Abstract
Mutations in PHEX (phosphate-regulating gene with homologies to endopeptidases on the X chromosome) and DMP1 (dentin matrix protein 1) result in X-linked hypophosphatemic rickets (HYP) and autosomal-recessive hypophosphatemic-rickets (ARHR), respectively. Specific binding of PHEX to matrix extracellular phosphoglycoprotein (MEPE) regulates the release of small protease-resistant MEPE peptides [acidic serine- and aspartate-rich MEPE-associated motif (ASARM) peptides]. ASARM peptides are potent inhibitors of mineralization (minhibins) that also occur in DMP1 [MEPE-related small integrin-binding ligand, N-linked glycoprotein (SIBLING) protein]. It is not known whether these peptides are directly responsible for the mineralization defect. We therefore used a bone marrow stromal cell (BMSC) coculture model, ASARM peptides, anti-ASARM antibodies, and a small synthetic PHEX peptide (SPR4; 4.2 kDa) to examine this. Surface plasmon resonance (SPR) and two-dimensional 1H/15N nuclear magnetic resonance demonstrated specific binding of SPR4 peptide to ASARM peptide. When cultured individually for 21 d, HYP BMSCs displayed reduced mineralization compared with wild type (WT) (−87%, P < 0.05). When cocultured, both HYP and WT cells failed to mineralize. However, cocultures (HYP and WT) or monocultures of HYP BMSCs treated with SPR4 peptide or anti-ASARM neutralizing antibodies mineralized normally. WT BMSCs treated with ASARM peptide also failed to mineralize properly without SPR4 peptide or anti-ASARM neutralizing antibodies. ASARM peptide treatment decreased PHEX mRNA and protein (−80%, P < 0.05) and SPR4 peptide cotreatment reversed this by binding ASARM peptide. SPR4 peptide also reversed ASARM peptide-mediated changes in expression of key osteoclast and osteoblast differentiation genes. Western blots of HYP calvariae and BMSCs revealed massive degradation of both MEPE and DMP1 protein compared with the WT. We conclude that degradation of MEPE and DMP-1 and release of ASARM peptides are chiefly responsible for the HYP mineralization defect and changes in osteoblast-osteoclast differentiation.
A MUTATED PHEX (phosphate-regulating gene with homologies to endopeptidases on the X chromosome) gene is responsible for the primary mineralization and renal phosphate homeostasis defects noted in X-linked hypophosphatemic rickets (HYP) in mice and humans (1). Over 250 human families and at least five mice models with diverse mutations in this conserved gene overwhelmingly support this conclusion (1,2). An extensive PHEX database website is also available at the web site http://www.phexdb.mcgill.ca/. PHEX belongs to a well-defined family of Zn metalloendopeptidases (M13 family; MA clan) involved in cancer, bone-renal diseases, cardiovascular disease, Alzheimer’s, arthritis, and inflammatory disorders (3,4). The prototypic member of this class of structurally complex proteins is neprilysin (CD10, CALLA). To date the physiological substrate and the precise molecular role for PHEX in mineralization and renal phosphate homeostasis remains unknown. Our previous work showed direct binding of PHEX to matrix extracellular phosphoglycoprotein (MEPE) (5), a protein expressed in bone, teeth, and renal proximal convoluted tubules (3,6). MEPE belongs to a group of extracellular matrix proteins [small integrin-binding ligand, N-linked glycoproteins (SIBLINGs)] involved in bone and teeth mineralization. These proteins all map to a tightly clustered region on chromosome 4q and include MEPE, dentin matrix protein 1 (DMP1), osteopontin (OPN), bone sialoprotein (BSP), enamelin, dentin sialo phosphoprotein (DSPP) and statherin. MEPE is a phosphate uptake inhibitory factor cloned from a tumor resected from a patient with tumor-induced osteomalacia and hypophosphatemia (7). A key feature of MEPE and several SIBLINGs including DMP1 is an acidic serine- and aspartate-rich MEPE-associated motif (ASARM motif) (3,7). This motif, when released as a protease-resistant phosphorylated peptide (ASARM peptide) negatively affects mineralization and phosphate uptake (3,5,8,9). We have shown indirectly that PHEX binds to MEPE via the ASARM motif (5) and also potently inhibits PHEX enzymatic hydrolysis of a nonphysiological synthetic peptide substrate (10). This interaction also prevents cathepsin B-mediated hydrolysis and release of protease-resistant ASARM peptide (5,8,11).
Without functional PHEX (HYP mice), an increase in both MEPE and osteoblastic protease expression occurs (3,8,10,11,12,13,14,15,16,17,18,19,20). This leads to excess ASARM peptides from MEPE and perhaps other SIBLINGs like DMP1 (3,5,8,14,17,21). Thus, bone accumulation of the protease-resistant ASARM peptides likely plays a key role in the defective mineralization or hyperosteoidosis in HYP (3,5,8,9). The precise relationship between PHEX and MEPE however remains unclear as well as the link between PHEX, MEPE, and phosphate handling. For example, one report describes MEPE-null mutations (mice) result in a marked age-dependent high bone mass phenotype with an increased in vivo mineral apposition rate (22). Also, this study and a second independent study report a marked and significant acceleration of in vitro mineralization of MEPE-null mutant bone cells in culture (22,23). Of note, although both studies reported a defective in vitro bone phenotype, the marked increase of in vivo bone mass reported by Gowen et al. (22) was not observed by Liu et al. (23). However, a key difference between the studies was the age of the mice used and the techniques for assessing the phenotypes. Specifically, Liu et al. (23) used significantly younger animals (12 wk) compared with the Gowen et al. (22) (4 months and 12 months). Because bone mass is the result of two distinct processes, modeling (early growth) and remodeling (mainly adult), these findings suggest MEPE action is more important for mature bone remodeling. Adult MEPE-null mice also have increased cancellous and cortical bone mass without changes in serum phosphate levels (22). Consistent with this, MEPE ASARM peptides are potent mineralization inhibitors in vitro and in vivo (3,5,8,9,10). Although the MEPE-null mutant mouse is normophosphatemic, recombinant MEPE introduced by bolus ip injections induces phosphaturia in rodents (3,9,24). Also, the HYP in vitro mineralization phenotype is corrected by transfer of HYP mice onto a MEPE-deficient mouse background, but the in vivo phenotype is not (23). This may be because of the continued proteolytic degradation of the extracellular protein matrix (increased osteoblast protease expression in HYP), release of SIBLING ASARM peptides (DMP1), and ongoing fibroblast growth factor 23 (FGF23)-mediated hypophosphatemia (3,8). More recently, reintroduction of the PHEX transgene into PHEX-defective HYP mice produced surprising results (13,25,26). Specifically, overexpression of human PHEX under the β-actin promoter in HYP mice rescued the bone phenotype almost completely but did not correct the hypophosphatemia or the increased FGF23 expression (13,25). The authors concluded that “different, possibly independent, pathophysiological mechanisms contribute to renal phosphate wasting and bone abnormalities in HYP” (25). Indeed, others suggested that PHEX may play dual roles (phosphate regulation and mineralization) dependent on the tissue of expression (13,25). Recently, we and others have shown protease treatment of HYP mice partially corrects the mineralization defect despite persistent hypophosphatemia, again suggesting a phosphate mineralization disconnect (8,15,19,20). Early and classic studies with HYP mice and human patients are consistent with this notion. These studies showed that although dietary phosphate supplementation corrects rickets and the endochondral calcification, the endosteal hyperosteoidosis remains (27,28,29,30,31,32,33). This is also true for the DMP1-null mouse, a murine model of autosomal recessive hypophosphatemic rickets (ARHR) (34). Of note, recent studies show hypophosphatemia is mainly responsible for the rickets (as opposed to the endochondral mineralization defect). This is because low serum phosphate impairs caspase-mediated apoptosis of hypertrophic chondrocytes (35). Consistent with this, intrinsic odontoblast defects that are independent of serum phosphate levels are chiefly responsible for the dentinal mineralization defects in the HYP mouse (36). Further confirmation comes from recent studies on teeth from patients with HYP (21). Specifically, massive degradation of MEPE and an increase in ASARM peptides occurs in HYP teeth. Bone resorption and osteoclastogenesis is also greatly altered in HYP mice and in normal mice fed with low-phosphate diets (37,38,39). In the HYP mouse, MEPE but not FGF23 reduces osteoclast number chiefly by impacting on RANKL (receptor activator for nuclear factor-κB ligand) expression (38). Consistent with these findings, abnormalities in cartilage (chondroclasts) and bone (osteoclasts), matrix protein expression, and matrix metalloproteinase-9 localization occur in HYP mice (39,40). Thus, these extensive studies support the notion that defective mineralization and bone abnormalities are not solely because of the hypophosphatemia and indicate SIBLING matrix proteins play important roles in mineralization.
In this study, we determined whether increased ASARM peptides released from MEPE and related SIBLING proteins (DMP1) contribute directly to the HYP mineralization defect and whether these peptides interact with PHEX and alter expression of key bone-related genes. To do this, we first designed small synthetic PHEX peptides using known structure data from related M13 Zn metalloendopeptidases. We then used surface plasmon resonance (SPR) and nuclear magnetic resonance (NMR) to characterize fully the binding sites between these novel peptides. These peptides and an in vitro bone marrow stromal cell (BMSC) culture model allowed us to study the biological roles of ASARM peptides and PHEX in HYP. We conclude that ASARM peptides are chiefly responsible for the in vitro mineralization defect in HYP and likely regulate mineralization and osteoblast-osteoclast differentiation.
Materials and Methods
Design of a small synthetic PHEX peptide (SPR4, 4.2 kDa) that binds to ASARM-PO4
After the cloning of the PHEX gene with the HYP Consortium (1), we characterized PHEX mutations from several HYP families (41). From these studies and work with M13 Zn metalloendopeptidases (neprilysin, endothelin-converting enzymes, and KELL) a substrate-binding site was deduced (3,41). Using this information with structural and bioinformatic modeling, several synthetic PHEX peptides were designed. Key considerations were the PHEX substrate-binding region with Zn-binding motif (HEFTH). Also important were residues for catalysis, specificity, Zn sequestration, stabilization of the transition state, and identity for the MA clan of M13 Zn metalloendopeptidases. Biomolecular software PHYRE and 3D-PSSM (http://www.sbg.bio.ic.ac.uk/∼phyre/) were used to model probable structure and optimal folding. Based on SPR, 1H/15N two-dimensional NMR and biological data (see below), a single PHEX peptide (4.2 kDa) called SPR4 was found to bind to ASARM-PO4 peptide (Table 1). PHEX has more than 99% sequence conservation across species and the Zn-binding motif region used to design the SPR4 peptide is identical across species. The human ASARM motif region (DSSESSDSGSSSESDGD) was incorporated into the ASARM peptides used in these studies. It must be noted that the rat and mouse ASARM motifs differ by two substitutions, and the rodent/human consensus sequence is DSSESSXSGSSSESXGD. In the rat both Xs are replaced by S, and in the mouse, an S and H, respectively. The physiochemical blueprint and secondary structures would not (predictably) be substantially altered between human and rodent ASARM motifs by these minor substitutions. In addition, the casein kinase II phosphorylation consensus sites remain. Also, the anti-ASARM antibodies used in this study cross-react with mouse and human MEPE ASARM epitopes providing further support for the interspecies applicability of our biological findings. The full-length ASARM peptide(s) used with the cathepsin cleavage site region (RDDSSESSDSGSSSESDGD) are shown in Table 1. The SPR4 peptide also has biological activity and is discussed in more detail in the following sections.
Table 1.
Sequences of peptides used for SPR, NMR, and cell culture experiments
| No. | Code | Sequence |
|---|---|---|
| SPR1 | Bio-RGD_MEPE | Biotin-(Ahx)-GYTDLQERGDNDISPFSGDGQPFKD-OH |
| SPR2 | Bio-Random-RGD-MEPE | Biotin-(Ahx)-SQGKDIFPDPDFGLYGDETRQNDGS-OH |
| SPR3 | PHEX-Zn region | NH2-GTEYPRSLSYGAIGVIVGHEFTHGFDNNGRKYDKNGNLD-OH |
| SPR4 | PHEX-Zn-designed-peptide | NH2-TVNAFYSASTNYPRSLSYGAIGVIVGHEFTHGFDNNGRGENIADNG-OH |
| SPR5 | Control randomer for SPR4 | NH2-LKGDHSGGNGNYTGLDYNIPGYFRSTIPFHGEVDKENVAR-OH |
| SPR6 | PHEX-ZN region peptide | NH2-AIGVIVGHEFTHGFDNNGRK-OH |
| SPR7 | Control randomer for SPR6 | NH2-GHIDEGGHNRAVITGVFNFK-OH |
| SPR8 | Bio-ASARM-PO4 | Biotin-(Ahx)-RDDSSESSDSGS(PO3H2)SS(PO3H2)ES(PO3H2)DGD-OH |
| SPR9 | Bio-ASARM | Biotin-(Ahx)-RDDSSESSDSGSSSESDGD-OH |
| ASARM-PO4 | NH2-RDDSSESSDSGS(PO3H2)SS(PO3H2)ES(PO3H2)DGD-OH | |
| ASARM | NH2-RDDSSESSDSGSSSESDGD-OH |
SPR4 peptide was also labeled with 15N isotope for all NMR experiments. Specifically, all SPR4 alanine (A) and glycine (G) residues were labeled except for the last C-terminal glycine. Ahx indicates an N-terminal 6-aminohexanoic acid linker for biotinylated peptides. Nonbiotinylated peptides were used for biological experiments, and biotinylated peptides were used as ligands for SPR.
SPR experiments
SPR was used to study the binding between soluble PHEX (sPHEX) and a biotinylated ASARM-PO4 peptide as well as to a number of biotinylated control peptides (ASARM and RGD-MEPE) (Table 1). Pure sPHEX protein was prepared as described previously (5,42,43). Neutravidin (Pierce, Rockford, IL) was immobilized on the surface of a CM5 sensor chip [4000–5000 response units (RU)] using standard amine coupling chemistry (5). The indicated biotinylated peptides were captured on individual flow cells, and the remaining unmodified neutravidin surface was used as the reference surface. Binding of the sPHEX concentration series (7.5, 15, 31.5, 62.5, 125, 250, and 500 nm) was studied at 25 C in 10 mm HEPES (pH 7.4), 150 mm NaCl, 2 mm ZnCl2, 0.005% surfactant P20 at a flow rate of 5 μl/min in a Biacore 3000 instrument (Biacore Inc., Piscataway, NJ). The experiment was done on two different CM5 chips with the following surface densities: bio-ASARM-PO4 (629 and 447 RU), bio-ASARM (447 and 407 RU), and bio-RGD-MEPE (831and 588 RU). The data were analyzed using BiaEvaluation 3.1 software (Biacore).
SPR was also used to study the inhibition of the sPHEX and ASARM-PO4 peptide interaction. For these experiments, a CM5 chip with neutravidin-captured biotinylated peptides (bio-ASARM-PO4 447 RU, bio-ASARM 407 RU, bio-RGD-MEPE 588 RU) was used (Table 1). The procedure indicated above was modified as follows: Immediately before analyte injection, a stock solution of experimental SPR4 peptide (or controls SPR3, SPR5; Table 1) dissolved in binding buffer [10 mm HEPES (pH 7.4), 150 mm NaCl, 2 mm ZnCl2, 0.005% surfactant P20] was added to a constant 50 nm PHEX analyte solution to give final peptide concentrations of either 0, 0.75, 1.5, 3, 6, 12.5, 25. 50, or 100 μm. In this way, a concentration-dependent inhibition curve was calculated for each of the peptides as described previously (5), although the SPR4 peptide was the only one to significantly inhibit the binding. Each Biacore Surface Plasmon Resonance (SPR) experiment was independently performed three times for all data points (n = 3).
Two-dimensional 1H/15N NMR
Demonstration of SPR4 binding to peptides was obtained after two-dimensional 1H/15N NMR. SPR4 peptide (1 mg) was first dissolved in 100 μl 25 mm acetic acid (pH 3.5) followed by 400 μl 50 mm Tris (pH 7) and 25 μm ZnCl2. Peptides dissolved in the same buffer were then added at different multiples of molar concentrations relative to SPR4 (1,2,3,4,5,6,7,8,9,10). Solutions were then made 6 mm relative to CaCl2 and incubated at 25 C for 1 h before NMR analysis. Samples were then transferred to 5 mm standard D2O-matched NMR tubes (Shigemi, Tokyo, Japan) for insertion in to the spectrometer. The 15N heteronuclear single quantum coherence experiments were carried out on an 800-MHz Bruker Avance NMR spectrometer with a cryogenically cooled triple-axis probe capable of delivering gradient pulses, using a pulse sequence containing WATERGATE for solvent suppression. All experiments were performed at 25 C. Each data set was collected using eight scans with 1024 points in 1H and 128 points in 15N. Samples contained 5% D2O, and referencing was performed relative to 2,2-dimethyl-2-silapentane-5-sulfonic acid, according to the procedure established by Wishart et al. (44). The data were then processed in nmrPipe using linear prediction with twice the number of collected data points and zero filling to 1600 points. All new peaks resulting from peptide interactions had a signal to noise ratio of at least 5:1, clearly distinguishing them from the noise using Sparky (45,46).
Animals
Five male wild-type (WT) and five male HYP mice were used; the animals in both groups were 6 wk old (The Jackson Laboratory, Bar Harbor, ME). Hind limbs were collected for primary cell culture, and calvariae were collected in proteases inhibitor cocktail for protein extraction and further SDS-PAGE analysis (see below).
Cell culture
Culture conditions.
Adherent BMSCs were cultured to assess the in vitro relative osteoblastic/osteoclastic potential of small peptides (ASARM and SPR4) based on a classical approach. The epiphyses of the bones (i.e. tibias and femurs) were removed, and whole marrow was flushed from the diaphyses by centrifugation at 11,500 rpm for 30 sec in standard α-MEM culture medium (Mediatech Inc., Manassas, VA) supplemented with 10% fetal bovine serum (Mediatech), 10 U/ml penicillin, and 100 μg/ml streptomycin. The supernatant was removed, and the cell pellet was resuspended in fresh standard medium. The cells were plated in standard 25-cm2 flasks and allowed to grow until confluence. They were passaged with 0.25% trypsin solution containing 0.02% EDTA (Sigma-Aldrich, St. Louis, MO). Cells were plated on 24-well plates for mineralization assays and on 96-well plastic culture plates for osteoclastic assessment at a concentration of 10 × 104 cells per well. Starting 72 h after seeding (referred to as d 0), nonadherent cells were discarded, and the medium was changed to one that supports osteoblast differentiation [standard medium plus 10 mm β-glycerophosphate (Sigma-Aldrich) and 50 mg/liter ascorbic acid (Sigma-Aldrich)] or osteoclastic development [50 ng/ml RANKL (Sigma-Aldrich, MO) and 50 ng/ml macrophage colony-stimulating factor (Sigma-Aldrich)]. These media were used throughout the rest of the culture period. To evaluate the involvement of SPR4 and ASARM-PO4 in the regulation of osteoblasts and osteoclasts growth/differentiation and activity, cells were treated during the culture period with either 5 μm ASARM-PO4 or 10 μm SPR4 or both. Untreated cells were used as controls. The compounds were added to the culture medium on d 0 and renewed every other day.
Mineralization assay.
Monolayers were washed with PBS and fixed in 10% (vol/vol) formaldehyde (Fisher Scientific, Pittsburgh, PA) at room temperature for 15 min. The monolayers were then washed twice with excess dH2O before addition of 1 ml of 10 mg/ml alizarin red S (ARS) solution in 0.2% ammonium (pH 6.36) per well. The plates were incubated at room temperature for 45 min with gentle shaking. After aspiration of the unincorporated dye, the wells were washed six times with 2 ml dH2O while shaking for 5 min. The plates were reaspirated and then stored at −20 C before dye extraction. For quantification of staining, 500 μl 10% (vol/vol) acetic acid was added to each well, and the plate was incubated at room temperature for 30 min while shaking. The monolayer was then scraped from the plate with a cell scraper and transferred with 10% (vol/vol) acetic acid to a 1.5-ml microcentrifuge tube. After vortexing for 30 sec, the slurry was heated to exactly 85 C for 10 min and transferred to ice for 5 min. The slurry was then centrifuged at 10,000 × g for 15 min, and 400 μl of the supernatant was removed to a new 1.5-ml microcentrifuge tube, and 150 μl 10% ammonium hydroxide was added to neutralize the pH down to 4.1–4.5. The 150-μl aliquots of the supernatant were read in triplicate at 450 nm in 96-well format using opaque-walled, transparent-bottomed plates.
Osteoblast alkaline phosphatase (ALP) staining.
After 2% formaldehyde fixation and rinsing, the activity of the plasma membrane-associated ALP was detected using an ALP leukocyte staining kit (Sigma-Aldrich) according to the manufacturer’s protocol. The cultures were then rinsed three times for 5 min each in deionized water, and nuclei were stained with 4′,6-diamidino-2-phenylindole. The percentage of ALP-positive cells were determined by counting cells in 10 contiguous fields per well after random starts.
Osteoclasts tartrate-resistant acid phosphatase (TRAcP) staining.
Monolayers were washed once with 100 μl PBS and fixed in 50 μl formalin for 5 min at room temperature. Each well was then washed three times with 250 μl dH2O before staining. Cells were incubated at 37 C for 60 min with 50 μl of a 150 μg/ml sodium tartrate solution containing 1 mg/ml naphthol-1-phosphate and 2.5 mg/ml Fast Violet (Sigma-Aldrich). After staining, cells were washed twice with dH2O water, and then enzymatic reaction was blocked with 50 μl 4.2 μg/ml sodium fluoride aqueous solution. The monolayers were then washed twice with excess dH2O and then air dried. The percentage of multinucleated TRAcP-positive osteoclasts was determined by counting cells in 10 contiguous fields per well after random starts.
RT-PCR
RNA extraction and RT.
RNA extraction was performed on three wells per experimental condition. Total RNAs were extracted using the RNeasy Mini Kit (QIAGEN, Valencia, CA) on each well according to the manufacturer instructions. The purity of total RNA prepared was monitored by the ratio of absorbance at 260 and 280 nm. Total RNAs were reverse transcribed into single-stranded cDNA using iScript cDNA synthesis kit (Bio-Rad Laboratories, CA) from 1 μg of total RNA.
Plasmids.
Escherichia coli bacteria expressing mouse 3594705 (Transferrin) plasmids (American Type Culture Collection, Manassas, VA), were cultured on LB medium supplemented with 50 μg/ml ampicillin, extracted, and purified using the QIAfilter Maxi kit (QIAGEN) and then resuspended in sterile endotoxin-free saline solution. Plasmid DNA concentration was measured at 260/280 nm. Plasmids were digested by the specific restriction endonucleases (Invitrogen, Carlsbad, CA), and the quality of DNA was assessed by 1% agarose gel electrophoresis. Plasmids were stocked at −80 C before use.
Real-time PCR.
The single-strand cDNA was diluted 1:10, and 20 μl was amplified in 50 μl PCR mixture containing 25 μl FastStart SYBR Green Master (FastStart SYBR Green Master PCR Kit; Roche, Mannheim, Germany) and 2.5 μl forward and 2.5 μl reverse oligo-primers (10 μm each). Amplifications were performed in the Bio-Rad iQ5 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA). The primer sequences and product sizes of each amplified marker are shown in Table 2. Quantified data were analyzed with the iCycler analysis software. Serial dilution of total cDNA was performed from and used as standards. Results were processed following the manufacturer’s instructions: 1) checking the PCR products specificity by using the melting temperature of each product followed by the product sequencing and 2) calculating the variation in PCR product concentration between experimental groups, normalized by transferrin (housekeeping gene) and expressed as a percentage of control condition mean values. The accuracy of cDNA amplification was assessed by amplifying in the same PCR the experimental sample cDNA and the same samples supplemented with a known plasmid concentration for transferrin housekeeping gene, plotted against a serial dilution plasmid standard curve.
Table 2.
Sequences of primers used for real-time PCR and relative product sizes
| Marker | Primer sequence
|
Product size (bp) | |
|---|---|---|---|
| Forward | Reverse | ||
| Transferrin | accatgttgtggtctcacga | acagaaggtccttggtggtg | 134 |
| PHEX | gtggtggtctgtggaatcag | agccggctttcttccaata | 91 |
| MEPE | cccaagagcagcaaaggtag | ccgctgtgacatccctttat | 215 |
| DMP1 | agtgaggaggacagcctgaa | gaggctctcgttggactcac | 86 |
| RUNX2 | ccgtggccttcaaggttgt | ttcataacagcggaggcattt | 117 |
| Osterix | tgaggaagaagcccattcac | acttcttctcccgggtgtg | 197 |
| MSX2 | accacgtcccagcttctagc | gctctgcgatggagaggtactg | 66 |
| Osteocalcin | agcaggagggcaataaggta | caagcagggttaagctcaca | 111 |
| Osteoprotegerin | gttcctgcacagcttcacaa | aaacagcccagtgaccattc | 120 |
| RANKL | agccgagactacggcaagta | gcgctcgaaagtacaggaac | 207 |
Immunohistochemistry.
For immunohistochemistry, cells were cultured on eight-well tissue culture-treated glass slides for 14 d (BD Biosciences, Billerica, MA). Before immunohistochemical screening, BMSCs were extensively washed with PBS to minimize contamination with endogenous peptides derived from cell culture media. After culture, cell slides were incubated with 0.03% H2O2 for 15 min to block endogenous peroxidase activity and then blocked with avidin/biotin blocking solution (Vector Laboratories Inc., Burlingame, CA) for 30 min and finally a 5% chicken/goat/horse serum solution for 45 min at room temperature. Samples were immunostained with either lab-designed rabbit polyclonal antibodies raised against MEPE ASARM peptide (C-terminal region) or MEPE mid-region RGD peptide (8,9,14) or DMP1 (LF-148) as described previously (6,47,48). Rabbit antibodies raised against the following peptide were used for screening PHEX: NH2-CMINQYSNYYWKKAGL-CONH2. Goat polyclonal anti-FGF23 (sc-16849) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). All samples were incubated with antisera for 1 h at room temperature. Negative control samples were incubated with rabbit IgG (5 μg/ml). Samples stained for MEPE and DMP1 were then incubated with antirabbit biotinylated secondary antibody (1/200) (Vector) for 30 min at room temperature and developed using the Vectastain ABC kit and stable diaminobenzidine (Vector). PHEX and FGF-23 staining were developed using Alexa Fluor 594 donkey antirabbit and Alexa Fluor 488 donkey antigoat fluorescent antibodies (Invitrogen). All samples were counterstained with DAPI, dehydrated and mounted with entellan (EMD Chemicals, NJ).
Total protein extraction and assay.
Total proteins from calvariae were extracted in 1 ml lysis buffer of T-PER tissue protein extraction reagent (Pierce). After centrifugation (10 min at 5000 rpm and 4 C), supernatants were stored at −80 C. Protein concentration was measured using the bicinchoninic acid protein assay kit (Pierce).
Western blotting
Protein lysates (2.5 mg/sample) were reduced and extracted in lithium dodecyl sulfate sample buffer (Invitrogen) heated for 10 min at 70 C, electrophoresed on NuPAGE Novex 4–12% bis-tris gels (Invitrogen), and then analyzed by Western blotting using the ECL Advance WB Detection Kit (GE Healthcare, Piscataway, NJ). Briefly, after SDS-PAGE, proteins were transferred to nitrocellulose membrane (Bio-Rad). After transfer, membranes were blocked overnight at 4 C using 5% nonfat dried milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T; pH 7.4) and incubated for 1 h at 22 C on a rocking platform with the rabbit polyclonal anti-ASARM (8,9,14) (1:2000), rabbit polyclonal anti-DMP1 (LF-148, 1:2000) (6,47,48), and goat polyclonal anti-GAPDH (sc-31914, 1:1000; Santa Cruz Biotechnology) antibodies. Membranes were washed three times in TBS-T and then incubated with secondary antibodies (peroxidase-conjugated goat antirabbit or peroxidase-conjugated rabbit antigoat, 1:20,000) for 1 h at room temperature. After washing three times with TBS-T, the immunoreactive bands were visualized using enhanced chemiluminescence detection reagents (GE Healthcare, UK) on a Fluor-S Multi Imager (Bio-Rad). Immunoblot were stripped by using mild antibody stripping solution (Pierce) and reprobed with another antibody. Band intensities were determined by densitometry using Quantity One software (Bio-Rad).
Statistical analysis
Statistical analysis was performed using commercially available statistical software STATISTICA (StatSoft Inc., Tulsa, OK). Differences between groups were initially analyzed by one-way ANOVA. When F values for a given variable were found to be significant, the sequentially rejecting Bonferroni-Holm test was subsequently performed using the Holm’s adjusted P values. Results were considered to be significantly different at P < 0.05.
Results
ASARM peptide binds directly to sPHEX as revealed by SPR
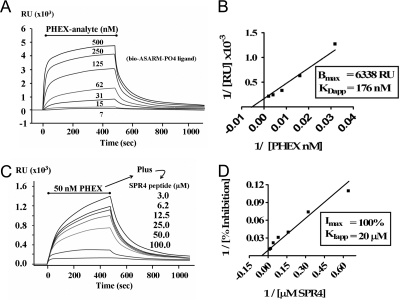
Our previous SPR studies indicate that MEPE binds to sPHEX in vitro (5). These studies also showed that ASARM-PO4 (a potent inhibitor of mineralization in vivo and in vitro) competitively inhibits the MEPE-sPHEX protein-protein interaction (3,5,9). Here, we extend these studies by demonstrating that there is a direct interaction between ASARM-PO4 peptide and sPHEX. To accomplish this, a short biotinylated ASARM-PO4 peptide (ligand) was captured onto a neutravidin-coated SPR chip surface. Varying concentrations of sPHEX (analyte) were flowed over the surface, and the binding was measured in real time. A saturable, concentration-dependent interaction was observed as the sPHEX analyte concentration was raised from 7.5 to 500 nm (Fig. 1A), which can be described by a KDapp of 176 nm± 40 and a Bmax of 6338 ± 616 (sd) RU (n = 3) (Fig. 1B). The interaction was specific because no binding was observed when either a biotinylated nonphosphorylated ASARM peptide or a biotinylated RGD-MEPE peptide was immobilized as control ligand or when proteins other than sPHEX (BSA and IgG) were used as control analytes. The interaction requires Zn, because no interaction could be measured when Zn was omitted from the reaction buffer.
Figure 1.
SPR experiments reveal that sPHEX interacts directly with ASARM-PO4 peptide (A and B) and that this interaction is inhibited by the PHEX SPR4 Peptide (C and D). A, The indicated concentrations of sPHEX were flowed over a captured ASARM-PO4 surface during the period indicated by the bar, and the SPR response was recorded as a function of time in the displayed sensorgram. Notice that the binding is concentration dependent and saturable. B, Linear transformation of sensorgram in A with x- and y-coordinates plotted as inverse values for plasmon RU and sPHEX protein concentration 1/[nm PHEX]. C, The indicated concentrations of SPR4 peptide were flowed over the ASARM-PO4 surface with a constant concentration of sPHEX (50 nm). A concentration-dependent inhibition of the ASARM-PO4-PHEX interaction by SPR4 is observed. Random control SPR5 peptide had no effect. D, Linear transformation of sensorgram C with x- and y-coordinates plotted as inverse values for plasmon RU and SPR4 peptide concentration 1/[μm SPR4]. Each Biacore SPR experiment was independently performed three times for all data points (n = 3). For graph A (PHEX binding to ASARM-PO4), a KDapp of 176 ± 40 nm and a Bmax of 6338 ± 616 (sd) occurs. For graph B (inhibition of PHEX ASARM-PO4 binding by SPR4), a KIapp of 20 ± 11 μm was measured.
The 4.2-kDa PHEX peptide 4 (SPR4) competitively inhibits ASARM peptide-sPHEX binding
To further delineate the region on sPHEX that interacts with the ASARM peptide, a number of peptides were designed, synthesized, and characterized. We found that one of these peptides, a 4.2 kDa PHEX synthetic peptide SPR4, specifically inhibits the binding of sPHEX to the captured ASARM-PO4 peptide (Fig. 1B). SPR4 inhibits the interaction in a concentration-dependent, saturable manner with a KIapp of 20 ± 11 μm (n = 3) (Fig. 1, C and D). A scrambled control peptide (SPR5) and a shortened form of the SPR4 peptide (SPR3) had no effect on the sPHEX-ASARM peptide interaction at concentrations as high as 100 μm (data not shown). All peptide sequences are indicated in Table 1.
Two-dimensional 1H/15N NMR demonstrates PHEX-SPR4 peptide (SPR4) binds directly to ASARM peptide
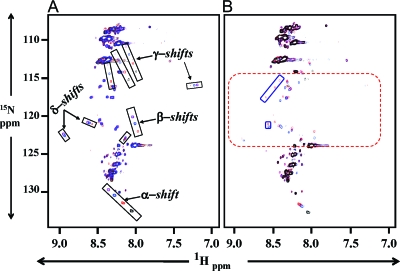
Our SPR experiments showed that the SPR4 peptide competitively inhibits the binding of sPHEX to ASARM-PO4 (Fig. 1). To directly measure the binding of the small synthetic SPR4 PHEX peptide to ASARM peptide we used two-dimensional 1H/15N NMR. Specifically, SPR4 peptide isotopically labeled with 15N on specific amide alanine and glycine residues was incubated with different concentrations of either ASARM-PO4 or ASARM peptides (see Table 1 for sequences). Figure 2A shows overlaid 1H/15N spectra of solutions containing isotopically labeled SPR4 peptide and nonisotopically labeled ASARM-PO4 peptide (see Materials and Methods). The overlaid spectra show perturbations only for SPR4 after peptide binding. The nonisotopic ASARM peptide ligand remains invisible. The overlaid spectrum in black represents SPR4 without ASARM-PO4. In contrast, red, blue, and purple spectra highlight SPR4 peptide in the presence of 1, 3, and 6 molar equivalents of ASARM-PO4, respectively. These spectra clearly show major perturbations demonstrating binding between SPR4 and ASARM-PO4. Figure 2A shows the migration of several resonances from SPR4 that are affected by increasing the amount of ASARM present (denoted as α-, β-, and γ-shifts). In addition, several unique peaks (δ-shifts) are observed as a result of the binding interaction. Specifically, peaks shift from black (SPR4 only) to red, blue, and finally purple as ASARM-PO4 concentration increases and SPR4 isotopic peptide remains constant. Thus, these data demonstrate SPR4 binds to MEPE ASARM peptide. Control peptides had no effect on the SPR4 NMR spectrum (data not shown). Also, overlapping spectra comparing SPR4 in the presence of nonphosphorylated and phosphorylated ASARM peptide revealed an important role for phosphoserines in the binding (Fig. 2B). Specifically, NMR demonstrates that peptide-peptide binding between SPR4 and ASARM chiefly determines the bound conformation. The phosphoserine moieties stabilize the interaction, increasing the binding strength.
Figure 2.
Two-dimensional 1H/15N NMR shows PHEX-SPR4 peptide (SPR4) binds directly to ASARM peptide. A, Overlaid NMR spectra of 15N isotopically labeled SPR4 peptide with no ASARM peptide (black), 1 molar equivalent ASARM-PO4 peptide (red), 3 molar equivalents ASARM-PO4 peptide (blue), and 6 molar equivalents (purple). The ASARM-PO4 peptide is nonisotopically labeled; thus, only the SPR4 peptide 1H/15N shifts are visible in the overlaid spectra. The major shifts are boxed and labeled as α, β, γ, and δ groups, respectively. A dose-dependent increase in shift occurs as illustrated by the black, red, blue, and purple peaks within each highlighted box. This demonstrates specific binding of SPR4 peptide to ASARM-PO4 peptide. B, The SPR4 PHEX peptide also binds nonphosphorylated ASARM peptide. An NMR spectrum of SPR4 in the presence of nonphosphorylated and phosphorylated ASARM peptide is shown in B. The overlaid spectra consist of SPR4 peptide without ASARM peptide (black spectrum), 1 molar equivalent of nonphosphorylated ASARM peptide (blue spectrum), and 1 molar equivalent of ASARM-PO4 peptide (red). The dashed red box highlights the region where many peaks appear as a result of the interaction between SPR4 and the peptide portion of ASARM. Solid red boxes enclose peaks unique to the phosphorylated peptide and reflect changes in SPR4 that are specific to interactions with the phosphate moiety at an equivalent 1:1 ratio. Increasing the concentration of ASARM peptide mimics the changes in the ppm positions observed with ASARM-PO4, but the same degree of change requires more ASARM to achieve compared with ASARM-PO4. The peaks in the blue boxes are considered unique because they were not observed at the highest concentration analyzed. The results indicate that peptide-peptide interactions between SPR4 and ASARM largely determine the bound conformation, whereas the phosphate moieties stabilize the interaction, increasing the binding strength.
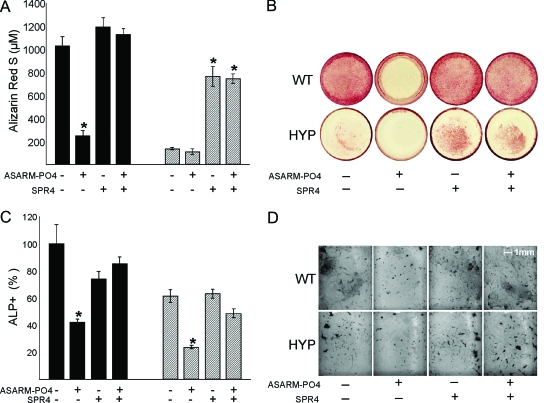
SPR4 (PHEX-related peptide) prevents ASARM-PO4 mediated inhibition of mineralization
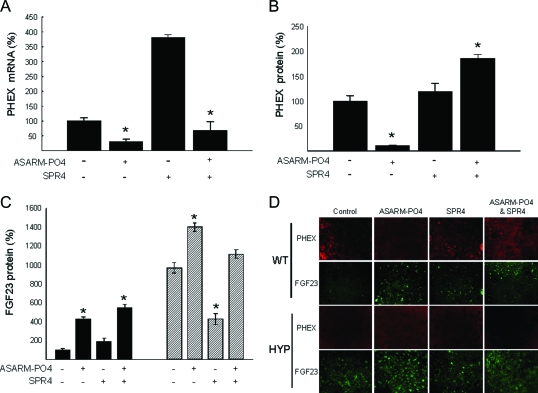
As previously reported, ASARM-PO4 peptide inhibits in vitro mineralization (3,5,8,9,10,14). Indeed, WT BMSCs cells incubated with this peptide display a mineralization defect similar to HYP BMSCs cultured in the absence of ASARM peptide. Consistent with the extensive binding experiments (NMR and SPR), SPR4 peptide was able to prevent ASARM peptide-mediated inhibition of mineralization in BMSC cultures. Furthermore, SPR4 significantly restores mineralization in HYP (2.5-fold, P < 0.05) compared with control and suppresses ASARM-PO4-induced mineralization defects in WT cells (Fig. 3, A and B). An identical dose-dependent reversal also occurs when sPHEX protein is coincubated with ASARM-PO4, as reported as abstracts (preliminary studies) at recent International Bone Mineral Society and American Society for Bone and Mineral Research meetings (49,50). Interestingly, in both cell models, ASARM-PO4 significantly alters the number of ALP-positive cells, suggesting that this peptide also greatly affects osteoblast development. Consistent with the mineralization findings, coincubation with SPR4 peptide prevented the ASARM-PO4-induced decrease in ALP-positive cells (Fig. 3, C and D).
Figure 3.
Effects of incubation with ASARM-PO4 and/or SPR4 peptides on BMSCs from WT (black bars) or HYP (striped bars) in monoculture. A, ARS concentration; B, Representative photomicrographs of ARS staining; C, number of ALP-positive cells, represented as a percentage of total surface and relative to the untreated WT; D, representative photomicrographs of cells stained for ALP. The data represent the mean ± sem of triplicates from a representative experiment of three separate experiments that were not significantly different (Mann-Whitney U test). *, P < 0.05 vs. matched control cells.
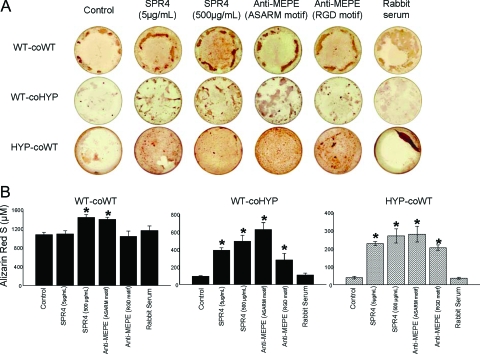
SPR4 or anti-ASARM antibodies corrects HYP BMSC mineralization defect
In agreement with previous studies reported by other investigators (51,52), HYP and WT cells failed to mineralize when cocultured (see Fig. 4). This is thought to be secondary to the action of an unknown diffusible factor (minhibin) secreted by HYP BMSCs. Control cocultures of WT cells with WT BMSCs mineralized normally. Also, HYP BMSCs did not mineralize in the presence of control or rabbit preimmune sera. We extended these studies to determine whether the unknown minhibin is ASARM peptide. Consistent with the monoculture experiments, SPR4 rescued the mineralization defect dose dependently in cocultures of WT and HYP BMSC cells. Anti-MEPE neutralizing antibodies also restored mineralization in both cell types, with a greater impact when using an antibody targeting the ASARM motif (Fig. 4). Taken together, the monoculture and coculture findings provide decisive evidence for the ASARM peptide(s) as the minhibin in HYP.
Figure 4.
Corrected mineralization after incubating BMSCs (cocultures; HYP and WT) with SPR4 peptide and anti-MEPE antibodies. WT cells were either cocultured with WT BMSCs (WT-coWT) or HYP (WT-coHYP), and HYP cells were cultured with WT cells (HYP-coWT). A, Representative photomicrographs of ARS staining; B, ARS concentration. The data represent the mean ± sem of triplicates from a representative experiment of three separate experiments that were not significantly different (Mann-Whitney U test). *, P < 0.05 vs. matched control.
ASARM-PO4 suppresses expression of osteoblastic genes
Consistent with the observed decrease in osteoblast number (Fig. 3C) ASARM-PO4 peptide dramatically altered osteoblast-specific gene marker expression. SPR4 peptide also prevented these changes (Table 3). Specifically, a decrease in runx2, osterix, and osteocalcin expression occurred with WT BMSCs supplemented with ASARM-PO4, suggesting that ASARM-PO4 peptide directly or indirectly suppresses expression of these osteoblastic markers. Coincubation with SPR4 and ASARM-PO4 corrected this effect in all cases except for msx2. Addition of SPR4 without ASARM-PO4 resulted in a dramatic and significant increase in gene expression in all cases, presumably by sequestering endogenous ASARM-PO4. In HYP, addition of ASARM-PO4 had no effect on expression of the same markers. This is likely due to endogenous ASARM-PO4 secretion by these cells and thus saturation. However, as observed with WT cells, addition of SPR4 peptide dramatically increased expression of all genes (Table 3). Thus, these in vitro findings are consistent with the preceding mineralization and binding experiments. Specifically, they support further the conclusion that ASARM-PO4 plays a major role in the bone and mineralization pathology in HYP.
Table 3.
Effects of incubation with ASARM-PO4 and/or SPR4 peptides of WT or HYP BMSCs on osteoblastic gene expression
| % of control
|
||
|---|---|---|
| WT | HYP | |
| runx2 | ||
| Control | 100.0 ± 9.3 | 100.0 ± 33.4 |
| ASARM-PO4 | 62.7 ± 9.5a | 106.1 ± 19.0 |
| SPR4 | 526.4 ± 175.7a | 340.0 ± 91.8a |
| ASARM-PO4+SPR4 | 181.5 ± 34.0 | 21.3 ± 10.3a |
| Osterix | ||
| Control | 100.0 ± 24.9 | 100.0 ± 42.7 |
| ASARM-PO4 | 52.0 ± 21.0 | 100.5 ± 10.2 |
| SPR4 | 372.5 ± 62.7a | 630.1 ± 321.8a |
| ASARM-PO4+SPR4 | 90.2 ± 11.5 | 189.6 ± 15.5 |
| msx2 | ||
| Control | 100.0 ± 37.4 | 100.0 ± 21.2 |
| ASARM-PO4 | 43.5 ± 9.0 | 77.2 ± 20.3 |
| SPR4 | 308.5 ± 77.3a | 269.6 ± 93.0a |
| ASARM-PO4+SPR4 | 59.3 ± 18.3 | 134.0 ± 4.6 |
| Osteocalcin | ||
| Control | 100.0 ± 48.5 | 100.0 ± 11.9 |
| ASARM-PO4 | 53.9 ± 6.4 | 109.3 ± 13.5 |
| SPR4 | 1105.9 ± 39.2a | 323.6 ± 33.1a |
| ASARM-PO4+SPR4 | 182.0 ± 77.6 | 248.3 ± 33.9a |
Gene expression was measured by real-time quantitative PCR.
P < 0.05 vs. matched control cells.
ASARM-PO4 decreases PHEX and increases FGF23 expression
ASARM-PO4 induced an early decrease in PHEX mRNA and protein expression at d 3 and protein levels after 14 d (Fig. 5, A, B, and D). In contrast, PHEX expression was significantly and markedly increased after SPR4 treatment (+280%, P < 0.002). Again, this suggests SPR4 binds ASARM-PO4 and neutralizes its activity. No other changes were observed at d 7 or 21 (data not shown). Note, as expected, no PHEX protein or mRNA was detected in HYP BMSCs. Consistent with our previous published results, FGF23 protein levels were increased in HYP (Fig. 5, C and D), and addition of ASARM-PO4 increased FGF23 protein levels in HYP and WT BMSCs (10).
Figure 5.
BMSCs treated with ASARM-PO4 and SPR4 peptides show altered expression of PHEX and FGF-23. Incubation with ASARM-PO4 and/or SPR4 peptides on BMSCs from WT (black bars) or HYP (striped bars) are shown. A, Phex mRNA levels at d 3; B, number of cells immunostained for PHEX at d 14; C, number of cells immunostained for FGF23 at d 14. The data represent the mean ± sem of triplicates from a representative experiment of three separate experiments that were not significantly different (Mann-Whitney U test). Results are expressed as percentage of values for WT untreated cells. *, P < 0.05 vs. matched control cells. D, Representative photomicrographs of PHEX and FGF23 immunostaining (×20).
SPR4 peptide rescues ASARM peptide-mediated decrease in DMP1 expression
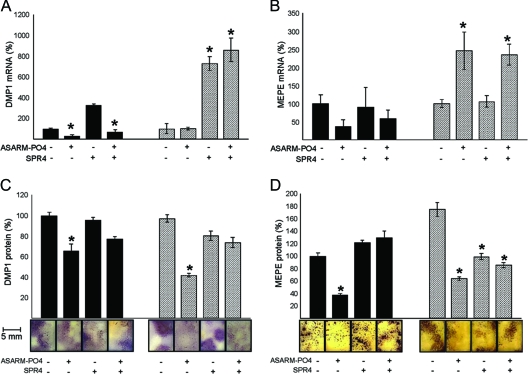
In WT BMSCs, DMP1 expression paralleled PHEX expression. Specifically, ASARM-PO4 peptide reduces expression of DMP1 mRNA and protein, and this was reversed by coincubation with SPR4 peptide (Fig. 6, A and B). As with PHEX, incubation with SPR4 in the absence of ASARM-PO4 results in a dramatic increase in DMP1 expression. With HYP cells, this pattern is quenched due to either endogenous overexpression of ASARM peptides or absence of PHEX ligand. ASARM-PO4 had no effect on DMP1 mRNA expression in HYP BMSCs, but SPR4 with or without ASARM-PO4 increased DMP1 immunoreactivity and mRNA expression (Fig. 6, A and B). Interestingly, although ASARM-PO4 peptide had no effect on DMP1 mRNA expression, a significant drop in DMP1 immunoreactivity was observed in HYP BMSCs, suggesting increased proteolytic degradation. The fact that coincubation of ASARM-PO4 and SPR4 peptides also increased DMP1 expression in HYP suggests stoichiometry and or binding affinities also play a role.
Figure 6.
BMSCs treated with ASARM-PO4 and SPR4 peptides show altered expression of DMP1 and MEPE. Incubation with ASARM-PO4 and/or SPR4 peptides of BMSCs from WT (black bars) or HYP (striped bars). A, DMP1 mRNA levels at d 3; B, DMP1 immunostained surfaces at d 14 (bar graphs and representative photomicrographs); C, MEPE mRNA levels at d 3; D, MEPE-immunostained surfaces at d 14 (bar graphs and representative photomicrographs). The data represent the mean ± sem of triplicates from a representative experiment of three separate experiments that were not significantly different (Mann-Whitney U test). Results are expressed as percentage of values for WT untreated cells *, P < 0.05 vs. matched control cells.
Reduced MEPE protein in HYP and ASARM-treated BMSCs is also rescued by SPR4
Addition of ASARM-PO4 peptide to WT BMSCs and HYP BMSCs caused a dramatic drop in MEPE protein immunoreactivity as screened using MEPE anti-ASARM antibodies (Fig. 6D). Also, BMSCs were screened with anti-MEPE antibodies raised against an N-terminal mid-region incorporating the MEPE RGD motif (anti-MEPE-RGD). With these antibodies, although WT BMSCs were strongly immunopositive, HYP BMSCs with and without ASARM peptide were completely negative and showed no immunoreactivity (data not shown). Again as with DMP1, this suggests increased proteolytic degradation of MEPE protein (small ASARM peptides not detectable). Addition of SPR4 peptide dramatically reversed this and increased MEPE protein immunoreactivity in both wild-type and HYP BMSCs (Fig. 6D). With WT BMSCs, addition of ASARM peptide or SPR4 peptide or coincubation with SPR4 and ASARM-PO4 had no effect on MEPE mRNA expression. In dramatic contrast, addition of ASARM-PO4 peptide to HYP BMSCs resulted in a dramatic increase in MEPE mRNA expression that was also reversed by SPR4 peptide (Fig. 6C).
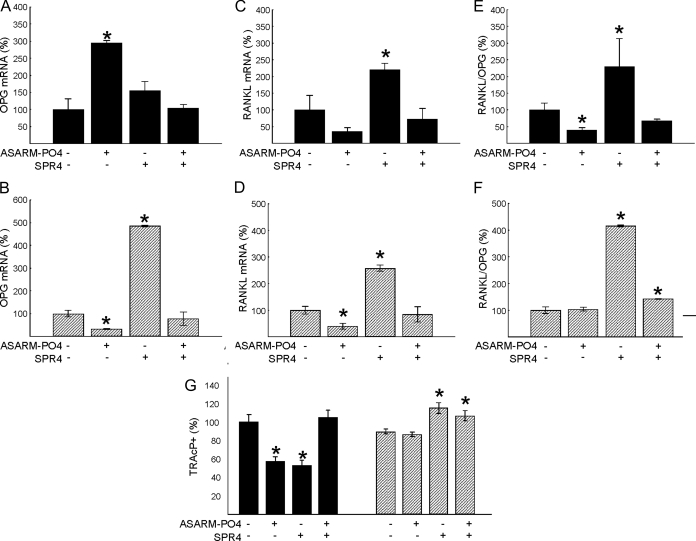
ASARM-PO4 differentially affects OPG/RANKL pathway in HYP and WT BMSCs
Osteoprotegerin (OPG) gene expression is significantly increased in WT cells incubated with ASARM-PO4. In marked contrast, a reverse pattern occurs with HYP cells, and OPG expression is decreased in the presence of ASARM-PO4 (Fig. 7, A and B). In both cases (HYP and WT), coincubation with SPR4 and ASARM-PO4 peptides neutralizes this effect. In the absence of ASARM-PO4, SPR4 peptide significantly increases OPG expression 5-fold in HYP cells presumably due to sequestration of endogenous ASARM peptides (Fig. 7B). An inhibition of RANKL expression occurs with HYP cells supplemented with ASARM-PO4, and a similar trend is observed in WT cells. However, RANKL expression is markedly increased in the presence of SPR4 peptide for both WT and HYP cells (Fig. 7, C and D). Again, consistent with endogenous overexpression of ASARM peptides in HYP cells, the RANKL/OPG ratio was unchanged in HYP BMSCs treated with ASARM-PO4 but markedly reduced in WT BMSCs. Interestingly, SPR4 peptide in the absence of ASARM-PO4 peptide dramatically increased the RANKL/OPG ratio in both WT and HYP cells. This implies the ASARM peptide alters RANKL/OPG ratio independent of PHEX (Fig. 7, E and F). Thus, in vitro ASARM-PO4 has no effect on HYP osteogenic or osteoclastogenic indices presumably due to endogenous and saturating overexpression of ASARM peptides and absence of PHEX in HYP BMSCs. In contrast, WT cell markers are dramatically altered by ASARM-PO4. The reduction in markers of osteoblastogenesis and osteoclastogenesis provides mounting evidence for a reduced bone turnover in the presence of ASARM-PO4. Conversely, SPR4 peptide had the opposite effects to the ASARM peptide on both WT and HYP cells, presumably by sequestering ASARM peptides. Thus, SPR4 peptide increases in vitro indices of both osteoblastogenesis and osteoclastogenesis, suggesting a potential for increased bone turnover in vivo.
Figure 7.
BMSCs incubated with ASARM-PO4 peptide or SPR4 peptide showed altered osteoclastogenesis. Incubation with ASARM-PO4 and/or SPR4 peptides of BMSCs from WT (black bars) or HYP (striped bars) on osteoclastogenesis are shown. A and B, OPG mRNA levels at d 3 for WT (A) and HYP (B); C and D, RANKL mRNA levels at d 3 for WT (C) and HYP (D); E and F, RANKL/OPG mRNA ratio at d 3 for WT (E) and HYP (F); G, TRAcP-immunostained surfaces. The data represent the mean ± sem of triplicates from a representative experiment of three separate experiments that were not significantly different (Mann-Whitney U test). Results are expressed as percentage of values for WT untreated cells. *, P < 0.05 vs. matched control cells.
SPR4 and ASARM-PO4 suppress osteoclastogenesis in WT BMSCs: evidence for a secondary PHEX ligand
In WT BMSCs, addition of ASARM-PO4 or SPR4 peptide significantly reduced TRAcP-positive staining. This was reversed by coincubation with SPR4 peptide and ASARM-PO4 (Fig. 7G). A marked decrease in cell proliferation was also noted (data not shown). In contrast, there was no significant change in the number of TRAcP-positive cells after addition of ASARM-PO4 to HYP cells. Again, this was likely due to overexpression of endogenous ASARM peptides by the HYP cells and signal saturation. Interestingly, SPR4 peptide in the absence of ASARM-PO4 peptide reduces osteoclast number in WT but increases it in HYP BMSCs (Fig. 7G). Coincubation with ASARM-PO4 and SPR4 restores basal levels in WT cells and produces a slight but significant increase in TRAcP-positive cells in HYP. Because SPR4 like ASARM-PO4 reduces TRAcP activity in only WT BMSCS, SPR4 peptide may compete with PHEX for a secondary PHEX ligand. In contrast, ASARM-PO4 may (by binding to PHEX directly) prevent this secondary PHEX-ligand interaction from occurring. This is further supported by the finding that coincubation of WT BMSCs with SPR4 and ASARM-PO4 peptides restores TRAcP to normal (Fig. 7G).
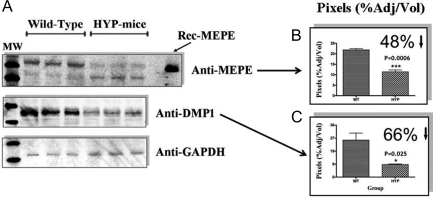
MEPE and DMP1 proteins are degraded in HYP calvariae
Proteins extracted from HYP and normal mice calvariae were separated on 4–12% SDS-PAGE gels and then serially immunoblotted (Western) with rabbit polyclonal antibodies against MEPE (ASARM region), DMP-1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), respectively (Fig. 8). Five calvariae were screened for all groups, and three representative screens for each group (MEPE, DMP1, and GAPDH) are shown in Fig. 8A. Figure 8, B and C, graphically shows chemiluminescent intensity (pixels percentage adjusted for volume) for MEPE (B) and DMP1 (C) after normalization for protein and GAPDH control (Quantity 1 Bio-Rad Software). Although MEPE mRNA expression is increased in HYP (Fig. 6) (10,11,13,18), major degradation of MEPE protein (>48%, P = 0.0006; n = 5) and DMP1 protein (>68%, P = 0.025; n = 5) was detected in HYP mice calvariae (Fig. 8). These findings are consistent with reports of increased degradation of MEPE protein in HYP patient teeth despite increased mRNA expression (21). Thus, increased degradation of these proteins (MEPE and DMP1) is likely responsible for the increased amounts of COOH-terminal, protease-resistant ASARM peptides in HYP because both proteins contain ASARM-like motifs (3,5,7,8,9,14). Also, protease levels in HYP mice osteoblasts are markedly increased (8,15,19,20). Degradation of matrix proteins is part of the mechanism whereby osteogenesis is inhibited in HYP.
Figure 8.
MEPE and DMP1 proteins are degraded in HYP mice calvariae. A, Chemiluminescent Western blots of HYP and WT calvariae screened with MEPE, DMP1, and GAPDH antibodies as indicated; B, adjusted pixel intensity of anti-MEPE blot; C, adjusted pixel intensity of anti-DMP1 blot. WT are left and HYP right on respective histograms. Calvariae from five male WT and HYP mice were used (n = 5; HYP and WT). Three separate representative calvariae are shown for WT and HYP.
Discussion
Our previous work demonstrated direct binding of PHEX to MEPE and showed ASARM-PO4 peptide competitively inhibits this binding (5). More recently, we extended this study and showed both ASARM peptide and MEPE protein potently inhibits PHEX enzymic hydrolysis of a nonphysiological fluorogenic peptide substrate (10). Both these studies provide indirect evidence that the PHEX-MEPE binding site involves the ASARM motif. For the first time, this study directly shows that binding between PHEX and MEPE occurs between the PHEX Zn-binding motif and a short COOH-terminal region of MEPE consisting of the ASARM motif. Indeed, not only does PHEX bind specifically to immobilized ASARM-PO4 peptide but a synthetic PHEX peptide (SPR4) comprising the Zn binding motif also directly and specifically binds free ASARM peptide (2.2 kDa). Both phosphorylated and nonphosphorylated forms of ASARM peptides bind SPR4 peptide (4.2 kDa) as demonstrated by NMR. The peptide sequence of both peptides control the bound conformation, and the phosphoserine moieties of the ASARM peptide stabilize the binding by increasing the binding strength. This may have physiological implications because a change in casein ectokinase expression and activity, present on the surface of osteoblasts, occurs in HYP (53,54).
Mature osteoblasts, osteocytes, odontoblasts, proximal convoluted tubules of the kidney (luminal side), and saliva all express MEPE (6,8,21,22,47,48,49,55,56,57). MEPE is also present in the circulation at high levels in normal subjects and age-dependently correlates with serum PO4, PTH, and bone mineral density (8,14,17). In HYP, a marked increase in MEPE expression (protein and mRNA) and ASARM peptides in serum, bone, and kidney occurs (3,8,10,11,12,13,14,18,26,50). SIBLING and MEPE ASARM peptides amass in bone in regions associated with hyperosteoidosis, and this is likely a key reason for the mineralization pathophysiology (3,8). Indeed, ASARM peptides are potent inhibitors of mineralization, and this is consistent with other similar peptides such as salivary statherin and osteopontin ASARM peptides (3,5,8,9,58,59,60). For the first time, we show in this study that ASARM peptide(s) are the primary HYP osteoblast factors inhibiting mineralization in vitro (minhibin). Anti-ASARM antibodies and SPR4 peptide that binds to ASARM peptides demonstrated this. Specifically, a correction of the mineralization defect occurred after incubation with SPR4 or anti-ASARM polyclonals in monocultures or cocultures of HYP and WT BMSCs.
Also in this study, we found that ASARM peptides induce a marked significant decrease in the expression of the osteoblastic markers (runx2, osterix, msx2, and osteocalcin) as well as in the number of osteoblasts and ALP-positive cells. Consistent with this, a decrease in osteoblast cell numbers and a significant decline in the number of ALP-positive cells also occurred. Coincubation with SPR4 reversed this. In HYP cells, because of constitutive overexpression of endogenous ASARM peptide, addition of synthetic ASARM peptide had no significant effect. However, as with WT BMSCs, SPR4 peptide dramatically increased expression of these markers by sequestering free ASARM peptides. Thus, ASARM peptides reduce osteoblastogenesis, and SPR4 synthetic PHEX peptide reverses this and potently up-regulates osteoblastic markers.
In HYP, a decrease in bone resorption, osteoclast number, and RANKL expression and a suppression of osteoblastic markers occurs (38,39,40,61). NPT2a−/− (Na-dependent phosphate cotransporter) mice also display a decrease in osteoclast numbers likely mediated in part directly or indirectly by hypophosphatemia (35,37). Of note, recombinant MEPE but not FGF23 inhibits osteoclast-like cell formation from induced WT BMSCs in culture (38). Bone formation and bone resorption are regulated through multiple mechanisms. One of the bone resorption mechanisms involves changes in the RANKL/OPG system and thus osteoclastogenesis (62,63). In this study, suppression of osteoclastogenesis and an increased RANKL/OPG ratio occurred in WT BMSCs treated with macrophage colony-stimulating factor and supplemented with ASARM peptides. SPR4 peptide induced an opposite effect and increased osteoclastogenesis by increasing RANKL expression. Thus, in vitro ASARM-PO4 and SPR4 peptides have opposite effects on osteoclastogenesis markers, and SPR4 binds and neutralizes ASARM peptide activity. Because of endogenous and constitutive overexpression of ASARM peptides in HYP, synthetic ASARM peptides had less of an effect but did decrease both RANKL and OPG mRNA expression. Our results are consistent with reports that recombinant MEPE inhibits osteoclast-like cell formation in BMSC cell cultures (38). Thus, like BMSCs from HYP mice, ASARM-PO4-treated BMSCs have decreased osteoclastogenic gene markers.
Because the ASARM peptide clearly binds to PHEX and has potent in vitro anti-mineralization effects and suppresses bone formation and resorption markers in vitro, we next focused on PHEX expression (mRNA and protein). Remarkably, WT BMSCs incubated with ASARM-PO4 displayed a marked decrease in PHEX protein and mRNA with increased FGF23 protein levels. The increase in FGF23 by ASARM-PO4 is consistent with our previously reported studies (10). Again co incubation with equimolar SPR4 peptide neutralized this effect likely because of sequestration and inactivation of ASARM peptide. A significant and marked increase in PHEX protein and mRNA occurred in cells cultured with SPR4 without ASARM peptide. Thus, this data supports the notion that ASARM peptide not only binds to PHEX but also suppresses PHEX expression (mRNA and protein) and this in turn also affects mineralization. The mechanism whereby ASARM peptide causes this (transcription, mRNA stability, or targeted proteolysis) is under investigation. Because loss of PHEX activity results in increased FGF23, this novel pathway is physiologically important.
ASARM peptides markedly affect mineralization and bone marker expression (osteoblastic and osteoclastic) and contribute to the HYP mineralization defect in vitro. In WT cultures, ASARM peptides likely inhibit mineralization by binding hydroxyapatite directly and suppressing osteoblastogenesis and osteoclastogenesis markers. However, as mentioned, the peptide also suppresses protein and mRNA expression of PHEX and coordinately increases expression of FGF23. This is consistent with our published in vitro and ex vivo findings that show both increased FGF23 expression and direct inhibition of PHEX enzymic fluorogenic hydrolysis by MEPE and ASARM peptides (10). Indeed, WT BMSC cultures, containing enhanced green fluorescence protein (GFP) driven by an FGF23 promoter, showed markedly increased GFP fluorescence compared with non-ASARM-peptide-treated cultures (10). Also, fluorescence remained constitutively high in control HYP BMSC cultures containing the same enhanced GFP FGF23-promoter construct. For the first time, this study shows a direct inhibition of the ASARM-PO4 peptide-induced increase in FGF23 expression by a small synthetic PHEX peptide SPR4. More importantly, SPR4 without ASARM-PO4 peptide decreases FGF23 protein expression in WT and HYP BMSCs. The implications are that SPR4 will have similar in vivo properties. If this is the case, the peptide may well be of therapeutic use in diseases where FGF23 expression is increased and mineralization is defective.
Another important feature of the ARHR and likely HYP phenotype is DMP1, an important matrix protein needed for normal mineralization and phosphate homeostasis. Indeed, DMP1-null mutants are phenotypically similar to HYP mice, and mutations in DMP1 are responsible for ARHR (34,64). Also, increased HYP osteoblast protease levels leads to excessive degradation of the extracellular matrix and increased protease-resistant ASARM peptides (3,8,15,19,20). We have shown that treatment of HYP mice with protease inhibitors partially corrects the mineralization defect with a commensurate decrease in ASARM peptides (8). Also marked degradation of MEPE protein occurs in HYP teeth (21). Consistent with this, our study demonstrates degradation of both DMP1 and MEPE protein in HYP calvarial cultures. Of further note, our data also show that in vitro addition of ASARM peptide increases degradation of MEPE and DMP1 protein in BMSC cultures (reversed by coincubation with SPR4 peptide). As mentioned, increased osteoblastic protease activity and expression is a key feature of the HYP phenotype (8,15,19,20). Because PHEX is defective in HYP and PHEX protein is suppressed by ASARM peptides in vitro, loss of PHEX is likely the primary link to increased protease activity and expression. Thus, we conclude loss of PHEX function leads to excess osteoblastic protease expression, increased MEPE expression, excessive degradation of both MEPE and DMP1, and release of protease-resistant ASARM peptides from both DMP1 and MEPE.
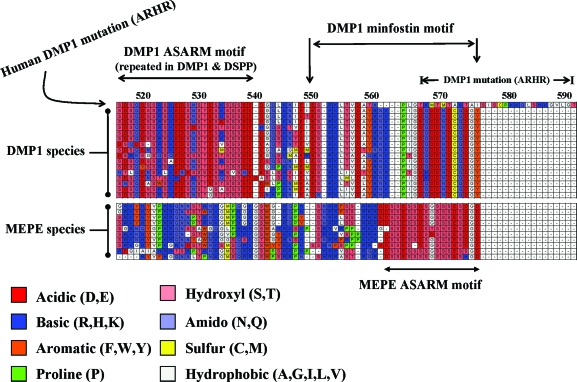
Figure 9 shows the remarkable biological conservation of the ASARM motif from DMP1 and MEPE across species and over millions of years of evolution. Species as diverse as the armadillo and duck-billed platypus still retain this feature. The motif also occurs in MEPE protein at the tip and terminal end of the protein COOH terminus. In DMP-1, the ASARM motif also occurs at the COOH terminus but unlike MEPE is repeated and more importantly capped by an additional 27–28 terminal residues. This DMP1 COOH region is labeled as the minfostin motif in Fig. 9. A frameshift mutation that extends this minfostin motif results in ARHR. Clearly, proteolytic degradation of both DMP1, a mineralization promoter (minfostin), and MEPE will generate excess amounts of protease-resistant ASARM peptides (minhibins) exacerbated by overexpression of MEPE in HYP. Of note, both DMP1 and MEPE retain consensus cleavage sites for cathepsin K and B in positions that would precisely release the ASARM peptides. Thus, the cathepsin cleavage sites in MEPE and DMP1 may be of physiological relevance because these sites are not conserved elsewhere and occur infrequently within these proteins.
Figure 9.
Clustal alignment of MEPE and DMP1 COOH-terminal regions with ASARM motifs. Alignments are ordered from top to bottom as follows: for DMP1, mutated human DMP1 (ARHR), normal human, chimpanzee, macaque-primate, bush baby, cat, elephant, cow, rabbit, hedgehog, tenrec, squirrel, opossum, duck-billed platypus, guinea pig, mouse, and rat; for MEPE, human, chimpanzee, macaque-primate, rhesus-primate, bat, dog, cow, squirrel, rat, and mouse. There are two key regions, an ASARM motif region (DMP1 and MEPE) and a minfostin motif region (DMP1). In both DMP1 and MEPE, the ASARM motif is highlighted and consists of acidic aspartate (D) and glutamate residues (E) plus hydroxyl serines (S). Several serine residues are likely phosphorylated and play key physiological roles. Note within the MEPE ASARM motif that there are also two highly conserved hydrophobic glycine residues (G). One of these residues is penultimate and occurs in all species. The DMP1 ASARM motif is repeated throughout the C-terminal region of the molecule. A region at the very COOH-terminal tip of DMP1 is highly conserved. This region is labeled as a minfostin motif in the diagram. This is because a mutation extending and altering the primary sequence of this motif results in ARHR. Cathepsin B and K sites are also highly conserved in DMP1 and MEPE and would result in the release of free protease-resistant ASARM peptides. Amino acid residues are colored according to chemical type, and a key is included below the figure. Numbers above the clustal alignment are amino acid residues for DMP1.
The experiments described in this paper and previous work by us and others allows us to propose a model for the role of MEPE, DMP1, ASARM peptides, PHEX, and FGF23 in bone renal phosphate metabolism. PHEX plays a central role by specifically binding accessible ASARM motifs or free ASARM peptides. There is evidence that PHEX may also hydrolyze and inactivate free ASARM peptides (43). These peptides are otherwise exquisitely resistant to proteolysis (3). The evidence for MEPE and ASARM peptides specifically binding to PHEX (a Zn-metalloendopeptidase) is unequivocal. In view of the similarities between DMP1 and MEPE ASARM peptides, binding experiments investigating interactions between DMP1 and PHEX may also prove informative. Intriguingly, matrix metalloproteinase-9, a known Zn-metalloendopeptidase DMP1-binding partner, is markedly reduced in HYP hypertrophic chondrocytes (39). Of note, there are two striking features in DMP1 that contrast with MEPE. The first is the presence of a 27-residue minfostin motif that terminates and caps the ASARM motif region of DMP1 (see Fig. 9). The second is the presence of multiple repeats of the ASARM motif N terminal and contiguous to the DMP1 minfostin motif. As mentioned previously, a frameshift mutation in the DMP1 minfostin motif that extends the sequence length by a few residues and alters the amino acid sequence causes ARHR (see Fig. 9) (34,64). Also, the mutated DMP1 protein, like the wild-type DMP1, is secreted. Thus, the DMP1 minfostin motif may play a signaling or epistatic role in coordinating mineralization. MEPE or ASARM peptides may regulate this process by competing for PHEX binding with other ligands or substrates. This model is also consistent with the finding that double MEPE/PHEX-null mutants retain a mineralization and phosphate defect (23).
In conclusion, this study demonstrates that PHEX binds to MEPE via the PHEX Zn-binding motif and the MEPE ASARM motif. ASARM peptides are responsible for the mineralization defect in HYP BMSCs in vitro and mimic the HYP phenotype. The ASARM peptide inhibition of mineralization is likely because of a combination of direct binding to hydroxyapatite crystals and decreased expression of PHEX. Although speculative, ASARM peptide or MEPE protein may also act by competitive prevention of PHEX binding to undefined ligands or substrates. The reduced PHEX expression induced by ASARM peptides is likely responsible for the increase in FGF23 expression. ASARM peptides also decrease both osteoblastic gene expression and osteoclast differentiation by decreasing the RANKL/OPG ratio. A small 4.2-kDa synthetic PHEX-related peptide (SPR4), incorporating the Zn-binding region, reverses the ASARM-mediated changes and dramatically reduces FGF23 protein expression in WT and HYP BMSCs. The SPR4 peptide may well be pharmaceutically useful to treat mineralization and hypophosphatemia disorders involving FGF23 and ASARM peptides. Also, based on these findings, it is likely that direct or indirect interactions between DMP1, MEPE, and PHEX are important for mineralization and phosphate homeostasis.
Acknowledgments
We acknowledge the very kind gift of pure sPHEX by Dr. Philippe Crine (Department of Biochemistry, University of Montreal, and Enobia Pharma). Also, we acknowledge the anti-DMP1 antibodies generously donated by Dr. Larry Fisher, National Institute of Dental and Craniofacial Research, Bethesda, MD.
Footnotes
We acknowledge the generous financial support from the National Institutes of Health to P.S.N.R. (RO-1 AR51598-01; National Institute of Arthritis and Musculoskeletal Diseases). Also, the SPR experiments were performed in the UTHSCSA Center for Macromolecular Interactions, which is supported by grants from the National Cancer Institute (CA54174) and UTHSCSA Executive Research Committee Research fund.
Disclosure Statement: The authors have nothing to disclose.
First Published Online December 27, 2007
Abbreviations: ALP, Alkaline phosphatase; ARHR, autosomal recessive hypophosphatemic rickets; ARS, alizarin red S; ASARM, acidic serine- and aspartate-rich MEPE-associated motif; BMSC, bone marrow stromal cell; DMP1, dentin matrix protein 1; FGF23, fibroblast growth factor 23; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescence protein; HYP, X-linked hypophosphatemic rickets; MEPE, matrix extracellular phosphoglycoprotein; NMR, nuclear magnetic resonance; OPG, osteoprotegerin; PHEX, phosphate-regulating gene with homologies to endopeptidases on the X chromosome; RANKL, receptor activator for nuclear factor-κB ligand; SIBLING, small integrin-binding ligand, N-linked glycoprotein; SPR, surface plasmon resonance; TBS-T, Tris-buffered saline containing 0.1% Tween 20; TRAcP, tartrate-resistant acid phosphatase; WT, wild type.
References
- Francis F, Hennig S, Korn B, Reinhardt R, de Jong D, Poustka A, Lehrach H, Rowe PS, Goulding JN, Summerfield T, Mountford RC, Read AP, Popowska E, Pronicka E, Davies KE, O’Riordan JL, Econs MJ, Nesbitt T, Drezner MK, Oudet C, Pannetier S, Hanauer A, Strom TM, Meindl A, Lorenz B, Cagnoli M, Mohnike KL, Murken J, Meitinger T 1995 A gene (PEX) with homologies to endopeptidases is mutated in patients with X-linked hypophosphatemic rickets. The HYP Consortium. Nat Genet 11:130–136 [Google Scholar]
- Prie D, Beck L, Urena P, Friedlander G 2005 Recent findings in phosphate homeostasis. Curr Opin Nephrol Hypertens 14:318–324 [DOI] [PubMed] [Google Scholar]
- Rowe PSN 2004 The wrickkened-pathways of FGF23, MEPE and PHEX. Crit Rev Oral Biol Med 15:264–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AJ, Tanzawa K 1997 Mammalian membrane metallopeptidases: NEP, ECE, KELL, and PEX. FASEB J 11:355–364 [DOI] [PubMed] [Google Scholar]
- Rowe PSN, Garrett IR, Schwarz PM, Carnes DL, Lafer EM, Mundy GR, Gutierrez GE 2005 Surface plasmon resonance (SPR) confirms MEPE binds to PHEX via the MEPE-ASARM motif: a model for impaired mineralization in X-linked rickets (HYP). Bone 36:33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbureke KU, Fisher LW 2005 Renal expression of SIBLING proteins and their partner matrix metalloproteinases (MMPs). Kidney Int 68:155–166 [DOI] [PubMed] [Google Scholar]
- Rowe PSN, de Zoysa P, Dong R, Wang H, White K, Econs M, Oudet C 2000 MEPE, a new gene expressed in bone-marrow and tumours causing osteomalacia. Genomics 67:54–68 [DOI] [PubMed] [Google Scholar]
- Rowe PS, Matsumoto N, Jo OD, Shih RN, Oconnor J, Roudier MP, Bain S, Liu S, Harrison J, Yanagawa N 2006 Correction of the mineralization defect in hyp mice treated with protease inhibitors CA074 and pepstatin. Bone 39:773–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe PSN, Kumagai Y, Gutierrez G, Garrett IR, Blacher R, Rosen D, Cundy J, Navvab S, Chen D, Drezner MK, Quarles LD, Mundy GR 2004 MEPE has the properties of an osteoblastic phosphatonin and minhibin. Bone 34:303–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Rowe PS, Vierthaler L, Zhou J, Quarles LD 2007 Phosphorylated acidic serine-aspartate-rich MEPE-associated motif peptide from matrix extracellular phosphoglycoprotein inhibits phosphate regulating gene with homologies to endopeptidases on the X-chromosome enzyme activity. J Endocrinol 192:261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Rowe PS, Liu S, Simpson LG, Xiao ZS, Quarles LD 2002 Inhibition of MEPE cleavage by Phex. Biochem Biophys Res Commun 297:38–45 [DOI] [PubMed] [Google Scholar]
- Argiro L, Desbarats M, Glorieux FH, Ecarot B 2001 Mepe, the gene encoding a tumor-secreted protein in oncogenic hypophosphatemic osteomalacia, is expressed in bone. Genomics 74:342–351 [DOI] [PubMed] [Google Scholar]
- Bai X, Miao D, Panda D, Grady S, McKee MD, Goltzman D, Karaplis AC 2002 Partial rescue of the Hyp phenotype by osteoblast-targeted PHEX (phosphate-regulating gene with homologies to endopeptidases on the X chromosome) expression. Mol Endocrinol 16:2913–2925 [DOI] [PubMed] [Google Scholar]
- Bresler D, Bruder J, Mohnike KL, Fraser D, Rowe PS 2004 Serum MEPE-ASARM peptides are elevated in X-linked rickets (HYP): implications for phosphaturia and rickets. J Endocrinol 183:R1–R9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois SG, Ruchon AF, Delalandre A, Boileau G, Lajeunesse D 2002 Role of abnormal neutral endopeptidase-like activities in Hyp mouse bone cells in renal phosphate transport. Am J Physiol Cell Physiol 283:C1414–C1421 [DOI] [PubMed] [Google Scholar]
- Guo R, Liu S, Spurney RF, Quarles LD 2001 Analysis of recombinant Phex: an endopeptidase in search of a substrate. Am J Physiol Endocrinol Metab 281:E837–E847 [DOI] [PubMed] [Google Scholar]
- Jain A, Fedarko NS, Collins MT, Gelman R, Ankrom MA, Tayback M, Fisher LW 2004 Serum levels of matrix extracellular phosphoglycoprotein (MEPE) in normal humans correlate with serum phosphorus, parathyroid hormone and bone mineral density. J Clin Endocrinol Metab 89:4158–4161 [DOI] [PubMed] [Google Scholar]
- Liu S, Guo R, Simpson LG, Xiao ZS, Burnham CE, Quarles LD 2003 Regulation of FGF23 expression but not degradation by phex. J Biol Chem 278:37419–37426 [DOI] [PubMed] [Google Scholar]
- Matsumoto N, Jo OD, Shih RN, Brochmann EJ, Murray SS, Hong V, Yanagawa J, Yanagawa N 2005 Increased cathepsin D release by Hyp mouse osteoblast cells. Am J Physiol Endocrinol Metab 289:E123–E132 [DOI] [PubMed] [Google Scholar]
- Matsumoto N, Jo OD, Shih RN, Yanagawa N 2005 Altered cathepsin D metabolism in PHEX antisense human osteoblast cells. Biochem Biophys Res Commun 332:248–253 [DOI] [PubMed] [Google Scholar]
- Boukpessi T, Septier D, Bagga S, Garabedian M, Goldberg M, Chaussain-Miller C 2006 Dentin alteration of deciduous teeth in human hypophosphatemic rickets. Calcif Tissue Int 79:294–300 [DOI] [PubMed] [Google Scholar]
- Gowen LC, Petersen DN, Mansolf AL, Qi H, Stock JL, Tkalcevic GT, Simmons HA, Crawford DT, Chidsey-Frink KL, Ke HZ, McNeish JD, Brown TA 2003 Targeted disruption of the osteoblast/osteocyte factor 45 gene (OF45) results in increased bone formation and bone mass. J Biol Chem 278:1998–2007 [DOI] [PubMed] [Google Scholar]
- Liu S, Brown TA, Zhou J, Xiao ZS, Awad H, Guilak F, Quarles LD 2005 Role of matrix extracellular phosphoglycoprotein in the pathogenesis of X-linked hypophosphatemia. J Am Soc Nephrol 16:1645–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbie H, Unwin RJ, Faria NJ, Shirley DG 23 Nov 2007 Matrix extracellular phosphoglycoprotein causes phosphaturia in rats by inhibiting tubular phosphate reabsorption. Nephrol Dial Transplant 10.1093/ndt/gfm535 [DOI] [PubMed] [Google Scholar]
- Erben RG, Mayer D, Weber K, Jonsson K, Juppner H, Lanske B 2005 Overexpression of human PHEX under the human β-actin promoter does not fully rescue the Hyp mouse phenotype. J Bone Miner Res 20:1149–1160 [DOI] [PubMed] [Google Scholar]
- Liu S, Guo R, Tu Q, Quarles LD 2002 Overexpression of Phex in osteoblasts fails to rescue the Hyp mouse phenotype. J Biol Chem 277:3686–3697 [DOI] [PubMed] [Google Scholar]
- Drezner MK 1984 The role of abnormal vitamin D metabolism in X-linked hypophosphatemic rickets and osteomalacia. Adv Exp Med Biol 178:399–404 [DOI] [PubMed] [Google Scholar]
- Drezner MK, Lyles KW, Haussler MR, Harrelson JM 1980 Evaluation of a role for 1,25-dihydroxyvitamin D3 in the pathogenesis and treatment of X-linked hypophosphatemic rickets and osteomalacia. J Clin Invest 66:1020–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher EM, Southard JL, Scriver CR, Glorieux FH 1976 Hypophosphataemia: Mouse model for human familial hypophosphatemic (vitamin D-resistant) rickets. Proc Natl Acad Sci USA 73:4667–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell RM, Lyles KW, Harrelson JM, Friedman NE, Drezner MK 1985 Healing of bone disease in X-linked hypophosphatemic rickets/osteomalacia. Induction and maintenance with phosphorus and calcitriol. J Clin Invest 75:1858–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles KW, Harrelson JM, Drezner MK 1982 The efficacy of vitamin D2 and oral phosphorus therapy in X-linked hypophosphatemic rickets and osteomalacia. J Clin Endocrinol Metab 54:307–315 [DOI] [PubMed] [Google Scholar]
- Marie PJ, Travers R, Glorieux FH 1982 Healing of bone lesions with 1,25-dihydroxyvitamin D3 in the young X-linked hypophosphatemic male mouse. Endocrinology 111:904–911 [DOI] [PubMed] [Google Scholar]
- Marie PJ, Travers R, Glorieux FH 1982 Bone response to phosphate and vitamin D metabolites in the hypophosphatemic male mouse. Calcif Tissue Int 34:158–164 [DOI] [PubMed] [Google Scholar]
- Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, Yu X, Rauch F, Davis SI, Zhang S, Rios H, Drezner MK, Quarles LD, Bonewald LF, White KE 2006 Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet 38:1310–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbagh Y, Carpenter TO, Demay MB 2005 Hypophosphatemia leads to rickets by impairing caspase-mediated apoptosis of hypertrophic chondrocytes. Proc Natl Acad Sci USA 102:9637–9642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Onishi T, Hayashibara T, Sakashita S, Okawa R, Ooshima T 2005 Dentinal defects in Hyp mice not caused by hypophosphatemia alone. Arch Oral Biol 51:58–63 [DOI] [PubMed] [Google Scholar]
- Gupta A, Tenenhouse HS, Hoag HM, Wang D, Khadeer MA, Namba N, Feng X, Hruska KA 2001 Identification of the type II Na+-Pi cotransporter (Npt2) in the osteoclast and the skeletal phenotype of Npt2−/− mice. Bone 29:467–476 [DOI] [PubMed] [Google Scholar]
- Hayashibara T, Hiraga T, Sugita A, Wang L, Hata K, Ooshima T, Yoneda T 2007 Regulation of osteoclast differentiation and function by phosphate: potential role of osteoclasts in the skeletal abnormalities in hypophosphatemic conditions. J Bone Miner Res 22:1743–1751 [DOI] [PubMed] [Google Scholar]
- Miao D, Bai X, Panda D, Karaplis AC, Goltzman D, McKee MD 2004 Cartilage abnormalities are associated with abnormal Phex expression and with altered matrix protein and MMP-9 localization in Hyp mice. Bone 34:638–647 [DOI] [PubMed] [Google Scholar]
- Miao D, Bai X, Panda D, McKee M, Karaplis A, Goltzman D 2001 Osteomalacia in Hyp mice is associated with abnormal Phex expression and with altered bone matrix protein expression and deposition. Endocrinology 142:926–939 [DOI] [PubMed] [Google Scholar]
- Rowe PSN, Oudet C, Francis F, Sinding C, Pannetier S, Econs MJ, Strom TM, Meitinger T, Garabedian M, David A, Macher MA, Questiaux E, Popowska E, Pronicka E, Read AP, Mokrzycki A, Glorieux FH, Drezner MK, Hanauer A, Lehrach H, Goulding J, O’Riordan JLH 1997 Distribution of mutations in the PEX gene in families with X-linked hypophosphataemic rickets (HYP). Hum Mol Genet 6:539–549 [DOI] [PubMed] [Google Scholar]
- Boileau G, Tenenhouse HS, Desgroseillers L, Crine P 2001 Characterization of PHEX endopeptidase catalytic activity: identification of parathyroid-hormone-related peptide107–139 as a substrate and osteocalcin, PPi and phosphate as inhibitors. Biochem J 355:707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Couture C, Hirata IY, Juliano MA, Loisel TP, Crine P, Juliano L, Boileau G, Carmona AK 2003 Human recombinant endopeptidase PHEX has a strict S1′ specificity for acidic residues and cleaves peptides derived from fibroblast growth factor-23 and matrix extracellular phosphoglycoprotein. Biochem J 373:271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD 1995 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6:135–140 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A 1995 NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG 2007 SPARKY3. University of California San Francisco: http://www.cgl.ucsf.edu/ [Google Scholar]
- Ogbureke KU, Fisher LW 2004 Expression of SIBLINGs and their partner MMPs in salivary glands. J Dent Res 83:664–670 [DOI] [PubMed] [Google Scholar]
- Ogbureke KU, Fisher LW 2007 SIBLING expression patterns in duct epithelia reflect the degree of metabolic activity. J Histochem Cytochem 55:403–409 [DOI] [PubMed] [Google Scholar]
- Addison WN, Nakano Y, Crine P, Rowe PS, McKee MD 2007 Regulation of mineralization by phosphorylated MEPE-ASARM peptide in MC3T3-E1 osteoblast cultures. Bone 40:S222–S222 [Google Scholar]
- Rowe PS, Hedge A, Schwarz P, Addison WN, McKee MD 2006 MEPE ASARM peptide(s) bind to PHEX protein, PHEX Zn-motif peptide(s) and hydroxyapatite, and inhibit mineralization of MC3T3–E1 osteoblast cultures. J Bone Miner Res 21:S11–S11 [Google Scholar]
- Nesbitt T, Fujiwara I, Thomas R, Xiao ZS, Quarles LD, Drezner MK 1999 Coordinated maturational regulation of PHEX and renal phosphate transport inhibitory activity: evidence for the pathophysiological role of PHEX in X-linked hypophosphatemia. J Bone Miner Res 14:2027–2035 [DOI] [PubMed] [Google Scholar]
- Xiao ZS, Crenshaw M, Guo R, Nesbitt T, Drezner MK, Quarles LD 1998 Intrinsic mineralization defect in Hyp mouse osteoblasts. Am J Physiol 275:E700–E708 [DOI] [PubMed] [Google Scholar]
- Rifas L, Cheng S, Halstead LR, Gupta A, Hruska KA, Avioli LV 1997 Skeletal casein kinase activity defect in the HYP mouse. Calcif Tissue Int 61:256–259 [DOI] [PubMed] [Google Scholar]
- Rifas L, Dawson LL, Halstead LR, Roberts M, Avioli LV 1994 Phosphate transport in osteoblasts from normal and X-linked hypophosphatemic mice. Calcif Tissue Int 54:505–510 [DOI] [PubMed] [Google Scholar]
- Amir LR, Jovanovic A, Perdijk FB, Toyosawa S, Everts V, Bronckers AL 2007 Immunolocalization of SIBLING and RUNX2 proteins during vertical distrac-ion osteogenesis in the human mandible. J Histochem Cytochem 55:1095–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluhak-Heinrich J, Pavlin D, Yang W, Macdougall M, Harris SE 2007 MEPE expression in osteocytes during orthodontic tooth movement. Arch Oral Biol 52:684–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueb B, Taeschler S, Schild C, Lang NP 2007 Expression of phosphoproteins and amelotin in teeth. Int J Mol Med 19:49–54 [PubMed] [Google Scholar]
- Goobes R, Goobes G, Campbell CT, Stayton PS 2006 Thermodynamics of statherin adsorption onto hydroxyapatite. Biochemistry 45:5576–5586 [DOI] [PubMed] [Google Scholar]
- Hoyer JR, Asplin JR, Otvos L 2001 Phosphorylated osteopontin peptides suppress crystallization by inhibiting the growth of calcium oxalate crystals. Kidney Int 60:77–82 [DOI] [PubMed] [Google Scholar]
- Schlesinger DH, Hay DI 1977 Complete covalent structure of statherin, a tyrosine-rich acidic peptide which inhibits calcium phosphate precipitation from human parotid saliva. J Biol Chem 252:1689–1695 [PubMed] [Google Scholar]
- Ono T, Tanaka H, Yamate T, Nagai Y, Nakamura T, Seino Y 1996 24R,25-Dihydroxyvitamin D3 promotes bone formation without causing excessive resorption in hypophosphatemic mice. Endocrinology 137:2633–2637 [DOI] [PubMed] [Google Scholar]
- Blair HC, Zaidi M 2006 Osteoclastic differentiation and function regulated by old and new pathways. Rev Endocr Metab Disord 7:23–32 [DOI] [PubMed] [Google Scholar]
- Hofbauer LC, Kuhne CA, Viereck V 2004 The OPG/RANKL/RANK system in metabolic bone diseases. J Musculoskelet Neuronal Interact 4:268–275 [PubMed] [Google Scholar]
- Lorenz-Depiereux B, Bastepe M, Benet-Pages A, Amyere M, Wagenstaller J, Muller-Barth U, Badenhoop K, Kaiser SM, Rittmaster RS, Shlossberg AH, Olivares JL, Loris C, Ramos FJ, Glorieux F, Vikkula M, Juppner H, Strom TM 2006 DMP1 mutations in autosomal recessive hypophosphatemia implicate a bone matrix protein in the regulation of phosphate homeostasis. Nat Genet 38:1238–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]