression of NRL and NR2E3 mRNA (4.0 and 3.3-fold, respectively, when compared with vehicle-treated samples). NRL and NR2E3 are present at extremely low levels in untransfected Y79 cells. Thus, CRX-induced rhodopsin transcription in Y79 cells could result from a synergistic action of CRX, NRL and NR2E3. It is important to mention that the ΔQ mutant is as potent as WT CRX to induce NRL and NR2E3 expression in Y79 cells based on qRT–PCR analysis [3.9 ±0.4-fold (ΔQ) versus 4.0 ±0.2-fold (WT) for NRL and 3.2 ±0.4-fold (ΔQ) versus 3.3 ±0.2-fold (WT) for NR2E3]. This is consistent with the previous report that the ΔQ mutant of CRX is able to bind to DNA targets and mediate transcriptional activation normally in vitro (70). The use of the ΔQ mutant has allowed us to separate Crx’s role in Gcn5-mediated histone H3 acetylation from its intrinsic transactivating activity. Since the peptide sequence altered in this mutant was previously demonstrated to be responsible for Gcn5 recruitment (70), these results have provided direct evidence that Gcn5 is important for opsin transcription, at least in Y79 cells.
Two other pieces of evidence also indirectly support an in vivo role of Gcn5 and Cbp in photoreceptor function. A poly-Q expanded mutant ataxin-7, which serves as a bridge protein between Crx and Gcn5, blocks Gcn5 nucleosomal HAT activity in vitro and causes SCA7 with retinal degeneration in humans and model mice (50,70,75). SCA7 mice showed impaired photoreceptor gene expression before the onset of photoreceptor degeneration (75,96,97). It has also been reported that some brain neurons of SCA7 patients had CBP, but not p300, sequestered in nuclear inclusion bodies with poly-Q expanded ataxin-7 (98), which could reduce the availability of CBP for its normal co-activator function. Because conventional knockouts of Cbp (99,100) and Gcn5 (101,102) are embryonic lethal, the exact role of these HATs in photoreceptor gene expression, development and maintenance in vivo remains to be determined.
The role of chromatin modifications in photoreceptor gene expression
This manuscript is a first report describing a comprehensive investigation of the role of histone modifications in photoreceptor gene transcription and development in the mammalian retina. Acetylation of histone H3 on K9 and K14 residues appears to correlate well with the transcription status of the opsin genes during retinal development in WT mice. AcH3 increases on the promoter and enhancer region of each opsin gene appear to have similar kinetics as transcription increases of the corresponding opsin gene. Changes in AcH3 levels have been detected in retinal disease models associated with transcriptional dysregulation of the photoreceptor genes. These models include Crx−/− mice (current study), a model for LCA linked to CRX mutations, SCA7-92Q transgenic mice (50) and Y79 retinoblastoma cells (current study) in which the RB1 gene is deleted or mutated (103). In all of these cases, low AcH3 levels correlated with the loss or significant reduction in levels of opsin transcripts. In the Crx−/− retina, significantly reduced AcH4 levels on the rhodopsin and S-opsin promoters may also contribute to the low expression levels of these two genes. These low AcH4 levels could be caused by the defective Cbp recruitment, as Cbp catalyzes acetylation of both histone H3 and H4. However, the relationship between AcH4 and opsin transcription during normal retinal development and maintenance remains to be determined.
Other pieces of evidence for the role of AcH3 in opsin gene transcription were obtained from ‘gain-of-function’ studies, in which histone H3 acetylation was enhanced by genetic or pharmacological approaches in Y79 cells. Three HDACi drugs TSA, SAHA and NaB significantly enhanced the expression of four opsin genes as well as several other rod and cone genes (data not shown), suggesting that these drugs may have therapeutic potential for treating retinal degeneration associated with Crx mutations. However, their effects on retinal gene expression in vivo remain to be determined. It is important to point out that the precise regulation of temporal and spatial expression of photoreceptor genes in the retina is expected to be contributed by the action of both HATs and HDACs, which establish a specific equilibrium between acetylated and deacetylated histones on each target gene at any given developmental stage. The AcH3 levels on opsin promoters/enhancers that we detected in the developing retina are the net result of this equilibrium. HDACi treatments could shift this equilibrium and produce differential effects on opsin transcription in vivo at different developmental stages. It was reported recently that TSA treatment of newborn mouse retina in explant cultures actually blocked transcription of rhodopsin as well as its key regulators Otx2, Crx and Nrl and induced cell death (56). Thus, premature hyperacetylation of histones during development could actually inhibit rod differentiation. On the other hand, promoting histone acetylation by HDACi treatment may enhance photoreceptor gene transcription in those cells that are already differentiated into photoreceptors. Our studies on the kinetics of HAT recruitment and AcH3 levels of opsin genes during retinal development, and dose response curves in Y79 cells would provide useful information for future studies on therapeutic potential of HDACi to treat diseases in which transcription dysregulation in differentiated photoreceptors leads to retinal degeneration.
Histone modification is a dynamic process that can include acetylation, methylation, phosphorylation, ubiquitylation and sumoylation at multiple sites (31). This dynamic process could be regulated by both cis-acting DNA sequences and trans-acting regulatory proteins. Thus, this study on histone H3 K9/K14 acetylation only touches the tip of an iceberg. Any factor that alters chromatin modifications could have a profound effect on transcription. On the other hand, variations in types and degrees of chromatin modifications among different genes could be encoded by specific regulatory sequences. For example, the chromatin configuration of the Rbp3 gene appears to be more accessible to Crx and other transcription factors than the opsin genes in early stages of mouse retinal development (data not shown) and in Y79 cells (Figs 5 and 6), when an extremely low level of Crx is present. This is consistent with hyperacetylation of histone H3 on the Rbp3 gene in those cases. The reason for this difference is unclear. Some other chromatin modifications or events prior to Crx action, such as Otx2 binding, could be responsible for chromatin remodeling and transcriptional activation of the Rbp3 gene during normal retinal development, as well as in Crx−/− retina and Y79 cells. The fact that Rbp3 expression begins prior to Crx expression during retinal development and is unaffected by a Crx null mutation (Crx−/−) has placed Rbp3 into the category of ‘Crx-independent genes’. However, this does not rule out the possibility that Crx contributes to the maintenance of Rbp3 expression after it is turned on. We have detected Crx binding to Rbp3 promoter in WT mouse retina and Y79 cells using ChIP analysis. In vitro protein-DNA binding (DNase I footprinting or electrophoretic mobility shift assays) and cell transfection assays also showed that both Crx and Otx2 can bind to and regulate the Rbp3 promoter (16,17,71,104). It is possible that Otx2 turns on Rbp3 expression during early retinal development via chromatin modifications, whereas Crx helps to maintain the Rbp3 chromatin configuration and transcription when the Otx2 level is reduced in differentiated photoreceptors. Studies of genetic interactions between Crx and Otx2 and epigenetic differences between Rbp3 and opsin genes may provide useful information on how various photoreceptor genes are differentially and coordinately regulated.
Model for photoreceptor gene expression and development
On the basis of the above results, we propose the model for photoreceptor-specific gene expression and development shown in Figure 8. Opsin gene transcription is controlled by two ongoing regulatory processes, (i) derepression via chromatin remodeling and (ii) classical transcriptional activation. In non-photoreceptors or retinal progenitor cells, opsin genes are not transcribed due to the absence of specific transactivators and a compacted chromatin configuration. During retinal development, some multi-potent progenitor cells exit mitotic cycling and begin to express Otx2 and subsequently Crx. Crx binds to target DNA regulatory elements and then recruits HAT-harboring co-activators (or co-activator complexes) to the opsin genes, triggering histone acetylation that results in a less repressive ‘relaxed’ chromatin configuration. These cells are now photoreceptor precursors. This change in chromatin configuration allows more Crx as well as other transcription factors and the RNA polymerase II-basal transcription machinery to access their targets to achieve transcriptional activation of the opsin genes, resulting in terminally differentiated photoreceptor cells. Thus, Crx is required for both derepression and activation processes, and some other photoreceptor transcription factors, such as Nrl, may act only in the later activation process once the chromatin is in the relaxed configuration. This study has demonstrated that Crx plays an initiator role in derepressing photoreceptor genes, which involves a mechanism by which Crx recruits HAT-containing co-activators for acetylation of the histones.
Figure 8.
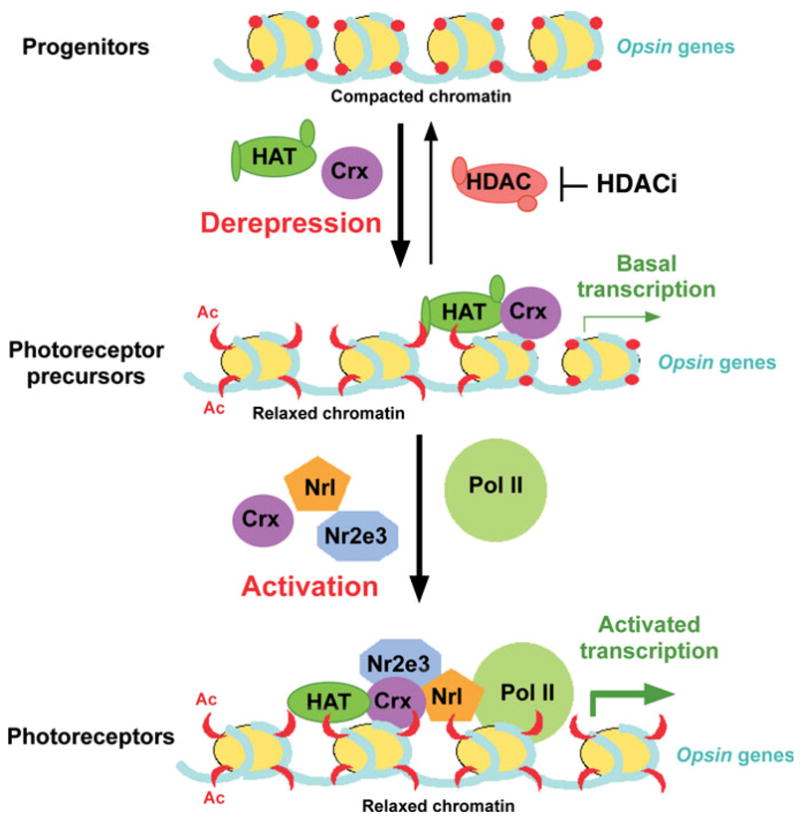
Model for the role of Crx in chromatin modification and transcription activation. In progenitor cells that have not committed to the photoreceptor lineage, chromatin associated with the opsin genes exists in a compacted state that excludes the transcription machinery and keeps these genes silenced. As these cells become committed to the photoreceptor lineage, they express Crx, which binds to opsin promoters. Bound Crx then recruits co-activators including HATs, which then modify lysine residues in the tails of histone H3 and H4, derepressing the chromatin associated with these genes and allowing basal levels of transcription. As additional transcription factors are expressed and recruited to these promoters, and as chromatin associated with enhancer elements becomes derepressed, transcription reaches high levels, allowing photoreceptors to build and maintain their outer segments and perform phototransduction.
MATERIALS AND METHODS
Mammalian expression vectors, antibodies and other reagents
pcDNA3.1/HisC (Invitrogen) based mammalian expression vectors carrying bovine Crx and its N- and C-terminal deletions were described previously (61). A mutant form of CRX, Q96/97WΔQ7, which does not interact with ataxin-7-containing STAGA complex was described previously (70). pcDNA3.1-based mammalian expression vectors containing mouse Cbp and human p300 coding cDNA were provided by the laboratory of Dr Kenneth Fischbeck (National Institute of Neurological Disorders and Stroke) (105).
Antibodies used in this study include: rabbit antibody p119b against Crx which was made against a peptide spanning mouse Crx amino acids 119–141 (Proteintech Group Inc., Chicago, IL, USA) and affinity purified; mouse monoclonal anti-Crx (Abnova, Taiwan, H00001406-M02); rabbit anti-Cbp (Upstate Biotechnology, Lake Placid, NY, USA; 06-297), mouse monoclonal anti-Cbp (Chemicon, Temecula, CA, USA; MAB1133); mouse monoclonal anti-p300 (Upstate, 05-257), rabbit anti-Gcn5 antibodies (Santa Cruz, sc20698 and sc8999), rabbit anti-di-acetylated (K9 and K14) histone H3 (AcH3, Upstate, 06-599) or anti-tetra-acetylated (K5, K8, K12 and K16) histone H4 (AcH4, Upstate, 06-598), rabbit anti-Nrl (Chemicon, AB5693), rabbit anti-Nr2e3 (29) and mouse monoclonal anti-RNA polymerase II CTD (Ser5p, Abcam, ab5408). Non-specific normal IgG from rabbit and mouse were purchased from Santa Cruz for negative controls in co-ip assays.
HDACi used in this study include sodium butyrate (NaB) (Sigma), TSA (Upstate), SAHA (BioVision, Mountain View, CA, USA) and Sirtinol (Sigma).
Mouse strains
Mouse strains used in this study include: C57BL/6J (‘wild-type’, The Jackson Laboratory); Crx knockout mouse (Crx−/−, kindly provided by Dr Constance Cepko, Harvard Medical School, Boston, MA, USA) (18); Nrl knockout mouse (Nrl−/−, provided by Anand Swaroop, University of Michigan) (19); C57BL/6J-Pdebrd1le/Pdebrd1le (rd1/rd1, The Jackson Laboratory) (106); Red-DT(A) (a gift of Dr Jeremy Nathans, Johns Hopkins University, Baltimore, MD, USA), a C57BL/6J transgenic line referred to as ‘coneless’ in adults (77) and Red-DT(A)/rd1/rd1, a strain resulting from breeding between C57BL/6J-Pdebrd1le/Pdebrd1le and Red-DT(A), referred to as ‘rodless/coneless’ in adults.
In vitro co-ip assays
Recombinant Crx, Cbp and p300 proteins with or without 35S-labeling were generated by in vitro translation of the corresponding mammalian expression vectors using TnT T7 Quick Coupled Transcription/Translation kit (Promega). Co-ip assays were performed as described previously (75) with minor modifications in the wash buffer [50 mM Tris–Cl (pH 7.5), 450 mM NaCl and 0.5% Triton X-100].
Cell cultures and transient transfections
Y79 retinoblastoma cell line was purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured on six-well plates in RPMI-1640 medium with 15% fetal bovine serum, according to ATCC instructions. Transient transfections were performed using DreamFect transfection reagent (OZ Bioscience, Marseille, France, US distributor: Boca Scientific Inc., Boca Raton, FL, USA) and 4 μg of CsCl gradient-purified plasmid DNA at a 2:1 ratio (μl reagent: μg DNA) in 200 μl final reaction volume. Typically, 30–50% transfection efficiency is achieved based on GFP+ cell counts when pEGFP-N1 (Clontech) is transfected as a control in each transfection. Normal human retina samples used as controls for Y79 cell RT–PCR and ChIP assays were from a 27-year-old male cadaver donor kindly provided by Dr David Beebe (Washington University, St Louis, MO, USA).
In vivo co-ip and western blot assays
Nuclear and cytoplasmic extracts were made from P14 mouse retinas using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL, USA) following manufacturer’s instructions. Protein concentration of each crude extract was measured using Bio-Rad Protein Assay (Bio-Rad 500-0006) with BSA as a standard, and an equal amount of protein (100 μg) was used for co-ip assays with 1 μg of rabbit anti-Crx antibody p119b coupled to Protein A beads (GE-Healthcare, Piscataway, NJ, USA) under similar conditions described for the in vitro co-ip assays. Proteins eluted from the beads were analyzed by SDS–PAGE/western blots as described previously (107). Ten micrograms of crude nuclear extract made from each mouse strain was used as input controls. Primary antibodies used in the western blots include mouse anti-Crx (p119b, 1:500), rabbit anti-Gcn5 (sc20698, 1:500), mouse anti-Cbp (1:2000), mouse anti-p300 (1:500) and mouse anti-β-actin (1:5000) for loading controls. The secondary antibodies used include horseradish-peroxidase coupled anti-mouse or anti-rabbit IgG (Santa Cruz) at 1:15 000–30 000 dilutions. The results were detected and visualized using the Millipore Western Blotting Detection System (Millipore, Billerica, MA, USA) and Amersham Hyperfilm (GE Healthcare).
ChIP assays
These were performed using six pooled mouse retinas, or 1 × 106 Y79 cells, essentially as described previously (70). The mouse strains used in ChIP assays include WT (C57BL/6J) at 6–8 weeks of age or at various developmental stages as noted, rodless/coneless [Red-DT(A)/rd1/rd1 at 80 days], coneless (Red-DT(A) at 56 days, Crx−/− (at 14 days) and Nrl−/− (at 14 days or as noted). Transcription regulators bound to target chromatin were immunoprecipitated by rabbit or mouse antibodies against specified antigens or normal rabbit/mouse IgG as negative controls. The immunoprecipitated chromatin DNA and input controls (without IP) were analyzed by PCR using specific primers for the promoter or enhancer region of Crx-regulated opsin genes and the Crx-independent gene Rbp3. Two non-Crx target genes in the retina, Thy1 and GluR6, were included as negative controls. PCR primer sequences for most mouse genes were described previously (71). The remaining primers are listed in Supplementary Material, Table S2. Quantifications of ChIP assays were performed as described previously (50). Briefly, quantitative real-time PCR analysis of regulatory sequences in input samples and immunoprecipitates was performed using SYBR Green JumpStart ReadyMix (Sigma) and iCycler PCR machine (Bio-Rad). The difference was calculated as: ΔCT = CTIP − CTinput, where CT, the threshold cycle, is the cycle number (in the exponential phase) at which enough amplified product has accumulated to yield a detectable fluorescent signal that is significantly above the baseline fluorescence level. Mean ΔCT values relative to a specified sample and standard deviation (STDEV) for each experiment were calculated from triplicate samples assayed in the same run, and statistical significance was determined using the paired Student’s t-test.
Quantitative real-time RT–PCR analysis
Total RNA was isolated from two retinas dissected from WT (C57BL/6J, Jackson Laboratory) mice (or embryos) at the indicated ages or from 1 × 107 Y79 cells cultured on 100 mM plates, using Versagene RNA Tissue Kit (Gentra Systems, Inc., Minneapolis, MN, USA). One microgram of total RNA was reverse transcribed using Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science, Indianapolis, IN, USA) and anchored-oligo-(dT)18 primer. The cDNA was diluted 10-fold and quantified by real-time PCR analysis in triplicate using SYBR Green JumpStart ReadyMix (Sigma) and iCycler PCR machine (Bio-Rad). β-actin was used as a loading control. Relative expression levels normalized to the β-actin levels were determined as described previously (29). Mean value and STDEV for each experiment from three repeats were calculated, and statistical significance was calculated using the paired Student’s t-test. RT–PCR primers not described previously (71) are listed in Supplementary Material, Table S2.
Immunohistochemistry studies
Paraffin-embedded eye sections from adult and neonatal C57BL/6J (WT) or Crx−/− mice were prepared and immunostained with specific antibodies as described previously (107). Antigen retrieval in a citrate buffer was performed as described previously (107). Primary antibodies used are as follows: rabbit anti-Crx-p119b (1:800); mouse anti-Cbp (Chemicon, 1:1000), mouse anti-p300 (Upstate, 1:600) and rabbit anti-Gcn5 (Santa Cruz, sc8999, 1:500). Goat anti-rabbit or mouse IgG antibodies coupled to Alexa Fluor A488 (Molecular Probes) were used as secondary antibody at a 1:400 dilution. The stained slides were cover slipped with Vectashield mounting medium with Propidium iodide (red, Vector Laboratories) and visualized using a fluorescence light microscope (Olympus BX51) with a Spot Cooled Color Digital Camera (Diagnostic Instrument Inc.).
Supplementary Material
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG Online.
Acknowledgments
We wish to thank Anne Hennig, Jianfeng Liu and Belinda McMahan for technical assistance, Anne Hennig for critical reading of the manuscript.
FUNDING
National Institutes of Health (EY12543 to S.C., EY02687 to WU-DOVS), Foundation Fighting Blindness (to S.C.), Prevent Blindness America (to S.C.), Knights Templar Foundation (to G.-H.P.) and Research to Prevent Blindness (to S.C. and WU-DOVS).
Footnotes
Conflict of Interest statement. None declared.
References
- 1.Humphries MM, Rancourt D, Farrar GJ, Kenna P, Hazel M, Bush RA, Sieving PA, Sheils DM, McNally N, Creighton P, et al. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat Genet. 1997;15:216–219. doi: 10.1038/ng0297-216. [DOI] [PubMed] [Google Scholar]
- 2.Olsson JE, Gordon JW, Pawlyk BS, Roof D, Hayes A, Molday RS, Mukai S, Cowley GS, Berson EL, Dryja TP. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron. 1992;9:815–830. doi: 10.1016/0896-6273(92)90236-7. [DOI] [PubMed] [Google Scholar]
- 3.Turner DL, Cepko CL. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- 4.Wetts R, Fraser SE. Multipotent precursors can give rise to all major cell types of the frog retina. Science. 1988;239:1142–1145. doi: 10.1126/science.2449732. [DOI] [PubMed] [Google Scholar]
- 5.Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM. Timing and topography of cell genesis in the rat retina. J Comp Neurol. 2004;474:304–324. doi: 10.1002/cne.20134. [DOI] [PubMed] [Google Scholar]
- 6.Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- 7.Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J Comp Neurol. 1979;188:263–272. doi: 10.1002/cne.901880205. [DOI] [PubMed] [Google Scholar]
- 8.Cepko CL. The patterning and onset of opsin expression in vertebrate retinae. Curr Opin Neurobiol. 1996;6:542–546. doi: 10.1016/s0959-4388(96)80062-6. [DOI] [PubMed] [Google Scholar]
- 9.Bibb LC, Holt JK, Tarttelin EE, Hodges MD, Gregory-Evans K, Rutherford A, Lucas RJ, Sowden JC, Gregory-Evans CY. Temporal and spatial expression patterns of the CRX transcription factor and its downstream targets. Critical differences during human and mouse eye development. Hum Mol Genet. 2001;10:1571–1579. doi: 10.1093/hmg/10.15.1571. [DOI] [PubMed] [Google Scholar]
- 10.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 11.Cayouette M, Poggi L, Harris WA. Lineage in the vertebrate retina. Trends Neurosci. 2006;29:563–570. doi: 10.1016/j.tins.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Cepko CL. The roles of intrinsic and extrinsic cues and bHLH genes in the determination of retinal cell fates. Curr Opin Neurobiol. 1999;9:37–46. doi: 10.1016/s0959-4388(99)80005-1. [DOI] [PubMed] [Google Scholar]
- 13.Hatakeyama J, Kageyama R. Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol. 2004;15:83–89. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Hennig AK, Peng G-H, Chen S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2007 June 30; doi: 10.1016/j.brainres.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- 16.Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 17.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 18.Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- 19.Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi M, Takezawa S, Hara K, Yu RT, Umesono Y, Agata K, Taniwaki M, Yasuda K, Umesono K. Identification of a photoreceptor cell-specific nuclear receptor. Proc Natl Acad Sci USA. 1999;96:4814–4819. doi: 10.1073/pnas.96.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng L, Hurley JB, Dierks B, Srinivas M, Salto C, Vennstrom B, Reh TA, Forrest D. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001;27:94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- 22.Srinivas M, Ng L, Liu H, Jia L, Forrest D. Activation of the blue opsin gene in cone photoreceptor development by retinoid-related orphan receptor beta. Mol Endocrinol. 2006;20:1728–1741. doi: 10.1210/me.2005-0505. [DOI] [PubMed] [Google Scholar]
- 23.Roberts MR, Hendrickson A, McGuire CR, Reh TA. Retinoid X receptor (gamma) is necessary to establish the S-opsin gradient in cone photoreceptors of the developing mouse retina. Invest Ophthalmol Vis Sci. 2005;46:2897–2904. doi: 10.1167/iovs.05-0093. [DOI] [PubMed] [Google Scholar]
- 24.Zhang CL, Zou Y, Yu RT, Gage FH, Evans RM. Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev. 2006;20:1308–1320. doi: 10.1101/gad.1413606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- 26.Rehemtulla A, Warwar R, Kumar R, Ji X, Zack DJ, Swaroop A. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc Natl Acad Sci USA. 1996;93:191–195. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005;25:118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corbo JC, Cepko CL. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 2005;1:e11. doi: 10.1371/journal.pgen.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14:747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- 30.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 31.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 35.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamagoe S, Kanno T, Kanno Y, Sasaki S, Siegel RM, Lenardo MJ, Humphrey G, Wang Y, Nakatani Y, Howard BH, et al. Interaction of histone acetylases and deacetylases in vivo. Mol Cell Biol. 2003;23:1025–1033. doi: 10.1128/MCB.23.3.1025-1033.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saha RN, Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13:539–550. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 39.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 40.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 41.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 42.Lin HY, Chen CS, Lin SP, Weng JR, Chen CS. Targeting histone deacetylase in cancer therapy. Med Res Rev. 2006;26:397–413. doi: 10.1002/med.20056. [DOI] [PubMed] [Google Scholar]
- 43.Marks PA, Richon VM, Miller T, Kelly WK. Histone deacetylase inhibitors. Adv Cancer Res. 2004;91:137–168. doi: 10.1016/S0065-230X(04)91004-4. [DOI] [PubMed] [Google Scholar]
- 44.Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. I Structural analysis using light and electron microscopy. J Comp Neurol. 1979;188:245–262. doi: 10.1002/cne.901880204. [DOI] [PubMed] [Google Scholar]
- 45.Johnson DA, Donovan SL, Dyer MA. Mosaic deletion of Rb arrests rod differentiation and stimulates ectopic synaptogenesis in the mouse retina. J Comp Neurol. 2006;498:112–128. doi: 10.1002/cne.21059. [DOI] [PubMed] [Google Scholar]
- 46.Helmlinger D, Hardy S, Abou-Sleymane G, Eberlin A, Bowman AB, Gansmuller A, Picaud S, Zoghbi HY, Trottier Y, Tora L, et al. PLoS Biol. Vol. 4. 2006. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction; p. e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Gray J, Wu L, Leone G, Rowan S, Cepko CL, Zhu X, Craft CM, Dyer MA. Rb regulates proliferation and rod photoreceptor development in the mouse retina. Nat Genet. 2004;36:351–360. doi: 10.1038/ng1318. [DOI] [PubMed] [Google Scholar]
- 48.Lebre AS, Brice A. Spinocerebellar ataxia 7 (SCA7) Cytogenet Genome Res. 2003;100:154–163. doi: 10.1159/000072850. [DOI] [PubMed] [Google Scholar]
- 49.Helmlinger D, Hardy S, Sasorith S, Klein F, Robert F, Weber C, Miguet L, Potier N, Van-Dorsselaer A, Wurtz JM, et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum Mol Genet. 2004;13:1257–1265. doi: 10.1093/hmg/ddh139. [DOI] [PubMed] [Google Scholar]
- 50.Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci USA. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donovan SL, Dyer MA. Developmental defects in Rb-deficient retinae. Vision Res. 2004;44:3323–3333. doi: 10.1016/j.visres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 52.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 53.Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 54.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 56.Chen B, Cepko CL. Requirement of histone deacetylase activity for the expression of critical photoreceptor genes. BMC Dev Biol. 2007;7:78. doi: 10.1186/1471-213X-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 58.Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 59.Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, Begemann M, Crabtree GR, Goff SP. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 60.Singh P, Coe J, Hong W. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 61.Chen S, Wang QL, Xu S, Liu Y, Lili YL, Wang Y, Zack DJ. Functional analysis of cone-rod homeobox (CRX) mutations associated with retinal dystrophy. Hum Mol Genet. 2002;11:873–884. doi: 10.1093/hmg/11.8.873. [DOI] [PubMed] [Google Scholar]
- 62.Freund CL, Gregory-Evans CY, Furukawa T, Papaioannou M, Looser J, Ploder L, Bellingham J, Ng D, Herbrick JA, Duncan A, et al. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell. 1997;91:543–553. doi: 10.1016/s0092-8674(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 63.Freund CL, Wang QL, Chen S, Muskat BL, Wiles CD, Sheffield VC, Jacobson SG, McInnes RR, Zack DJ, Stone EM. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat Genet. 1998;18:311–312. doi: 10.1038/ng0498-311. [DOI] [PubMed] [Google Scholar]
- 64.Rivolta C, Berson EL, Dryja TP. Dominant Leber congenital amaurosis, cone-rod degeneration, and retinitis pigmentosa caused by mutant versions of the transcription factor CRX. Hum Mutat. 2001;18:488–498. doi: 10.1002/humu.1226. [DOI] [PubMed] [Google Scholar]
- 65.Sohocki MM, Daiger SP, Bowne SJ, Rodriquez JA, Northrup H, Heckenlively JR, Birch DG, Mintz-Hittner H, Ruiz RS, Lewis RA, et al. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum Mutat. 2001;17:42–51. doi: 10.1002/1098-1004(2001)17:1<42::AID-HUMU5>3.0.CO;2-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Swain PK, Chen S, Wang QL, Affatigato LM, Coats CL, Brady KD, Fishman GA, Jacobson SG, Swaroop A, Stone E, et al. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron. 1997;19:1329–1336. doi: 10.1016/s0896-6273(00)80423-7. [DOI] [PubMed] [Google Scholar]
- 67.Swaroop A, Wang QL, Wu W, Cook J, Coats C, Xu S, Chen S, Zack DJ, Sieving PA. Leber congenital amaurosis caused by a homozygous mutation (R90W) in the homeodomain of the retinal transcription factor CRX: direct evidence for the involvement of CRX in the development of photoreceptor function. Hum Mol Genet. 1999;8:299–305. doi: 10.1093/hmg/8.2.299. [DOI] [PubMed] [Google Scholar]
- 68.Blackshaw S, Fraioli RE, Furukawa T, Cepko CL. Comprehensive analysis of photoreceptor gene expression and the identification of candidate retinal disease genes. Cell. 2001;107:579–589. doi: 10.1016/s0092-8674(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 69.Livesey FJ, Furukawa T, Steffen MA, Church GM, Cepko CL. Microarray analysis of the transcriptional network controlled by the photoreceptor homeobox gene Crx. Curr Biol. 2000;10:301–310. doi: 10.1016/s0960-9822(00)00379-1. [DOI] [PubMed] [Google Scholar]
- 70.Chen S, Peng GH, Wang X, Smith AC, Grote SK, Sopher BL, La Spada AR. Interference of Crx-dependent transcription by ataxin-7 involves interaction between the glutamine regions and requires the ataxin-7 carboxy-terminal region for nuclear localization. Hum Mol Genet. 2004;13:53–67. doi: 10.1093/hmg/ddh005. [DOI] [PubMed] [Google Scholar]
- 71.Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis Neurosci. 2005;22:575–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- 72.Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J Biol Chem. 2000;275:29794–29799. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- 73.Lerner LE, Peng GH, Gribanova YE, Chen S, Farber DB. Sp4 is expressed in retinal neurons, activates transcription of photoreceptor-specific genes, and synergizes with Crx. J Biol Chem. 2005;280:20642–20650. doi: 10.1074/jbc.M500957200. [DOI] [PubMed] [Google Scholar]
- 74.Wang QL, Chen S, Esumi N, Swain PK, Haines HS, Peng G, Melia BM, McIntosh I, Heckenlively JR, Jacobson SG, et al. QRX, a novel homeobox gene, modulates photoreceptor gene expression. Hum Mol Genet. 2004;13:1025–1040. doi: 10.1093/hmg/ddh117. [DOI] [PubMed] [Google Scholar]
- 75.La Spada AR, Fu YH, Sopher BL, Libby RT, Wang X, Li LY, Einum DD, Huang J, Possin DE, Smith AC, et al. Polyglutamine-expanded ataxin-7 antagonizes CRX function and induces cone-rod dystrophy in a mouse model of SCA7. Neuron. 2001;31:913–927. doi: 10.1016/s0896-6273(01)00422-6. [DOI] [PubMed] [Google Scholar]
- 76.Yanagi Y, Masuhiro Y, Mori M, Yanagisawa J, Kato S. p300/CBP acts as a coactivator of the cone-rod homeobox transcription factor. Biochem Biophys Res Commun. 2000;269:410–414. doi: 10.1006/bbrc.2000.2304. [DOI] [PubMed] [Google Scholar]
- 77.Soucy E, Wang Y, Nirenberg S, Nathans J, Meister M. A novel signaling pathway from rod photoreceptors to ganglion cells in mammalian retina. Neuron. 1998;21:481–493. doi: 10.1016/s0896-6273(00)80560-7. [DOI] [PubMed] [Google Scholar]
- 78.Daniele LL, Lillo C, Lyubarsky AL, Nikonov SS, Philp N, Mears AJ, Swaroop A, Williams DS, Pugh EN., Jr Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest Ophthalmol Vis Sci. 2005;46:2156–2167. doi: 10.1167/iovs.04-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oh EC, Khan N, Novelli E, Khanna H, Strettoi E, Swaroop A. From the Cover: Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc Natl Acad Sci USA. 2007;104:1679–1684. doi: 10.1073/pnas.0605934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.del Cerro M, Notter MF, Seigel G, Lazar E, Chader G, del Cerro C. Intraretinal xenografts of differentiated human retinoblastoma cells integrate with the host retina. Brain Res. 1992;583:12–22. doi: 10.1016/s0006-8993(10)80005-8. [DOI] [PubMed] [Google Scholar]
- 81.Denu JM. The Sir 2 family of protein deacetylases. Curr Opin Chem Biol. 2005;9:431–440. doi: 10.1016/j.cbpa.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 82.Nishihara E, O’Malley BW, Xu J. Nuclear receptor coregulators are new players in nervous system development and function. Mol Neurobiol. 2004;30:307–325. doi: 10.1385/MN:30:3:307. [DOI] [PubMed] [Google Scholar]
- 83.Kishimoto M, Fujiki R, Takezawa S, Sasaki Y, Nakamura T, Yamaoka K, Kitagawa H, Kato S. Nuclear receptor mediated gene regulation through chromatin remodeling and histone modifications. Endocr J. 2006;53:157–172. doi: 10.1507/endocrj.53.157. [DOI] [PubMed] [Google Scholar]
- 84.Chen Q, Dowhan DH, Liang D, Moore DD, Overbeek PA. CREB-binding protein/p300 co-activation of crystallin gene expression. J Biol Chem. 2002;277:24081–24089. doi: 10.1074/jbc.M201821200. [DOI] [PubMed] [Google Scholar]
- 85.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 86.He L, Campbell ML, Srivastava D, Blocker YS, Harris JR, Swaroop A, Fox DA. Spatial and temporal expression of AP-1 responsive rod photoreceptor genes and bZIP transcription factors during development of the rat retina. Mol Vis. 1998;4:32. [PubMed] [Google Scholar]
- 87.Grimm C, Wenzel A, Hafezi F, Reme CE. Gene expression in the mouse retina: the effect of damaging light. Mol Vis. 2000;6:252–260. [PubMed] [Google Scholar]
- 88.Wenzel A, Grimm C, Marti A, Kueng-Hitz N, Hafezi F, Niemeyer G, Reme CE. c-fos controls the ‘private pathway’ of light-induced apoptosis of retinal photoreceptors. J Neurosci. 2000;20:81–88. doi: 10.1523/JNEUROSCI.20-01-00081.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hever AM, Williamson KA, van Heyningen V. Developmental malformations of the eye: the role of PAX6, SOX2 and OTX2. Clin Genet. 2006;69:459–470. doi: 10.1111/j.1399-0004.2006.00619.x. [DOI] [PubMed] [Google Scholar]
- 90.Chen D, Livne-bar I, Vanderluit JL, Slack RS, Agochiya M, Bremner R. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell. 2004;5:539–551. doi: 10.1016/j.ccr.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 91.Yanagisawa J, Kitagawa H, Yanagida M, Wada O, Ogawa S, Nakagomi M, Oishi H, Yamamoto Y, Nagasawa H, McMahon SB, et al. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol Cell. 2002;9:553–562. doi: 10.1016/s1097-2765(02)00478-1. [DOI] [PubMed] [Google Scholar]
- 92.Oishi H, Kitagawa H, Wada O, Takezawa S, Tora L, Kouzu-Fujita M, Takada I, Yano T, Yanagisawa J, Kato S. An hGCN5/TRRAP histone acetyltransferase complex co-activates BRCA1 transactivation function through histone modification. J Biol Chem. 2006;281:20–26. doi: 10.1074/jbc.M510157200. [DOI] [PubMed] [Google Scholar]
- 93.Lang SE, McMahon SB, Cole MD, Hearing P. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J Biol Chem. 2001;276:32627–32634. doi: 10.1074/jbc.M102067200. [DOI] [PubMed] [Google Scholar]
- 94.Liu X, Tesfai J, Evrard YA, Dent SY, Martinez E. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J Biol Chem. 2003;278:20405–20412. doi: 10.1074/jbc.M211795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kumar R, Chen S, Scheurer D, Wang QL, Duh E, Sung CH, Rehemtulla A, Swaroop A, Adler R, Zack DJ. The bZIP transcription factor Nrl stimulates rhodopsin promoter activity in primary retinal cell cultures. J Biol Chem. 1996;271:29612–29618. doi: 10.1074/jbc.271.47.29612. [DOI] [PubMed] [Google Scholar]
- 96.Yoo SY, Pennesi ME, Weeber EJ, Xu B, Atkinson R, Chen S, Armstrong DL, Wu SM, Sweatt JD, Zoghbi HY. SCA7 knockin mice model human SCA7 and reveal gradual accumulation of mutant ataxin-7 in neurons and abnormalities in short-term plasticity. Neuron. 2003;37:383–401. doi: 10.1016/s0896-6273(02)01190-x. [DOI] [PubMed] [Google Scholar]
- 97.Abou-Sleymane G, Chalmel F, Helmlinger D, Lardenois A, Thibault C, Weber C, Merienne K, Mandel JL, Poch O, Devys D, et al. Polyglutamine expansion causes neurodegeneration by altering the neuronal differentiation program. Hum Mol Genet. 2006;15:691–703. doi: 10.1093/hmg/ddi483. [DOI] [PubMed] [Google Scholar]
- 98.Takahashi J, Fujigasaki H, Zander C, El Hachimi KH, Stevanin G, Durr A, Lebre AS, Yvert G, Trottier Y, de The H, et al. Two populations of neuronal intranuclear inclusions in SCA7 differ in size and promyelocytic leukaemia protein content. Brain. 2002;125:1534–1543. doi: 10.1093/brain/awf154. [DOI] [PubMed] [Google Scholar]
- 99.Kung AL, Rebel VI, Bronson RT, Ch’ng LE, Sieff CA, Livingston DM, Yao TP. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 100.Oike Y, Takakura N, Hata A, Kaname T, Akizuki M, Yamaguchi Y, Yasue H, Araki K, Yamamura K, Suda T. Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood. 1999;93:2771–2779. [PubMed] [Google Scholar]
- 101.Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, Westphal H, Ozato K, Nakatani Y. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc Natl Acad Sci USA. 2000;97:11303–11306. doi: 10.1073/pnas.97.21.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- 103.Madreperla SA, Bookstein R, Jones OW, Lee WH. Retinoblastoma cell lines Y79, RB355 and WERI-Rb27 are genetically related. Ophthalmic Paediatr Genet. 1991;12:49–56. doi: 10.3109/13816819109023085. [DOI] [PubMed] [Google Scholar]
- 104.Fong SL, Fong WB. Elements regulating the transcription of human interstitial retinoid-binding protein (IRBP) gene in cultured retinoblastoma cells. Curr Eye Res. 1999;18:283–291. doi: 10.1076/ceyr.18.4.283.5360. [DOI] [PubMed] [Google Scholar]
- 105.McCampbell A, Taylor JP, Taye AA, Robitschek J, Li M, Walcott J, Merry D, Chai Y, Paulson H, Sobue G, et al. CREB-binding protein sequestration by expanded polyglutamine. Hum Mol Genet. 2000;9:2197–2202. doi: 10.1093/hmg/9.14.2197. [DOI] [PubMed] [Google Scholar]
- 106.Carter-Dawson LD, LaVail MM, Sidman RL. Differential effect of the rd mutation on rods and cones in the mouse retina. Invest Ophthalmol Vis Sci. 1978;17:489–498. [PubMed] [Google Scholar]
- 107.Wang X, Xu S, Rivolta C, Li LY, Peng GH, Swain PK, Sung CH, Swaroop A, Berson EL, Dryja TP, et al. Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J Biol Chem. 2002;277:43288–43300. doi: 10.1074/jbc.M207952200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG Online.