Abstract
Array based comparative genomic hybridisation (aCGH) is a powerful technique for detecting clinically relevant genome imbalance and can offer 40 to > 1000 times the resolution of karyotyping. Indeed, idiopathic learning disability (ILD) studies suggest that a genome-wide aCGH approach makes 10–15% more diagnoses involving genome imbalance than karyotyping. Despite this, aCGH has yet to be implemented as a routine NHS service. One significant obstacle is the perception that the technology is prohibitively expensive for most standard NHS clinical cytogenetics laboratories. To address this, we investigated the cost-effectiveness of aCGH versus standard cytogenetic analysis for diagnosing idiopathic learning disability (ILD) in the NHS. Cost data from four participating genetics centres were collected and analysed. In a single test comparison, the average cost of aCGH was £442 and the average cost of karyotyping was £117 with array costs contributing most to the cost difference. This difference was not a key barrier when the context of follow up diagnostic tests was considered. Indeed, in a hypothetical cohort of 100 ILD children, aCGH was found to cost less per diagnosis (£3,118) than a karyotyping and multi-telomere FISH approach (£4,957). We conclude that testing for genomic imbalances in ILD using microarray technology is likely to be cost-effective because long-term savings can be made regardless of a positive (diagnosis) or negative result. Earlier diagnoses save costs of additional diagnostic tests. Negative results are cost-effective in minimising follow-up test choice. The use of aCGH in routine clinical practice warrants serious consideration by healthcare providers.
Keywords: Microarrays, Comparative genomic hybridisation, Cost-effectiveness, Learning disability
Introduction
Learning disability (LD) is a common condition affecting 1–3% of individuals worldwide (Roeleveld et al. 1997). Most with moderate to severe LD (intelligence quotient (IQ) under 50) require life long support and half of those with mild LD (IQ 50–70) are significantly impaired throughout life (Department of Health 2001; Mencap 2001). Despite the clinical, social and psychological challenges associated with LD, up to 80% of cases have no specific causal diagnosis.
Standard testing to detect constitutional anomalies (present at or before birth) is chromosome analysis (karyotyping) at the 450–500 G-band level. Karyotyping can detect large genomic imbalances (losses or gains of DNA) in LD conditions such as Down, Turner and Edwards Syndromes. However, the resolution is insufficient to routinely detect rearrangements smaller than 5 million base pairs (5 Mb) and even abnormalities of 15 Mb may be missed where the banding pattern is indistinct.
As smaller genomic imbalances can be clinically important, demand has increased for higher resolution assays to detect them. This is particularly true for idiopathic (without known cause) LD (ILD) cases, that represent ∼15% of referrals to clinical genetics and paediatrics clinics. Despite ILD being incurable, a diagnosis is important for many reasons including, providing accurate prognostic information and genetic counselling, directing appropriate clinical care and educational needs, considering future preventative and therapeutic regimes and finally helping clinicians to answer the parents’ question “why?”. The clarification of genetic risk for both the immediate and wider family is particularly important because it enables meaningful reproductive choice. For example, a negative result can substantially reduce risk whereas a positive result can open an avenue for prenatal diagnosis (in appropriate cases).
A major advance in diagnosing ILD through genetics was the discovery that cytogenetically invisible genome imbalances involving chromosome tips (telomeres) account for many ILD cases (Flint et al. 1995). Subsequently, a test assaying every telomere of an individual by fluorescence ‘in situ’ hybridisation to chromosomes (‘multi-telomere FISH’) was developed and widely adopted in diagnostic laboratories (Knight et al. 1997). Further technological advances led to a new approach, array comparative genome hybridisation (aCGH), that identifies cryptic genome imbalances at the genome-wide level (Knight and Regan 2006).
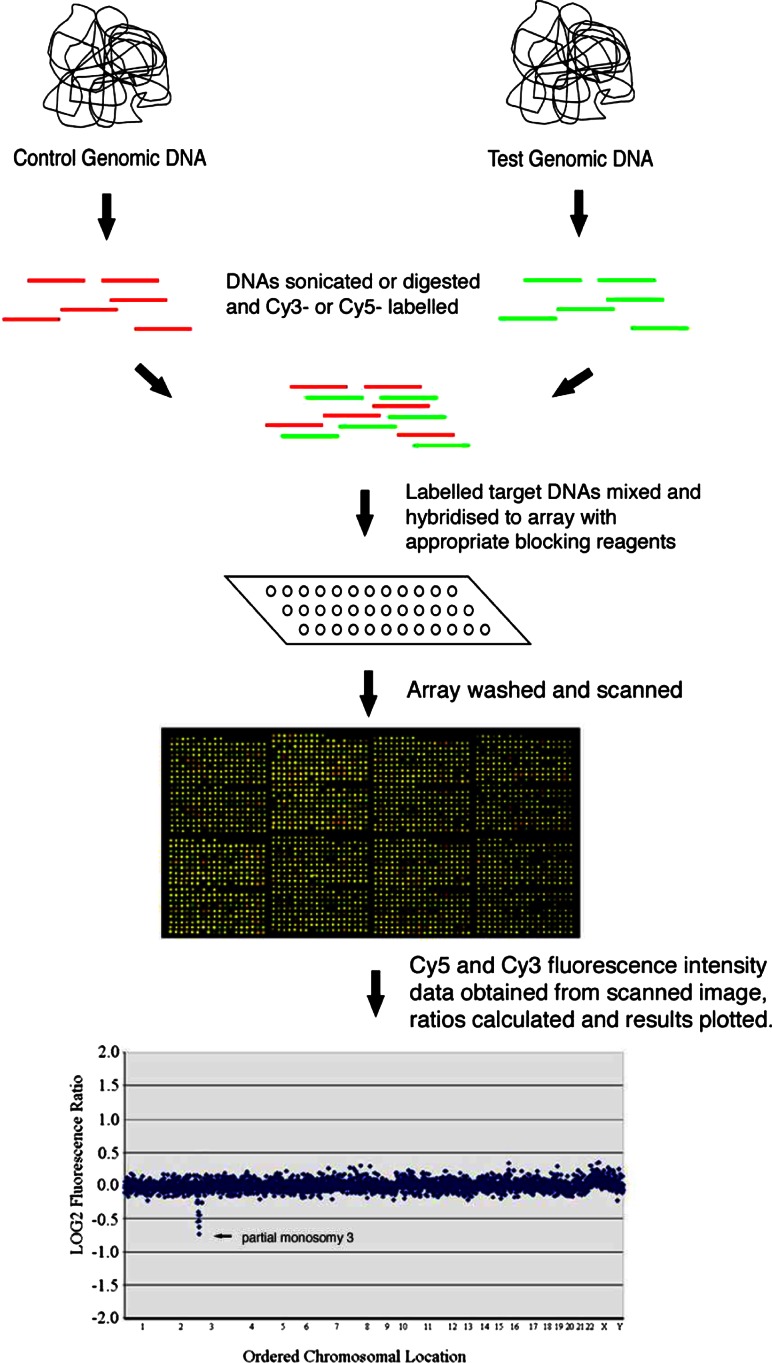
Microarrays have received considerable attention in the scientific research community. An array (microarray or chip) is a solid surface, often a microscope slide, onto which control DNA, cDNA (complementary DNA) or short single stranded sequences (oligonucleotides) are spotted (Aitman 2001). In aCGH, an array is used to compare a control versus a test genome searching for differences in the test genome (Fig. 1). When the test genome is a patient DNA sample, such differences signpost DNA sequences that might be implicated in the patient’s phenotype. aCGH has application in many genetic conditions, proving particularly useful in diagnosing ILD. Indeed, research indicates that at least 10–15% more diagnoses are made compared with standard cytogenetic analysis (Knight et al. 2006).
Fig. 1.
Overview of aCGH protocol (reproduced from Knight and Regan (2006) with permission from S. Karger and AG. Basel)
Despite this, aCGH is not implemented widely in the NHS. One obstacle is the lack of consensus regarding ‘platform choice’, that is, the best combination of array type, experimental methodology and analysis system. Another obstacle is concern over the proportion of confirmed genome imbalances where the significance of the positive result is unknown e.g. very small ‘de novo’ imbalances and some inherited imbalances. However, the most significant obstacle to date is the perception that the technology is prohibitively expensive for most NHS clinical cytogenetics laboratories. Local commissioners are unable to endorse implementation without considering the clinical utility and economic implications of technology adoption. Whilst the clinical and scientific utility of aCGH in ILD is impressive, information on its economic viability in routine clinical practice is lacking. Therefore, our study aimed to estimate the cost-effectiveness of aCGH compared with standard cytogenetic analysis in ILD.
Methods
Cost-effectiveness analysis
The costs and effects (number of additional diagnoses) of an aCGH test versus standard cytogenetic analysis using karyotyping, were compared. A cost per diagnosis detected was used rather than a cost per life year gained or quality adjusted life year (QALY), as testing is unlikely to save lives and evaluating QALY’s is problematic in children, especially those with LD.
An NHS perspective was adopted and to make the results generally applicable to UK laboratories, four laboratories currently investigating ILD using aCGH, karyotyping or both contributed to data collection: (i) The Wellcome Trust Centre for Human Genetics, University of Oxford (arrays), (ii) Oxford Regional Cytogenetics Laboratory (karyotyping), (iii) Birmingham Regional Genetics Laboratory (arrays and karyotyping) and (iv) South East Scotland Cytogenetics Laboratory, Edinburgh (arrays). These were selected because they employ slightly different testing procedures (e.g. different staff grades or level of automation).
Testing pathways and resource use
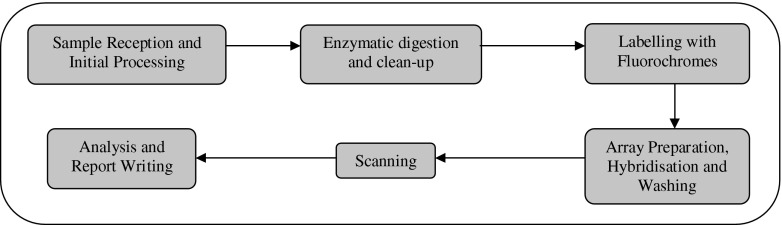
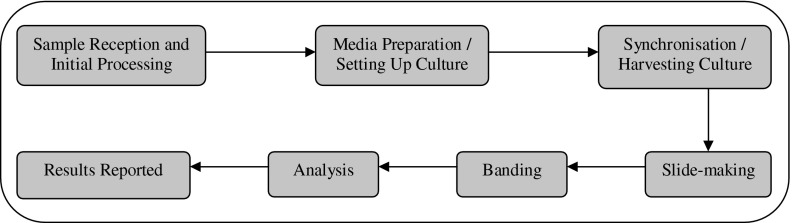
This information was obtained through laboratories completing cost questionnaires (available from authors). Detailed information, from a blood sample arriving at the laboratory through to result reporting was collected (Figs. 2, 3).
Fig. 2.
Array-CGH testing process
Fig. 3.
Karyotyping testing process
Resource information on staff times, consumables and capital was derived from the questionnaires. Salary costs were attached to these based on NHS Agenda for Change figures (Department of Health 2005) and unit costs were attached to equipment and resource information from laboratory price lists including 17.5% VAT, with maintenance and service costs being included under the equipment warranty. For capital items (e.g. array scanners), the cost was spread over the items predicted lifetime and depreciated using equivalent annual costing, discounted at 3.5% (Drummond et al. 2005; HM Treasury 2006). Overheads, including electricity were calculated as a percentage of total costs (around 20%).
The costs of routine cytogenetics analysis include karyotype analysis (see http://www.oup.co.uk/pdf/pas/12–7–1.pdf for standard protocol). Array costs were based on Agilent Technologies Inc. 4 × 44 K genome-wide oligonucleotide multi-sample format arrays, with four different patient DNAs per slide (see www.chem.agilent.com/temp/radAAF6F/00060479.pdf for protocols).
Testing scenarios
Most cases require additional tests to establish the clinical relevance of a putative genomic imbalance identified by an initial aCGH test. This is because a positive aCGH result may be due to several reasons including:
-
(i)
Imbalance is real and clinically relevant; without a positive family history, this would generally be expected to be ‘de novo’ (absent in clinically normal parents) and may or may not have been reported before in similarly affected individuals. However, the imbalance may also be inherited from a clinically normal parent, the phenotype due to a recessive condition, incomplete penetrance or genomic imprinting, for example.
-
(ii)
Imbalance is real, but not clinically relevant; it may be a benign polymorphism inherited from a clinically normal parent or a ‘de novo’ benign variant that may or may not have been reported before.
-
(iii)
Imbalance is not real; it is a false positive that a different test fails to confirm.
By contrast, additional tests undertaken after karyotyping are most often to find a diagnosis, rather than to understand the clinical relevance of an abnormality.
Several testing and reporting scenarios were identified. For arrays, additional tests such as testing parents using arrays or FISH were included. For karyotyping, testing parents and using feasible follow-up tests of multi-telomere FISH and multi-telomere MLPA, were costed. Expert opinion (Laboratory Directors) and laboratory records developed these scenarios. Average test throughput was determined by annual laboratory figures, equipment and staff availability. Additional targeted tests e.g. those for specific gene mutations and biochemical tests were not costed, as they apply to both karyotyping and aCGH approaches when negative.
Sensitivity analysis
This explored the impact that changing individual costs has on total costs. The costs varied included: arrays and scanner, percentage used to calculate overheads, array labelling, different staff grades, karyotyping probe costs and test throughput. Ranges were based on expert opinion. Data analyses were conducted in Microsoft Excel 2003 and costs reported in Pounds sterling (£), using 2006 prices. As costs were derived from different laboratories, the results presented are averages of the four laboratories.
To create a cost per diagnosis, cost data were combined with information on the predicted number of diagnoses for 100 hypothetical ILD cases referred via genetics clinics for genomic imbalance testing. Costs were assigned to the karyotyping route (factoring in one additional genome imbalance test, a telomere assay, for karyotypically normal samples) and for aCGH (where few, if any, additional genome imbalance tests are required). The number of diagnoses expected and the testing scenarios were derived from clinical diagnostic laboratory records (karyotyping), research experience (testing scenarios and 44 K aCGH results to date) and published data (karyotyping, subtelomeric studies and aCGH ILD studies) (de Vries et al. 2005; Knight 2005; Knight and Regan 2006; Menten et al. 2006; Miyake et al. 2006; Rauch et al. 2006; Ravnan et al. 2006; Rosenberg et al. 2005; Schoumans et al. 2005; Shaw-Smith et al. 2004; Tyson et al. 2005; Vissers et al. 2003).
Results
Table 1 presents staff time and costs for a single aCGH and karyotype test. For aCGH, the average staff time per sample is 142 min, at an average sample cost of £42 (range £36–£48). For karyotyping, the average staff time per sample is 210 min, at an average sample cost of £85 (range £73–£96).
Table 1.
Staff costs for aCGH and Karyotyping
| Medical technical officer | Clinical scientist | Consultant grade scientist | Secretarial staff | Total time | |
|---|---|---|---|---|---|
| Array CGH | |||||
| Cost per hour range (£.p)a | 13.01–16.83 | 18.34–24.71 | 40.17–49.73 | N/A | N/A |
| Median cost per hour (£.p) | 14.69 | 21.65 | 44.09 | N/A | N/A |
| Hands-on time (minutes) | 61.00 | 76.00 | 5.00 | N/A | 142.00 |
| Cost per sample range (£.p) | 9.67–12.51 | 23.45–31.44 | 3.35–4.14 | N/A | 36.47–48.09 |
| Cost per sample (£.p)b | 10.92 | 27.55 | 3.67 | N/A | 42.14 |
| Karyotyping | |||||
| Cost per hour range (£.p)a | 15.46–19.35 | 23.00–24.56 | 43.78–44.40 | 12.98 | N/A |
| Hands-on time range (minutes) | 40 –113 | 5–192 | 10–45 | 0–15 | 178–242c |
| Hands-on time mid-point (minutes) | 76.50 | 98.50 | 27.50 | 7.50 | 210.00 |
| Cost per sample range (£.p) | 10.31–34.51 | 1.92–78.71 | 7.05–33.30 | 0.00–3.24 | 73–96c |
| Median Cost per sample (£.p)b | 22.41 | 40.32 | 20.18 | 1.62 | 84.53 |
a Includes superannuation and national insurance
b Cost per sample does not always equal cost per hour multiplied by time spent on one test due to batching
c The time ranges are based on several different labs, hence the ‘minimums’ are not all referring to the same lab, nor are all the ‘maximums’. The lab with the shortest process, for example, used 113 min of MTO time, 5 min of Clinical Scientist time, 45 min of Consultant Grade Scientist time, and 15 min of Secretarial time, making 178 min in total. The cost per sample range is calculated in a similar manner
Table 2 presents the costs for a straightforward aCGH test (no sample quality or quantity issues). The total cost of £442 is the baseline cost excluding reporting or any other investigations. Array slides account for 42% of total test cost. Table 3 shows the costs for each testing stage when a single karyotype is performed. The total cost is £117 (range £103–£131), 73% of which is staffing.
Table 2.
Array CGH cost breakdown
| Stagea | Cost |
|---|---|
| Sample reception and initial processing | £45 |
| Digestion/Reference Sample Processing | £15 |
| Cleaning | £4 |
| Labelling | £78 |
| Arrays, plus preparation and washing b | £188 |
| Scanning | £14 |
| Analysis and report writing | £24 |
| General resources (e.g. PC and printer) | £1 |
| Overheads | £73 |
| Total c | £442 |
a Cost of obtaining blood sample not included
b Cost of array: £500 for four patients, £125 each
c Baseline of 25 tests per week (1,150 per annum)
Table 3.
Karyotyping cost breakdown
| Stagea | Cost | Range |
|---|---|---|
| Sample reception and initial processing | £4.53 | £4.48–£4.58 |
| Media preparation/setting up culture | £2.39 | £1.62–£3.16 |
| Synchronisation/harvesting culture | £3.81 | £2.93–£4.69 |
| Slide-making | £1.93 | £1.05–£2.81 |
| Banding | £3.36 | £2.28–£4.45 |
| Analysis and checking | £47.11 | £39.15–£55.08 |
| Reporting results and authorisation | £29.39 | £27.94–£30.84 |
| Clinical liaison | £1.27 | £0.37–£2.18 |
| General resources (e.g. PC and printer) | £2.00 | £2.00–£2.00 |
| Overheads | £21.29 | £21.28–£21.30 |
| Total b | £117 | £103–£131 |
a Cost of obtaining blood sample not included
b Baseline of 61 tests per week (2,800 per annum)
Karyotyping costs associated with different testing and reporting scenarios
Table 3 presents a breakdown of the typical cost of karyotyping. Table 4 shows the cost differences associated with different karyotype reporting scenarios and the costs of one further test, a test of subtelomeric integrity either by FISH or by MLPA, that may be requested following a negative karyotyping result. Note that the identification of an abnormality with unknown clinical relevance always requires an additional parental testing stage to help delineate pathogenic from non-pathogenic anomalies. The cheapest scenarios, 1 and 2, are where genome imbalances already known to be clinically benign or clinically relevant are found (£117). In cases where an anomaly is ‘de novo’, the cheapest scenario is Scenario 3: here, the genome imbalance is cytogenetically visible, but of unknown clinical relevance and parental samples are karyotyped to determine whether the anomaly is benign or inherited (£351). For cases where the initial karyotyping result is ‘normal’ and an additional assay of telomere integrity reveals a clinically relevant (or clinically benign) genome imbalance, the most expensive testing scenario is Scenario 7 (£724); here a multi-telomere FISH test is employed and follow-up tests of parental samples are performed using targeted FISH. In the directly comparable scenario (Scenario 6) where multi-telomere MLPA is used for the patient and parental testing and targeted FISH for patient confirmation, the cost is £472. The most common outcome of karyotyping plus an additional multi-telomere assay testing is a negative result i.e. no genome imbalance is found. In this case, Scenario 4 (multi-telomere MLPA), is significantly cheaper than Scenario 5 (Multi-telomere FISH), with tests costs of £162 and £400 respectively.
Table 4.
Karyotyping––Cost of reporting different scenarios
| Scenarioa | Cost of Karyotypingb | Cost of Multi-telomere MLPAb | Cost of Multi-telomere FISHb | Cost of Targeted telomere FISHb | Total costa |
|---|---|---|---|---|---|
| 1 No genome imbalance found. | £117 (£103–£131) | – | – | – | £117 (£103–£131) |
| 2 Genome imbalance found of known clinical relevance. | £117 (£103–£131) | – | – | – | £117 (£103–£131) |
| 3 Genome imbalance found of unknown clinical relevance. Parental samples karyotyped. Imbalance confirmed as inherited or ‘de novo’. | £351 (£309– £393) (3 tests) | – | – | – | £351 (£309–£393) (3 tests) |
| 4 No genome imbalance found. Multi-telomere MLPA test performed. No imbalance found. | £117 (£103–£131) | £45 (£40–£50) | – | – | £162 (£143–£181) |
| 5 No genome imbalance found. Multi-telomere FISH test performed. No imbalance found. | £117 (£103–£131) | – | £283 (£254–£313) | – | £400 (£357–£444) |
| 6 No genome imbalance found. Multi-telomere MLPA test performed. Genome imbalance found. Parental samples tested by Multi-telomere MLPA. Imbalance identified as clinically benign (normal parent carries identical anomaly) or imbalance confirmed ‘de novo’ by FISH and likeky to be clinically relevant. | £117 (£103–£131) | £45 (£40–£50) | – | £220 (£200–£240) (max. 2 tests) | £472 (£423–£521) |
| 7 No genome imbalance found. Multi-telomere FISH test performed. Genome imbalance found. Parental samples tested by Targeted telomere FISH. Imbalance identified as clinically benign (normal parent carries identical anomaly) or clinically relevant. | £117 (£103–£131) | – | £283 (£254–£313) | £324 c (£304–£344) (max. 4 tests) | £724 (£661–£788) |
a Scenarios 1–3: stand-alone karyotyping. Scenarios 4–7: karyotyping plus multi-telomere testing approach (FISH or MLPA)
b Cost ranges given in parentheses
The cost of a single telomere test (£110) is significantly more than testing the same telomere again (£52) due to the initial cost of growing glycerols and preparing the probe ‘in house’.
aCGH costs associated with different testing and reporting scenarios
When an initial aCGH test reveals a putative genomic imbalance, additional tests are needed to help establish the clinical relevance. Table 5 shows a number of possible testing scenarios and associated costs (the baseline cost of £442 derived in Table 2 differs from that given in the Table 5 scenarios by £4–£16 due to different staff reporting times). The cheapest scenarios, 1 and 2, are where genome imbalances already known to be clinically benign or clinically relevant are found (£446). In cases where an anomaly is ‘de novo’ and therefore more likely to be clinically relevant, the cheapest scenario is Scenario 3: here targeted FISH tests on the patient and parental samples are performed (£672). In the case of ‘de novo’ duplications that cannot be seen by FISH, then the cheapest scenario is Scenario 5: here, an initial FISH test reveals that the duplication is not visible by FISH and therefore targeted patient and parental MLPA tests (that can detect duplications) are performed (£858). Importantly, FISH is the cheapest follow-up test approach for all patients revealing a putative genome imbalance by aCGH because for non-commercially available probes, a single targeted FISH test is less expensive than a single targeted MLPA test. In addition, the working resolution of the 44 K arrays is sufficient to allow confirmatory FISH tests. By means of contrast, the most expensive scenarios, 6 (£1,232) and 7 (£1,358), both arise when the initial patient aCGH test reveals an imbalance of unknown clinical relevance and the follow-up testing strategy involves aCGH testing of parental samples.
Table 5.
aCGH–Cost of different reporting scenarios
| Scenarioa | Cost of patient aCGH test | Cost of Targeted FISH test | Cost of MLPA test | Cost of 2 parental aCGH testsb | Total cost |
|---|---|---|---|---|---|
| 1 No genome imbalance found; only known benign variants/polymorphisms | £446 | – | – | – | £446 |
| 2 Genome imbalance (deletion or duplication) found of known clinical relevance. | £446 | – | – | – | £446 |
| 3 Genome imbalance (deletion or duplication) found of unknown clinical relevance. Targeted FISH test performed on patient sample. Genome imbalance confirmed. Targeted FISH test on parental samples. Imbalance confirmed as inherited or ‘de novo’. | £458 | £214 c | – | – | £672 |
| 4 Genome imbalance (duplication only) found of unknown clinical relevance. Targeted FISH test performed on patient sample. Genome imbalance not confirmed. Targeted MLPA test on parental samples. Imbalance confirmed as inherited. | £458 | £110 | £245d | – | £813 |
| 5 Genome imbalance (duplication only) found of unknown clinical relevance. Targeted FISH test performed on patient sample. Genome imbalance not confirmed. Targeted MLPA test on parental samples negative. Targeted MLPA test on patient sample performed. Imbalance confirmed as ‘de novo’. | £458 | £110 | £290d | – | £858 |
| 6 Genome imbalance (deletion or duplication) found of unknown clinical relevance. Parental samples tested by aCGH. Imbalance confirmed as inherited | £442 | – | – | £790 | £1,232 |
| 7 Genome imbalance (deletion or duplication) found of unknown clinical relevance. Parental samples tested by aCGH and negative. Targeted FISH test performed on patient sample. Imbalance confirmed as ‘de novo’ | £458 | £110 | – | £790 | £1,358 |
a Scenarios 1 and 2 give the cost for a single patient sample only; the remaining scenarios account for the testing of both a patient and their two parents by aCGH, targeted FISH or MLPA
b The cost of testing each parent sample is slightly less than the cost of testing the patient sample, since some of the preliminary work and therefore a proportion of the cost is contained within the patient test cost
c Three FISH tests in total; the cost of testing a single sample (£110) is significantly more than testing each additional sample (£52) due to the initial cost of growing glycerols and preparing the probe ‘in house’
d The cost of testing a single sample (£200) is significantly more than testing each additional sample (£45) due to the initial cost of designing and purchasing targeted MLPA probe sets
Sensitivity analysis
For karyotyping, staff time required to perform the test or grade of staff used was the area most likely to impact upon total costs. For instance, substituting a clinical scientist with an MTO reduced the total cost to £95, a difference of £22. Other costs had limited impact upon karyotyping total costs. For aCGH, varying array (slide) costs had the greatest impact upon total cost. Changing the array to £25 per patient, reduced total test cost to £342. By comparison, equipment and staff costs had limited impact.
Comparing the costs of aCGH versus standard karyotyping
Table 6 directly compares aCGH with karyotyping by resource category. The basic total cost difference is £325, with array costs accounting for the largest cost difference between tests. By contrast, karyotyping is more labour intensive, with staff costs almost double those of array testing. Overheads for aCGH are higher as they are calculated as a percentage of overall costs (∼20%). Finally, there is little difference in equipment costs; even the cost of the array scanner (average £40,000) calculated over its predicted life and divided by test throughput, is small.
Table 6.
Cost comparison of aCGH and karyotyping per sample
| Cost category | ACGH | Karyotyping | Cost difference |
|---|---|---|---|
| Staff | £42 | £85 | −£43 |
| Equipment | £15 | £3 | +£12 |
| Consumables | £275 | £6 | +£269 |
| Overheads | £74 | £21 | +£53 |
| Other costs | £36 | £2 | +£34 |
| Total | £442 | £117 | £325 |
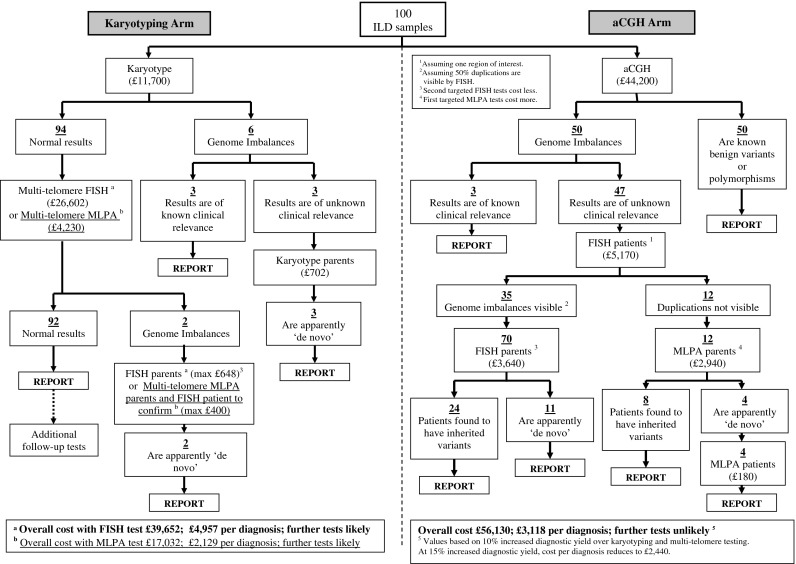
Figure 4 shows an example of the testing pathways, predicted number of diagnoses and associated cost implications of testing the same 100 ILD genetics clinic referrals for genomic imbalance via the routine karyotyping route (factoring in one additional test, either a multi-telomere FISH test or a multi-telomere MLPA test, for karyotypically normal samples) and via the most cost-effective aCGH route. For both approaches, most results are negative; there is no diagnosis in 92% of cases using karyotyping and multi-telomere tests and none in 82% of cases using aCGH. Interestingly, the cost of karyotyping plus one multi-telomere FISH test (£400) is comparable to a single 44 K aCGH test where no putative abnormality is found (£442).
Fig. 4.
Flowchart example of the testing pathways, predicted number of diagnoses and associated cost implications of testing the same 100 ILD genetics clinic referrals for genome imbalance via the routine karyotyping route (factoring in FISH and MLPA based telomere tests, for karyotypically normal samples) and via the aCGH route. The numbers of diagnoses expected via the karyotyping and aCGH routes and the testing scenarios are derived directly from published data, clinical diagnostic laboratory records and our own research experience. The aCGH route is expected to yield 10–15% more diagnoses than the karyotyping and multi-telomere testing route. The costings given in the flowchart are based on the conservative estimate of 10% more diagnoses, but an overall cost per diagnosis is calculated both for 10% and 15% more diagnostic yields
For positive results (diagnoses), Fig. 4 shows that with karyotyping and multi-telomere testing, 8/100 diagnoses are expected, costing £39,652 using multi-telomere FISH and £17,032 using multi-telomere MLPA. With aCGH, the most conservative estimate of at least 18 diagnoses is used (10% more than the karyotyping route). Here, the least expensive testing strategy (aCGH followed by patient targeted FISH and parental targeted FISH or targeted MLPA) gives an overall cost of £56,130. Thus, karyotyping with just one additional test of multi-telomere FISH equates to spending £4,957 to obtain a single diagnosis with 92% cases requiring further tests to reach a diagnosis at a later stage. Using multi-telomere MLPA the figure is reduced to £2,129 per diagnosis, again with 92% cases requiring further tests to reach a diagnosis. By contrast, the aCGH route equates to £3,118 per single diagnosis (assuming 10% more diagnoses than karyotyping plus multi-telomere testing combined), with no further tests for genomic imbalance required. This reduces to £2,440 per diagnosis if the diagnostic yield of aCGH is 15% more than karyotyping plus multi-telomere testing.
Discussion
This paper has reported a cost-effectiveness analysis comparing aCGH with karyotyping for detecting genomic imbalances that diagnose ILD. The average cost of aCGH was £442 per single (patient) sample and the average cost of karyotyping was £117 per sample. The majority of the cost-difference was accounted for by the array cost. Thus, from a single test perspective, aCGH is more expensive than karyotyping, explaining, in part, the hesitation by commissioners to fund aCGH in NHS diagnostic laboratories.
In reality, the situation is more complex because information regarding subsequent tests for genomic imbalance must be considered before the true cost-effectiveness can emerge. We have shown that the overall cost per diagnosis of the karyotyping route, including a single multi-telomere FISH assay (£4,957) is more expensive than that of the aCGH route (£3,118) that yields 10% more diagnoses. However, if the less conservative yield of 15% more diagnoses is correct, then the aCGH cost reduces to £2,440 per diagnosis, a figure more comparable to karyotyping plus the alternative multi-telomere assay, MLPA (£2,129 per diagnosis). Importantly, 92% of cases tested by karyotyping and a multi-telomere assay will require further tests for an eventual diagnosis. By contrast, the aCGH route, which effectively represents karyotyping, multi-telomere testing and not one, but ∼34,000 interstitial FISH tests as well as assaying the entire human genome at higher resolution is unlikely to require further genome-wide tests for genome imbalance.
Stand-alone karyotyping is the cheapest test when considered per diagnosis (£2,067), but this is at the sacrifice of missing ∼75% (12/18) diagnoses achievable by aCGH (Fig. 4). Thus, the crux of the aCGH versus karyotyping argument in ILD comes down to diagnostic capability versus cost; how much is it acceptable to spend and how many diagnoses is it acceptable to miss? aCGH clearly offers the greatest diagnostic capability, providing 10–15% more diagnoses over all other available tests.
One limitation of our study is that we do not know the full magnitude of the cost for additional follow-up tests after karyotyping. However, we do know that such costs would rapidly escalate and even then the majority of diagnoses achievable by aCGH would be missed; clinically relevant genomic imbalances found by genome-wide aCGH are rarely recurrent and therefore targeted approaches are unlikely to improve diagnostic yield (Veltman and de Vries 2006). Even if multi-telomere FISH or multi-telomere MLPA are not the tests chosen following karyotyping, costing in only five targeted tests for genomic imbalance at £100/test for every sample with a normal karyotype would raise the overall cost of testing 100 patients to ∼£59,402 (compared with £56,130 for aCGH) and offer negligible improvement in resolution overall. Thus, even without precise costing of follow-up tests, our results suggest that aCGH is the most cost-effective testing strategy in the long-term for testing ILD patients.
A further study limitation is the use of a simple outcome measure, diagnosis, rather than the more usual cost per quality adjusted life year promoted by the National Institute for Health and Clinical Excellence. However, describing and valuing health states in children is difficult and well documented (Petrou 2003). Current methodological work in health economics may give useable health states for children, but is unlikely to be suited for those with LD.
In this study, one scenario that we were unable to cost either for karyotyping or array CGH was that of further researching apparently inherited genome imbalances for which clinical relevance cannot be excluded. Such cases may reflect benign variants or may cause disease through unmasking recessive mutations, through variable penetrance or through imprinting, for example. Currently these account for up to 32% cases (see Fig. 4) and therefore follow-up tests such as sequencing would be prohibitively expensive (not all inherited imbalances are small). However, as more and more studies are performed and more data regarding benign/relevant genome imbalances and genotype/phenotype correlations are added to databases, it is anticipated that the clinical relevance of a significant number of these cases will be defined earlier, thereby minimising the need for parental testing or additional follow up tests. This in turn will lead to reduced aCGH testing costs and costs arising from doctor/counsellor time taken to discuss uncertain results. Furthermore, it may become possible to reduce costs more by employing better defined clinical ‘gatekeeping’ criteria to help clinical geneticists direct testing (thereby minimising total tests done). Databases such as The Database of Genomic Variants (http://projects.tcag.ca/variation/), The Human Structural Variation database, (http://humanparalogy.gs.washington.edu/structuralvariation/), ECARUCA (http://agserver01.azn.nl:8080/ecaruca/ecaruca.jsp) and the DatabasE of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER http://www.sanger.ac.uk/PostGenomics/decipher/) have all been designed to expedite these processes. Another possibility may be to use targeted arrays as an initial screening test for paediatrician referrals, though currently these offer no cost advantage over genome-wide arrays and utility will depend on a high diagnostic pick-up rate.
In the meantime, it will continue to be important for families to be counselled in possible outcomes before taking up the test, for both parental samples to be available for testing and for any outgoing laboratory reports to be carefully designed with clearly defined results e.g. array batch, controls used, imbalances found, confirmatory method and database search results that might help inform clinical relevance.
In conclusion, our study demonstrates that in the context of ILD, genome-wide aCGH is viable for NHS diagnostic use. Indeed, where possible, it may be appropriate to replace karyotyping with aCGH as the first-line test for genomic imbalance in ILD. If needed, samples that are normal by aCGH could then be karyotyped in order to identify truly balanced rearrangements or further characterise genome imbalances. Additionally, aCGH is expected to be useful for clarifying previous equivocal karyotyping results (e.g. enabling definition of a cryptic translocation in a family where only one of two unbalanced outcomes is cytogenetically visible).
In the future, improved diagnostic yields of aCGH and reduced follow-up tests will lower the costs of clinical follow-up and additional investigations. Advances in technology will also reduce costs (e.g. automation, increased probe density, multi-sample and cheaper array production and hybridisation methods) and software improvements may reduce analysis time.
Finally, it is important to note that potential applications of microarray technology extend beyond the genetic diagnosis of ILD to include a range of other conditions with suspected genome imbalance and/or aberrant gene expression e.g. haematological malignancies, colorectal cancer and other fields including oncology, immunology, neurology and pathology. The UK Department of Health is keen for the NHS to adopt new technologies (Department of Health 2003), yet commissioners are unable to endorse implementation without considering the clinical utility and economic implications of technology adoption. Our costing, with the results divided into different testing stages provides a framework for costing array implementation in different settings. Not least, it is intended that the study will be useful for healthcare providers faced with the decision of introducing aCGH testing into NHS laboratories before the availability of substantial effectiveness information.
Acknowledgements
The authors are grateful for financial support from the Oxford Genetics Knowledge Park (Dept of Health and Dept of Trade and Industry: RR, SK and JT). SW is supported by a UK Department of Health Fellowship. We acknowledge support from the UK Microarray Working Party, especially Hilary Burton, John Barber and Phillipa Brice.
Footnotes
Copyright statement
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd, and its Licensees to permit this article (if accepted) to be published in BMJ editions and any other BMJPGL products and to exploit all subsidiary rights, as set out in our licence (bmj.com/advice/copyright.shtml).
Authorship
The authors included on this paper fulfil the criteria of authorship and no one who fulfils the criteria has been excluded from authorship. The authors made a substantial contribution to the conception, design, analysis and interpretation of data. They were involved in drafting the article or revising it critically for important intellectual content and approving the version to be published.
Contributorship
Sarah Wordsworth (Guarantor): Planning, conducting and reporting work, interpretation of data, drafting and revising article.
James Buchanan: Conducting and reporting work, interpretation of data, revising article.
Regina Regan: Completing costing questionnaire, providing protocol details, other costing information, interpretation of data, information about learning disability and genome imbalance and revising article.
Val Davison: Completing costing questionnaire, providing protocol details, sharing overall laboratory experience and drafting article.
Kim Smith: Completing costing questionnaire, providing protocol details, drafting article.
Sara Dyer: Completing costing questionnaire and providing protocol details.
Carolyn Campbell: Completing costing questionnaire and providing protocol details.
Edward Blair: Critical appraisal of article for clinical content and revising article.
Eddy Maher: Completing costing questionnaire, providing protocol details, sharing overall laboratory experience and drafting article.
Jenny Taylor: Planning and facilitating work between centres. Drafting and revising article.
Samantha JL Knight: Completing costing questionnaire, providing protocol details, other costing information, interpretation of data, providing information about learning disability and genome imbalance, drafting and revising article.
Jenny Taylor and Samantha JL Knight contributed equally to the work presented.
References
- Aitman Science, medicine, and the future: DNA microarrays in medical practice. BMJ. 2001;323:611–615. doi: 10.1136/bmj.323.7313.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries B, Pfundt R, Leisink M, Koolen D. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Department of Health (2001) A new strategy for learning disability for the 21st Century. (http://www.archive.official-documents.co.uk/document/cm50/5086/5086.pdf) [PMC free article] [PubMed]
- Department of Health (2003) Genetics white paper: our inheritance, our future–realising the potential of genetics in the NHS. (http://www.dh.gov.uk/PublicationsAndStatistics/Publications/PublicationsPolicyAndGuidance/PublicationsPolicyAndGuidanceArticle/fs/en?CONTENT_ID = 4006538&chk = enskFb)
- Department of Health (2005) NHS reference costs. (www.doh.gov.uk/nhsexec/refcosts.htm)
- Drummond M, Sculpher M, Torrance G, O’Brien B et al. (2005) Methods for the economic evaluation of health care programmes. Oxford University Press, Oxford
- Flint J, Wilkie A, Buckle V, Winter R. The detection of subtelomeric chromosomal rearrangements in idiopathic mental retardation. Nat Genet. 1995;9:132–140. doi: 10.1038/ng0295-132. [DOI] [PubMed] [Google Scholar]
- HM Treasury (2006) Green book, appraisal and evaluation in central government. (http://greenbook.treasury.gov.uk/)
- Knight S (2005) Subtelomeric rearrangements in unexplained mental retardation. In: Fuchs P (eds) Encyclopedia of medical genomics and proteomics. Marcel Dekker Inc, New York, pp 1246–1252
- Knight S, Horsley S, Regan R, Lawrie N. Development and clinical application of an innovative fluorescence in situ hybridisation technique which detects submicroscopic rearrangements involving telomeres. Eur J Hum Gen. 1997;5:1–8. [PubMed] [Google Scholar]
- Knight S, Regan R. Idiopathic learning disability and genome imbalance. Cytogen Gen Res. 2006;115:215–224. doi: 10.1159/000095917. [DOI] [PubMed] [Google Scholar]
- Mencap (2001) No ordinary life: The support needs for families caring for children and adults with profound and multiple learning difficulties. (http://www.mencap.org.uk/download/no_ordinary_life.pdf)
- Menten B, Maas N, Thienpont B, Buysse K et al (2006) Emerging patterns of cryptic chromosomal imbalances in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of the literature. Dig J Med Genet: doi:10.1136/jmg.2005.039453 (http://jmg.bmj.com/cgi/rapidpdf/jmg.032005.039453v039451) [DOI] [PMC free article] [PubMed]
- Miyake N, Shimokawa O, Harada N, Sosonkina N. BAC array CGH reveals genomic aberrations in idiopathic mental retardation. Am J Med Genet Part A. 2006;140A:205–211. doi: 10.1002/ajmg.a.31098. [DOI] [PubMed] [Google Scholar]
- Petrou S. Methodological issues raised by preference-based approaches to measuring the health status of children. Health Eco. 2003;12:697–702. doi: 10.1002/hec.775. [DOI] [PubMed] [Google Scholar]
- Rauch A, Hoyer J, Guth S, Zweier C. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet Part A. 2006;140A:2063–2074. doi: 10.1002/ajmg.a.31416. [DOI] [PubMed] [Google Scholar]
- Ravnan JB, Tepperberg JH, Papenhausen P, Lamb AN. Subtelomere FISH analysis of 11,688 cases: an evaluation of the frequency and pattern of subtelomere rearrangements in individuals with developmental disabilities. J Med Genet. 2006;43:478–489. doi: 10.1136/jmg.2005.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeleveld N, Zielhuis G, Gabreels F. The prevalence of mental retardation: a critical review of recent literature. Develop Med Child Neurol. 1997;39:125–132. doi: 10.1111/j.1469-8749.1997.tb07395.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg C, Knijnenburg J, Chauffaille M, Brunoni D. Array CGH detection of a cryptic deletion in a complex chromosome rearrangement. Human Genet. 2005;116:390–394. doi: 10.1007/s00439-004-1248-x. [DOI] [PubMed] [Google Scholar]
- Schoumans J, Ruivenkamp C, Holmberg E, Kyllerman M. Detection of chromosomal imbalances in children with idiopathic mental retardation by array based comparative genomic hybridisation (array-CGH) J Med Genet. 2005;42:699–705. doi: 10.1136/jmg.2004.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw-Smith C, Redon R, Rickman L, Rio M. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet. 2004;41:241–248. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson C, Harvard C, Locker R, Friedman J. Submicroscopic deletions and duplications in individuals with intellectual disability detected by array-CGH. Am J Med Genet Part A. 2005;139A:173–185. doi: 10.1002/ajmg.a.31015. [DOI] [PubMed] [Google Scholar]
- Veltman J, de Vries B. Diagnostic genome profiling: unbiased whole genome or targeted analysis? J Mole Diag. 2006;8:534–537. doi: 10.2353/jmoldx.2006.060131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers L, deVries B, Osoegawa K, Janssen I. Array-based comparative genomic hybridization for the genome-wide detection of submicroscopic chromosomal abnormalities. Am J Human Genet. 2003;73:1261–1270. doi: 10.1086/379977. [DOI] [PMC free article] [PubMed] [Google Scholar]