Abstract
Some inroads have been made into characterizing histone variants and posttranslational modifications of histones in Trypanosoma brucei. Histone variant H2BV lysine 129 is homologous to Saccharomyces cerevisiae H2B lysine 123, whose ubiquitination is required for methylation of H3 lysines 4 and 79. We show that T. brucei H2BV K129 is not ubiquitinated, but trimethylation of H3 K4 and K76, homologues of H3 K4 and K79 in yeast, was enriched in nucleosomes containing H2BV. Mutation of H2BV K129 to alanine or arginine did not disrupt H3 K4 or K76 methylation. These data suggest that H3 K4 and K76 methylation in trypanosomes is regulated by a novel mechanism, possibly involving the replacement of H2B with H2BV in the nucleosome.
Keywords: Chromatin structure, Histone methylation, Histone modification, Histone ubiquitination, Histone variant, Mass spectrometry, Nucleosome, Trypanosoma brucei
Introduction
Histones form the building blocks of the nucleosome, the fundamental unit of chromatin. Posttranslational modifications (PTMs) of histones, including acetylation, methylation, phosphorylation, and ubiquitination, have been widely studied for their role in gene expression, DNA replication and repair, and nucleosome assembly [1]. Causal relationships between the various histone modifications led to the ‘trans-histone’ cross-talk hypothesis, which states that PTMs on one histone may prohibit or be required for PTMs on the same or another histone [2]. In one famous instance of cross-talk, ubiquitination of histone H2B lysine 123 in S. cerevisiae was shown to be required for methylation of H3 lysine 4 and 79 [3, 4]. Furthermore, these modifications are linked to transcription elongation, as it has been shown that the elongating RNA polymerase II and its binding partners form a platform for the recruitment of H3 K4 and K79 methyltransferases Set1 and Dot1, respectively, following H2B ubiquitination (reviewed in [5]).
Replacement of canonical histones by histone variants is another mechanism for expanding the functional diversity of the nucleosome, affecting gene expression, DNA repair, and centromere assembly (reviewed in [6]). Variants may be incorporated into the nucleosome outside of S phase by ATP-dependent nucleosome remodeling enzymes [7, 8].
Many PTMs of histones in T. brucei, the causative agent of human African Sleeping Sickness, have been identified [9, 10]. Histone modifications have been implicated in some biological processes in trypanosomes, including cell-cycle regulation, life-cycle progression, and, possibly, antigenic variation [11–13]. Novel variants of the four major histones have also been identified in trypanosomes [14, 15]. Sequence comparison of T. brucei H2B and H2BV with histone H2B from other organisms suggests that a homologue of yeast H2B K123 is absent from the trypanosome H2B but can be clearly identified in H2BV. Although we found that H2BV K129 is not ubiquitinated, this variant specifically associates with H3 that is trimethylated at K4 and K76. The data suggest that trypanosomes have a novel mechanism involving H2BV in H3 K4 and K76 methylation.
Materials and methods
Cell lines
Cell lines were derived from the ‘single marker’ (sm) clone of the T. brucei Lister 427 bloodstream forms that constitutively expresses T7 RNA polymerase and the tet repressor, allowing for inducible expression of ectopic genes downstream of a T7 promoter [16]. Plasmid pJEL69, which is used to introduce an inducible ectopic copy of H2BV-FLAG, was made by PCR amplifying the H2BV coding sequence, adding the FLAG epitope (DYKDDDDK) at the N-terminus (J. Lowell, unpublished data). pJEL69 was linearized with NotI and transfected in sm cells. The endogenous copies of H2BV were then replaced by puromycin and hygromycin drug resistance markers, using plasmids pJEL74 and pJEL75 respectively, to create the cell line BFJEL18 [15]. Mutations in H2BV-FLAG were generated in pJEL69, using the QuikChange™ site-directed mutagenesis kit (Stratagene). Lysine 129 (AAA) was mutated to alanine (GCA) or arginine (AGA). The H2BVΔCTD mutation was made by PCR amplifying H2BV 1–127 with a premature stop codon from pJEL69 and using it to replace the wildtype copy of H2BV in the same plasmid. FLAG-tagged H2BV mutants were used to replace endogenous H2BV alleles in sm cells as described above. A cell line (BFJEL8) containing an inducible ectopic copy of H2B-FLAG derived from single marker was also used [15].
Purification and analysis of histone H2BV
Histones were purified from BFJEL18 as previously [9], up to the stage prior to HPLC separation, then incubated with anti-FLAG M2 affinity gel (Sigma) with rotation for 2 hr at 4°C. Solutions were treated with 10 mM iodoacetamide. Bound material was washed three times with Tris Buffered Saline (TBS) and eluted by boiling the gel in 2x SDS loading buffer for 3 min. The eluate was analyzed by SDS-PAGE and Coomassie staining. Gel slices were digested with trypsin and analyzed by Liquid Chromatography/Electrospray tandem mass spectrometry (LC/ESI-MS/MS) using a QSTAR XL Quadropole-quadropole time-of-flight (QqTOF) mass spectrometer (Applied Biosystems) as described in [9].
Anti-H3K4me3 antibody
Polyclonal antibodies were raised by immunizing rabbits (Sigma) with KLH-conjugated peptide RTKme3ETARTC (H3 2–9). The antibody was purified by depleting cross-reacting components to peptide VSGAQKme3EGLRFC (H3 71–81), coupled to Sulfolink gel (Pierce), followed by affinity purification on H3 2–9, coupled to Sulfolink, as described [11]. The following peptides were used to verify antibody specificity: RTKETARTC (H3K4me0), RTKme3ETARTC (H3K4me3), VSGAQKEGLRFC (H3K76me0), and VSGAQKme3EGLRFC (H3K76me3).
Preparation of mononucleosomes and co-immunoprecipitation
Mononucleosomes were prepared as described [15] from cell lines encoding the following inducible fusion proteins: H2B-FLAG, H2BV-FLAG, H2BV K129A-FLAG, and H2BV K129R-FLAG. For co-immunoprecipitation, the mononucleosome preparation was incubated with anti-FLAG M2 affinity gel (Sigma) with rotation for 2 hr at 4°C. Bound material was washed once with isotonic buffer (100 mM KCl; 10 mM Tris, pH 8.0; 10 mM CaCl2; 5% glycerol) with 200 mM NaCl for 10 min with rotation. Bound material was then washed twice with isotonic buffer for 10 min with rotation then eluted by boiling the gel in 2 x SDS loading buffer for 3 min.
Results
Sequence comparison of T. brucei histones H2B and H2BV
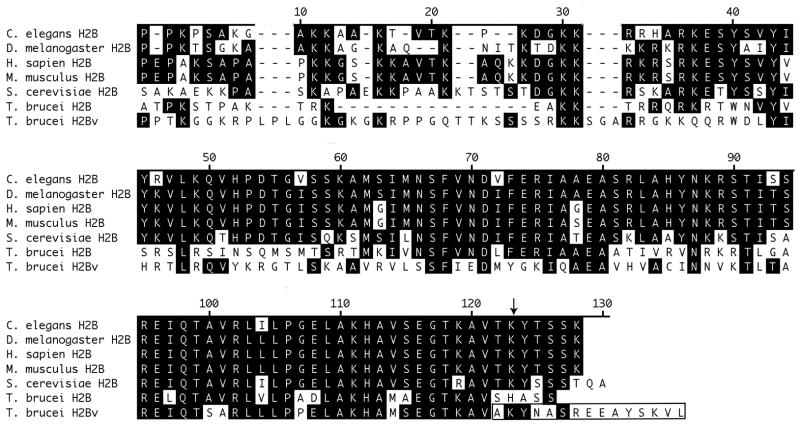
T. brucei histone variant H2BV shares 38% sequence identity with H2B [15]. The region of greatest similarity is the C-terminus, where both histones also have significant identity with H2B from other organisms (Fig. 1). The C-terminal tails of H2B and H2BV are different, however. H2BV is more similar to the H2B consensus sequence from other organisms, including the presence of lysine at position 129 that is homologous to lysine 123 in S. cerevisiae, which is well known as a site of ubiquitination that is required for methylation of H3 K4 and K79 [3, 4]. We therefore investigated whether T. brucei H2BV might be ubiquitinated (H2B was not), and if the presence of H2BV in the nucleosome might affect H3 K4 and K76 methylation.
Fig. 1.
Sequence alignment of T. brucei histones H2B and H2BV with H2B from other organisms. Identical residues are shaded. The sequence ruler is numbered according to the S. cerevisiae sequence. An arrow points to T. brucei H2BV lysine 129, which is homologous to the ubiquitinated lysine 123 from S. cerevisiae. H2BV residues 128–142 are boxed.
Purification of H2BV and identification of PTMs
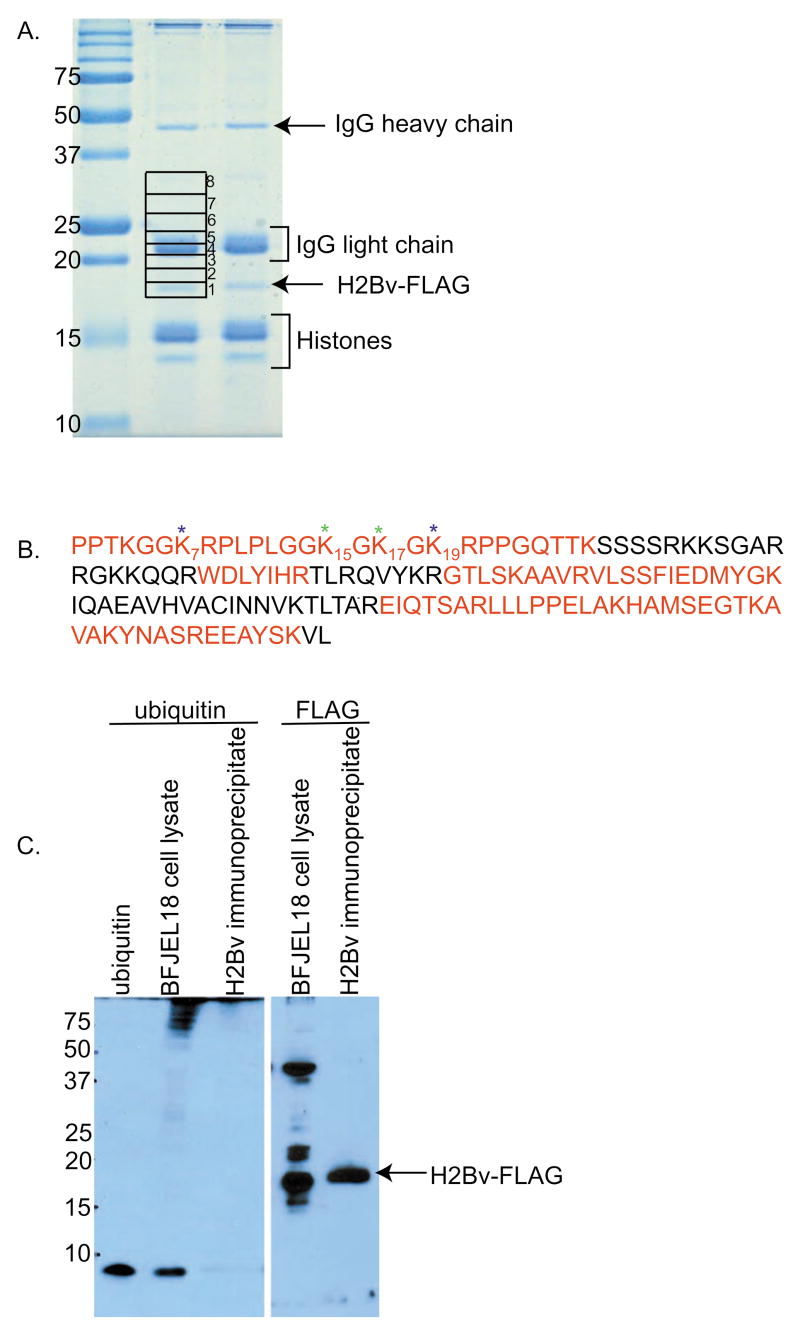
To investigate whether H2BV K129 is ubiquitinated, FLAG-tagged H2BV was affinity purified from 1 × 1010 T. brucei. After SDS-PAGE of the purified material (Fig. 2A), several bands were revealed, all of which were identified by MS, including H2BV-FLAG (16.7 kDa), the heavy (50 kDa) and light (25 kDa) chains of the immobilized M2 antibodies that dissociated during the elution process, and histones (11–15 kDa) that co-immunoprecipitated with H2BV-FLAG. In order to look for ubiquitinated or otherwise modified H2BV-FLAG, which would migrate more slowly than the unmodified H2BV-FLAG, the region of the gel between ~17 and 35 kDa was cut into 8 gel slices, trypsinized, and analyzed by tandem MS. Gel slices 1–7 contained H2BV-FLAG tryptic peptides and provided adequate sequence coverage by MS (Fig. 2B). Several PTMs were identified at the H2BV N-terminus, including acetylation of lysines 7 and 19 and methylation of lysines 15 and 17 (data not shown). Ubiquitination of lysine residues can be identified by MS by searching for a GG dipeptide linkage (114 Da) to lysine, which results from tryptic cleavage of ubiquitin. Although a targeted search for the ubiquitinated H2BV peptide 126–134 (AVAKYNASR) containing lysine 129 was performed within the data from all 8 gel slices, no evidence of H2BV K129 ubiquitination was found. A broader search revealed no evidence for ubiquitination elsewhere on H2BV.
Fig. 2.
Histone H2BV is not ubiquitinated. (A) H2BV-FLAG was purified from BFJEL18 cells. The Coomassie-stained bands representing H2BV-FLAG, light and heavy chains of the α-FLAG M2 antibody, and histones are indicated. Eight gel slices were excised from the gel, trypsinized, and analyzed by MS. (B) Histone H2BV sequence, with regions for which tandem MS data were acquired highlighted in red. Acetylated and methylated lysines are indicated with a blue or green asterisk, respectively. (C) H2BV-FLAG was purified from 2 × 108 BFJEL18 cells and analyzed with ubiquitin antibody (left panel). 100 ng ubiquitin (8.4 kDa, Sigma) and cell lysate from 2 × 106 BFJEL18 cells are included to verify the activity of the antibody. BFJEL18 cell lysate and purified H2Bv-FLAG from 2 × 107 cells were reacted with the FLAG antibody (Sigma), to confirm that the purification was successful (right panel).
To confirm that H2BV-FLAG was not ubiquitinated, purified material was analyzed on a western blot using antibody to ubiquitin (Fig. 2C). The blot shows no evidence of ubiquitinated proteins in the purified H2BV-FLAG. A western blot of a cell lysate from BFJEL18 demonstrates that the antibody recognizes T. brucei ubiquitin.
H3 K4 and K76 methylation is enriched in H2BV-containing mononucleosomes
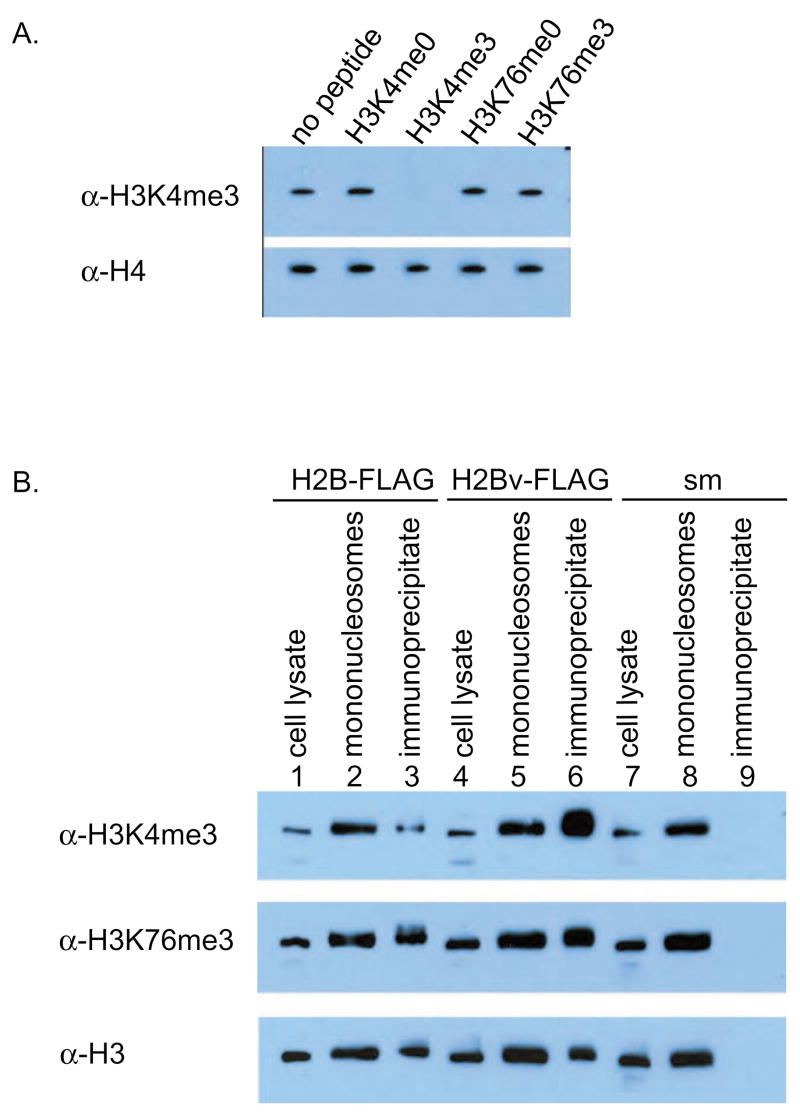
The sequence similarity between yeast H2B and trypanosome H2BV was intriguing, despite the absence of H2BV ubiquitination. To address the possibility of ubiquitin-independent H3 methylation, the effects of H2BV incorporation into a nucleosome on H3 K4 and K76 methylation were assessed. A specific antibody was raised against H3K4me3. The specificity of the antibody was confirmed by peptide competition (Fig. 3A). H3K76 methylation was evaluated using an H3K76me3-specific antibody, which was characterized previously [11].
Fig. 3.
Histone H2BV co-immunoprecipitates with histone H3 that is trimethylated at lysines 4 and 76. (A) A specific antibody was raised against H3K4me3. Pre-incubation of the antibody without peptide or 10 ng/ml H3K4me0, H3K4me3, H3K76me0, or H3K76me3 peptides shows that the antibody only binds the H3K4me3 peptide. An antibody to H4 (Sigma) was used as a loading control. (B) Mononucleosomes containing either H2B-FLAG or H2BV-FLAG were immunoprecipitated with FLAG antibody affinity gel and analyzed by Western blot with H3K4me3 and H3K76me3 antibodies. An antibody to H3 (Abcam ab1791) was used as a loading control.
To investigate whether H2BV is predominantly incorporated into nucleosomes that are methylated at H3 K4 and K76, we prepared mononucleosomes from cell lines containing FLAG-tagged H2B or H2BV. Mononucleosomes were immunoprecipitated with FLAG affinity resin, and H3 K4 and K76 methylation levels in H2B- and H2BV-containing mononucleosomes were analyzed using specific antibodies (Fig. 3B). H2BV-containing mononucleosomes were highly enriched for H3 K4 methylation (lane 6) compared to H2B-containing mononucleosomes (lane 3). H2BV-containing mononucleosomes were also slightly, but very reproducibly, enriched for H3 K76 methylation (lane 6).
Mutation of H2BV K129 does not affect H3 K4 or K76 methylation levels
To investigate whether H2BV K129, the yeast H2B K123 homologue, played a role in the enrichment of H3 K4 and K76 methylation in H2BV-containing mononucleosomes, K129 was replaced by alanine (K129A) or arginine (K129R). Lysates from cell lines in which two endogenous h2bv alleles were replaced with FLAG-tagged H2BV K129A or K129R were prepared and analyzed by western blot using H3K4me3- and H3K76me3-specific antibodies (Fig. 4A). Mutation of H2BV K129 appeared to have no effect on the level of H3 K4 and K76 methylation, compared to wild-type H2BV. Next, we prepared mononucleosomes containing FLAG-tagged H2BV K129A and K129R and immunoprecipitated them with FLAG affinity gel. Mutation of H2BV K129 did not prevent enrichment of H3 K4 and K76 methylation in H2BV-containing nucleosomes (Fig. 4B). These results show that H2BV K129 is neither a prerequisite for H3 K4 or K76 methylation nor is it involved in incorporation of H2BV into nucleosomes that are enriched in H3 K4 or K76 methylation.
Fig. 4.
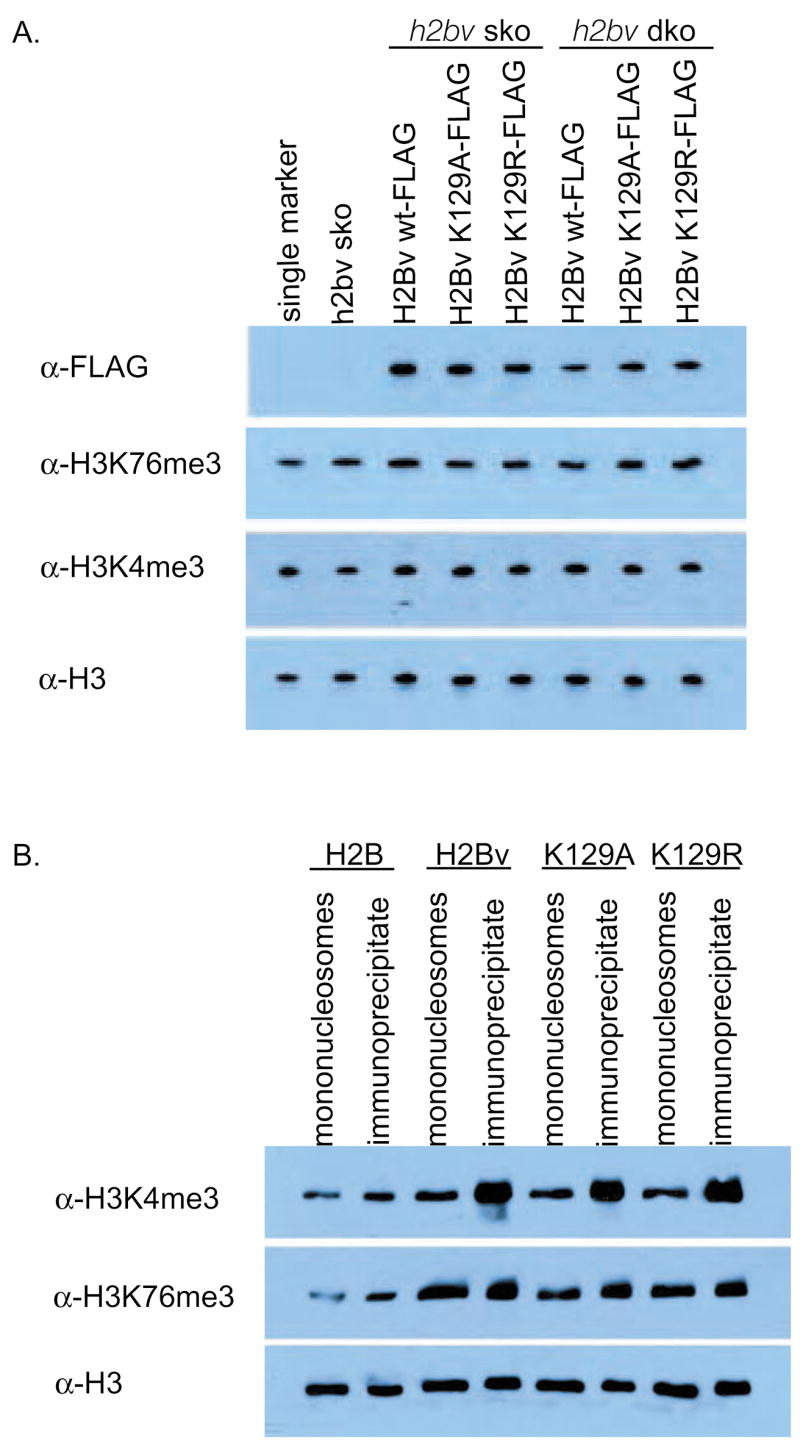
Mutation of H2BV lysine 129 does not interfere with H3 lysine 4 or 76 methylation. (A) Lysate from cells expressing ectopic copies of FLAG-tagged H2BV, H2BV K129A, or H2BV K129R in H2BV+/− heterozygous or homozygous deletion backgrounds. Lysates were analyzed by western blot with antibodies to FLAG, H3K4me3, H3K76me3, and H3. (B) Co-immunoprecipitation with mononucleosomes containing the H2BV K129 mutation.
To determine whether the C-terminal tail of H2BV affected the relationship between H2BV and H3 methylation, a FLAG-tagged H2BV mutant was made in which the C-terminal amino acids 128–142 were removed (H2BVΔCTD). When the FLAG-tagged H2BVΔCTD mutant replaced one allele of endogenous H2BV, however, the FLAG fusion protein was not detected (data not shown). The second allele of H2BV could not be replaced, indicating that the C-terminal tail of H2BV is essential.
Discussion
The sequence homology of T. brucei H2BV with the H2B consensus sequence led to our initial hypothesis that lysine 129 was ubiquitinated and required for H3 K4 and K76 methylation. Ubiquitinated histones are usually present in low abundance, so they may be overlooked when performing a survey of histone PTMs by MS. We performed a targeted search for ubiquitinated H2BV K129 by MS, as well as western blot analysis with an anti-ubiquitin antibody, which enables us to conclude that H2BV K129 is not ubiquitinated. This is supported by the observation that mutation of K129 does not disrupt H3 K4 or K76 methylation. These experiments show that H2BV K129 does not play a role homologous to yeast H2B K123, despite the sequence similarities.
Although our original hypothesis proved to be incorrect, we were able to show, using specific antibodies, that H2BV is present in mononucleosomes that are enriched for H3 K4 and K76 methylation. H3 K4 and K76 methylation are typically associated with transcriptionally active chromatin [1, 17], although this relationship has not been demonstrated in trypanosomes. Based on our results, we hypothesize that H2BV replaces H2B in nucleosomes, which permits H3 K4 and K76 to be methylated. This may be a mechanism for relieving transcriptional repression and would be in keeping with the role of histone variants in other organisms. In Drosophila, for example, histone variant H3.3 replaces major H3 in a replication-independent manner and is deposited at sites of active transcription [8]. This provides a possible mechanism for relieving transcriptional repression by modified canonical histones.
Previous work on T. brucei H2BV and H2AZ showed that the two variants co-immunoprecipitate in the same nucleosome, so either or both may be involved in promoting H3 methylation [15]. H2AZ does not co-immunoprecipitate with canonical H2A or H2B, suggesting that nucleosomes containing H2BV consist of two H2AZ/H2BV dimers. The same study showed that H2AZ and H2BV did not co-localize with sites of active transcription that were visualized by BrUTP incorporation. However, the authors suggested that H2AZ/H2BV-containing nucleosomes are localized to the promoter regions of active genes and are displaced during transcription by RNA polymerase II, so the variants may not be readily detectable in actively transcribed chromatin. This phenomenon may be analogous to what is observed in yeast, where H2A.Z is poised at inactive promoters, and, upon activation, H2A.Z is evicted from the nucleosome, enabling active transcription complexes to be assembled at the promoter [18, 19].
H2BV is an essential gene, but deletion of the first 23 amino acids did not affect viability, indicating that the N-terminus is not essential [15]. A FLAG-tagged H2BV mutant in which C-terminal residues 128–142 were removed was not expressed in an H2BV+/− heterozygous background and the second allele could not be deleted from this cell line, which illustrates that residues 128–142 are essential. It is difficult to draw any conclusions concerning the role of a mutation when the phenotype is lethality, but we hypothesize that residues 128–142 are involved in the enrichment of H3 methylation in H2BV-containing nucleosomes based on the conservation of some of these residues with the H2B C-terminus in other organisms.
In conclusion, we have identified a potentially novel mechanism for H3 K4 and K76 methylation in trypanosomes that utilizes the incorporation of H2BV into nucleosomes rather than lysine ubiquitination. Our analysis of H2BV shows that no lysines are ubiquitinated. We have not determined if H2AZ is ubiquitinated, so it is possible that ubiquitination of one of the H2AZ lysines acts as a ‘master switch’ in the pathway leading to H3 K4 and K76 methylation. Our efforts to study chromatin in trypanosomes illustrate that, while some histone sequences and PTMs are conserved, the functional relationships between marks may not be. Work in divergent organisms will help to define some of the general principles underlying chromatin biology.
Acknowledgments
We thank Joanna Lowell for constructs and cell lines and the Proteomics Resource Center at The Rockefeller University for their contributions to this study, which was supported by the National Institutes of Health (Grants AI021729 and GM07739).
Abbreviations
- MALDI-TOF
matrix-assisted laser-desorption/ionization time of flight
- MS
mass spectrometry
- PTM
posttranslational modification
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–83. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 3.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 4.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–8. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 5.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–69. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein E, Hake SB. The nucleosome: a little variation goes a long way. Biochem Cell Biol. 2006;84:505–517. doi: 10.1139/o06-085. [DOI] [PubMed] [Google Scholar]
- 7.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–8. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 8.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 9.Mandava V, Fernandez JP, Deng H, Janzen CJ, Hake SB, Cross GAM. Histone modifications in Trypanosoma brucei. Mol Biochem Parasitol. 2007;156:41–50. doi: 10.1016/j.molbiopara.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janzen CJ, Fernandez JP, Deng H, Diaz R, Hake SB, Cross GAM. Unusual histone modifications in Trypanosoma brucei. FEBS Lett. 2006;580:2306–10. doi: 10.1016/j.febslet.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 11.Janzen CJ, Hake SB, Lowell JE, Cross GAM. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol Cell. 2006;23:497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Horn D, Cross GAM. A developmentally regulated position effect at a telomeric locus in Trypanosoma brucei. Cell. 1995;83:555–561. doi: 10.1016/0092-8674(95)90095-0. [DOI] [PubMed] [Google Scholar]
- 13.Horn D, Cross GAM. Position-dependent and promoter-specific regulation of gene expression in Trypanosoma brucei. EMBO J. 1997;16:7422–7431. doi: 10.1093/emboj/16.24.7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowell JE, Cross GAM. A variant histone H3 is enriched at telomeres in Trypanosoma brucei. J Cell Sci. 2004;117:5937–47. doi: 10.1242/jcs.01515. [DOI] [PubMed] [Google Scholar]
- 15.Lowell JE, Kaiser F, Janzen CJ, Cross GAM. Histone H2AZ dimerizes with a novel variant H2B and is enriched at repetitive DNA in Trypanosoma brucei. J Cell Sci. 2005;118:5721–30. doi: 10.1242/jcs.02688. [DOI] [PubMed] [Google Scholar]
- 16.Wirtz E, Leal S, Ochatt C, Cross GAM. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 17.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Zanton SJ, Pugh BF. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 2006;20:2250–65. doi: 10.1101/gad.1437506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–31. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]