Abstract
The occurrence of antimicrobial resistance in generic Escherichia coli can serve as an indicator of the pool of resistance genes potentially available for transfer to pathogenic organisms. This study was conducted on 29 volunteer beef farms in Ontario to describe the prevalence and patterns of antimicrobial resistance in E. coli, and to describe changes in the prevalence of resistance during the feedlot stage of production. From the pooled fecal samples on 28 of the 29 farms, 31% of isolates from feedlots (n = 993) and 12% of isolates from cow-calf farms (n = 807) were resistant to one or more of 16 antimicrobials tested. No isolates were resistant to ceftriaxone, ciprofloxacin, gentamicin, or nalidixic acid, and < 1% of the pooled isolates were resistant to ceftiofur. Two percent of both feedlot and cow-calf isolates were resistant to ≥ 5 antimicrobials. Cow-calf farms were at significantly lower risk than feedlots for having E. coli isolates that were resistant to streptomycin, sulfamethoxazole, and tetracycline. On average, the prevalence of sulfamethoxazole resistant E. coli isolates was significantly higher in calves than in cows. No resistance was observed to ceftriaxone or ciprofloxacin among isolates (n = 1265) obtained from individually sampled feedlot animals on 2 farms. Less than 1% of these isolates were resistant to gentamicin, nalidixic acid, and ceftiofur. From the individually sampled feedlot animals, resistance to streptomycin (on 1 farm), sulfamethoxazole, and tetracycline increased significantly from arrival to mid-point during the feeding period, and these levels persisted until market-readiness.
Résumé
La fréquence de résistance antimicrobienne chez les Escherichia coli génériques peut servir d’indicateur du pool de gènes de résistance potentiellement disponible pour le transfert à des agents pathogènes. La présente étude a été effectuée sur 29 fermes ontariennes de bovins d’embouche, participant de façon volontaire, afin de décrire la prévalence et les patrons de résistance aux antimicrobiens chez E. coli, et de décrire les changements dans la prévalence de résistance durant la période de production en parc d’engraissement. À partir des pools d’échantillons fécaux de 28 des 29 fermes, 31 % des isolats des parcs d’engraissement (n = 993) et 12 % des isolats des élevages vache-veau (n = 807) étaient résistants à au moins un des 16 antimicrobiens testés. Aucun isolat n’était résistant au ceftriaxone, au ciprofloxacin, à la gentamycine ou à l’acide nalidixique, et < 1 % des isolats était résistant au ceftiofur. Deux pourcents des isolats provenant des parcs d’engraissement de même que des élevages vache-veau étaient résistants à ≥ 5 antimicrobiens. Les élevages vache-veau étaient significativement moins à risque que les parcs d’engraissement d’avoir des isolats d’E. coli qui étaient résistants à la streptomycine, au sulfaméthoxazole et à la tétracycline. En moyenne, la prévalence d’isolats d’E. coli résistants au sulfaméthoxazole étaient significativement plus élevée chez les veaux que chez les vaches. Aucune résistance n’a été observée au ceftriaxone ou au ciprofloxacin parmi les isolats (n = 1265) obtenus d’animaux individuels dans des parcs d’engraissement sur 2 fermes. Moins de 1 % de ces isolats étaient résistants à la gentamycine, à l’acide nalidixique et au ceftiofur. À partir des animaux échantillonnés individuellement, la résistance à la streptomycine (sur 1 ferme), au sulfaméthoxazole et à la tétracycline a augmenté de manière significative du moment de l’arrivée jusqu’à la mi-période de la période d’engraissement, et ces niveaux ont persisté jusqu’au moment où les animaux étaient prêts pour le marché.
(Traduit par Docteur Serge Messier)
Introduction
The knowledge of prevalence and patterns of antimicrobial resistance (AMR) in generic Escherichia coli might indicate the pool of resistance elements that are available for transfer to other bacterial species including pathogens (1). The transfer of resistance genes to human pathogens is of primary concern to public health, but there is the additional concern of the transfer of resistance genes to animal pathogens and associated subsequent loss of therapeutic options for veterinary medicine.
Generic Escherichia coli are used to monitor changes in prevalence and patterns of resistance as these commensal bacteria are regularly found in the gastrointestinal tract of animals and humans (2,3) and their recovery is easy and cost-effective (4). Globally, there are few organizations that routinely monitor AMR in commensal and pathogenic enteric bacteria from apparently healthy animals. Examples are the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS), the Danish Integrated Antimicrobial Resistance Monitoring and Research Program, and the Swedish Veterinary Antimicrobial Resistance Monitoring Program (5–7). Although much of the monitoring is conducted at the abattoir level, it is also important to understand the patterns of change in the prevalence of AMR throughout the animal-production stages in order to identify risk factors for resistance development or amplification. Only a few studies have investigated farm level resistance in E. coli in beef cattle in Canada (8–13) and no other study has investigated this topic in Ontario.
The objectives of this study were to describe the prevalence and patterns of AMR among generic E. coli recovered from 29 beef cattle operations, and to describe temporal changes in E. coli resistance in individual feedlot animals during the feeding period.
Materials and methods
Farm enrolment
This study is part of a larger study that included documenting antimicrobial use practices on the same farms. Producers were invited to participate in the study through presentations to the Ontario Cattlemen’s Association (August 1999) and to the Ontario Cattle Feeders Association (September 1999), and via a poster presentation of the project at the Agriculture’s Role in Managing Antimicrobial Resistance Conference in 1999 (14). Eleven feedlot operations, 8 cow-calf operations, and 5 “combination” farms (farms with both operation types; yielding a total of 29 farms; 16 feedlots and 13 cow-calf farms) were voluntarily enrolled in the prospective longitudinal study. Veal farms and farms that kept young cattle only for short periods before entry to feedlots (backgrounded cattle) were excluded from the study.
Sample collection
Pooled fecal pat samples were collected from 28 of the 29 farms, and rectal fecal samples were collected from individual animals on 2 of the feedlots. One of the feedlots in the individual animal sampling did not participate in the pooled sampling.
Pooled fecal sampling
For the 28 farms, pooled fecal pat samples were collected 3 times per farm, approximately 3 to 4 mo apart, from January to August 2001. Five groups of animals were selected for sampling from each farm, stratified by animal type as described in the following text. In cow-calf farms, samples were taken from 5 cow-calf groups and 3 sets of samples were proportioned from cows and 2 from calves. These were all nursing calves and a wide age range was sampled, from neonates to just pre-weaning; a particular age of calf was not targeted. In feedlots, 5 separate pens or barns were sampled if available. For feedlots with > 5 separate pens or barns, the sampled sites were chosen to distribute the samples as evenly across the various pens/barns of the farm as possible. Four fresh fecal pats were collected from the 5 different locations/animal groups using sterile gloves, and samples were stored in sterile plastic bags in chilled coolers until being delivered to the laboratory the same day. This yielded a total of 60 samples per farm.
Individual animal sampling
Two feedlots were selected, based upon the safety of the handling facilities and producer compliance, for repeated sampling of individual cattle (hereafter called “individual animal samples”, and “farm 1” or “farm 2”). Sampling took place between July 2001 and May 2002. The producers were asked if 50 animals could be followed over time at their feedlots. Rectal fecal samples were collected from cattle on 3 separate occasions: the day the cattle arrived at the feedlot, mid-way during the feeding period (at producer convenience), and as close as possible to the time of shipment for slaughter (at producer convenience and within 2 wk of shipping). This yielded a total of 300 samples.
Fecal processing and storage
For the pooled samples, 5 g of feces from each of the 4 sampled pats were mixed into a sterile Seward Stomacher bag (VWR Canlab, Mississauga, Ontario), and from the intensively sampled feedlots 20 g of feces per animal were placed into sterile Seward stomacher bags (VWR Canlab). Then, 40 mL of sterile 0.9% saline solution was added to the feces in each bag. Bags were stomached for 20 s, and then 0.6 mL aliquots of the filtered homogenate were added to 1.25 mL cryotubes (Sarstedt, Montreal, Quebec) containing 0.6 mL of sterile tryptic soy broth with 50% glycerol. The tubes were inverted several times to mix and then frozen at −70°C.
Escherichia coli isolation
Escherichia coli isolation was conducted at the University of Guelph, Guelph, Ontario. A small amount (10 μL) of each frozen fecal sample was thawed and streaked onto a MacConkey agar plate (Difco, Becton, Dickson, Oakville, Ontario) and incubated for 18–24 h at 37°C. Five isolated colonies with the typical color and appearance of E. coli were streaked out further on MacConkey agar plates as a purification step and incubated for 18 to 24 h at 37°C. One isolated colony from each of the 5 putative E. coli colonies was streaked onto Luria-Bertani Miller agar (Difco, Becton, Dickson) and incubated for 18 to 24 h at 37°C. One colony was picked from each of these plates for the Indole Spot Test. The test was conducted by smearing the colony onto Whatman filter paper (VWR Canlab, Mississauga, Ontario) wetted with Indole Spot Reagent (PML Microbiologicals, Mississauga, Ontario), and p-dimethylaminobenzaldehyde (1% in 10% HCl w/w). If the isolate was an indole producer (blue color reaction), then a small amount was picked from the same colony and inoculated onto a Simmons citrate slant (Difco, Becton, Dickson). The isolate was considered to be an E. coli strain if it appeared negative on the citrate slant. Isolates were incubated in Mueller Hinton (Difco, Becton, Dickson) broth for 1 h at 37°C, then 0.6 mL of the broth was mixed with 0.6 mL of Mueller Hinton broth containing 50% glycerol in a 2-mL Sarstedt tube (Sarstedt, St. Leonard, Quebec) and frozen at −70°C.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was conducted at the Laboratory for Foodborne Zoonoses, Guelph, Ontario, using the Sensititre (Trek Diagnostic Systems, Cleveland, Ohio, USA) automated broth microdilution system, using the Sensititre custom panel CMV7CNCD (Trek Diagnostic Systems), as per CIPARS 2002 (5). Escherichia coli ATCC 25922 and E. coli ATCC 35218 were used as quality control organisms.
Bacteria were tested for their susceptibility to the following antimicrobials with respective breakpoints (as described by the Clinical and Laboratory Standards Institute) (15,16): amikacin (≥ 64 μg/mL), amoxicillin-clavulanic acid (≥ 32/16 μg/mL), ampicillin (≥ 32 μg/mL), cefoxitin (≥ 32 μg/mL), ceftiofur (≥ 8 μg/mL), ceftriaxone (≥ 64 μg/mL), cephalothin (≥ 32 μg/mL), chloramphenicol (≥ 32 μg/mL), ciprofloxacin (≥ 4 μg/mL), gentamicin (≥ 16 μg/mL), kanamycin (≥ 64 μg/mL), nalidixic acid (≥ 32 μg/mL), sulfamethoxazole (≥ 512 μg/mL), tetracycline (≥ 16 μg/mL), and trimethoprim-sulfamethoxazole (≥ 4/76 μg/mL). For streptomycin, which had no established breakpoint for the CMV7CNCD plate, 64 μg/mL was used, as this is the breakpoint identified for the CMV6CNCD plate and is used by the US National Antimicrobial Resistance Monitoring System (NARMS) (17). Antimicrobial abbreviations and the range of concentrations tested are as described for CIPARS 2002 (5). Based on these breakpoints, isolates were classified susceptible or resistant (intermediate isolates were classified as susceptible). The antimicrobials were additionally classified according to their importance to human health using a system proposed by the Veterinary Drugs Directorate (VDD), Health Canada (18).
The following antimicrobial resistance patterns as per CIPARS 2003 are reported: A3C, which indicates phenotypic resistance to amoxicillin-clavulanic acid, cefoxitin, ceftiofur, and cephalothin; and AC(K)SSuT which indicates resistance to ampicillin, chloramphenicol, (kanamycin), streptomycin, sulfamethoxazole and tetracycline (19).
Data management and analysis
Data were entered into Microsoft Office Excel 2000 (Microsoft, Redmond, Washington, USA) spreadsheets. All statistical procedures were performed using SAS software, version 8.02 (SAS Institute, Cary, North Carolina, USA).
The pooled sample data were structured into 3 levels of hierarchy: isolate, pool, and farm. Similarly, for the intensively sampled feedlots, the data were structured as: isolate, individual animal, and farm. For prevalence of resistance to individual antimicrobial drugs, binomial proportions were computed using exact probability distributions (20).
The SAS GLIMMIX macro was used to compare whether farm type (cow-calf farm versus feedlot) was associated with risk of resistance. In addition, for the cow-calf data, cows and calves were compared to see if the class of animal was associated with risk of resistance. Resistance to antimicrobials to which > 5% of the isolates were resistant was modeled, using a binomial error structure and method as REML. The variables “pool”, “farm” and “visit” were treated as random effects (visit was nested within farm; pool was nested within visit and farm), despite not being randomly sampled, as they would not be consistent factors if this study were to be repeated and E. coli isolates cannot be resampled. For all models, the random effects were removed if they were not significant at the 5% level of significance (tested in a null model). Similarly, for the individual animal samples, the variable “animal” was considered a random effect as individual animals would not be consistent factors if this study were to be repeated and the specific E. coli isolates could not be resampled. The prevalence of resistance to specific antimicrobials (> 5% prevalence of resistance) was compared between the different visits, treating the variable “farm” as a fixed effect.
Results
Farm description
The feedlots on average produced 2219 cattle per year (range 55 to 10 000). The cow-calf herds ranged from 9 to 170 cows and on average had 83 cows.
For the intensively sampled feedlots, Farm 1 produced approximately 900 cattle per year, whereas Farm 2 produced approximately 5000 cattle per year. Farm 1 purchased cattle that weighed between 220 and 270 kg, and Farm 2 purchased cattle that weighed between 320 and 455 kg. Both farms purchased cattle primarily from Ontario and kept new arrivals isolated. Farm 1 did not often buy prevaccinated calves, whereas Farm 2 preferred prevaccinated calves. Farm 1 administered oxytetracycline routinely to all new arrivals, whereas Farm 2 did not. Farm 1 administered tylosin and monensin in the feed, whereas Farm 2 administered oxytetracycline, tylosin, and monensin in the feed. Cattle were housed for 194 and 145 d on Farms 1 and 2, respectively. For Farm 1, the middle visit was exactly halfway during the feeding period, whereas the middle visit to Farm 2 occurred only 19 days before the last visit.
The total number of isolates tested differed from the planned 2100 isolates for pooled samples and 1500 isolates for individually sampled animals (pooled samples n = 1800 isolates; individually sampled animal n = 1265 isolates). For the pooled samples, there were not enough animals available on some visits to fulfill the sampling plan. For the individual animal samples, some animals were lost to follow-up (due to lost ear-tags), thus there were fewer animals sampled on the 2nd and 3rd visits.
Prevalence of resistance — pooled samples
The minimum inhibitory concentrations (MICs) and prevalences of resistance to individual antimicrobial drugs are found in Table I. Specific AMR patterns are found in Table II. For amikacin, the resistance breakpoint was beyond the tested range, thus 37 isolates with MICs > 4 μg/mL were non-interpretable. At the farm level, all farms had at least 1 isolate resistant to tetracycline, and most farms had at least 1 isolate resistant to sulfamethoxazole (89%), cephalothin (89%), and streptomycin (86%). For antimicrobials of highest importance to human medicine (18) 7/1800 (0.39%) of the total pooled isolates were resistant to ceftiofur and 14/1800 (0.8%) were resistant to amoxicillin-clavulanic acid. No isolates in the study were resistant to the other antimicrobials of highest importance to human medicine (ceftriaxone and ciprofloxacin) (18).
Table I.
Distribution of the minimum inhibitory concentrations (MICs) and prevalence of resistance in generic Escherichia coli recovered from pooled samples (feedlot n = 993 isolates, cow-calf farms n = 807 isolates)a
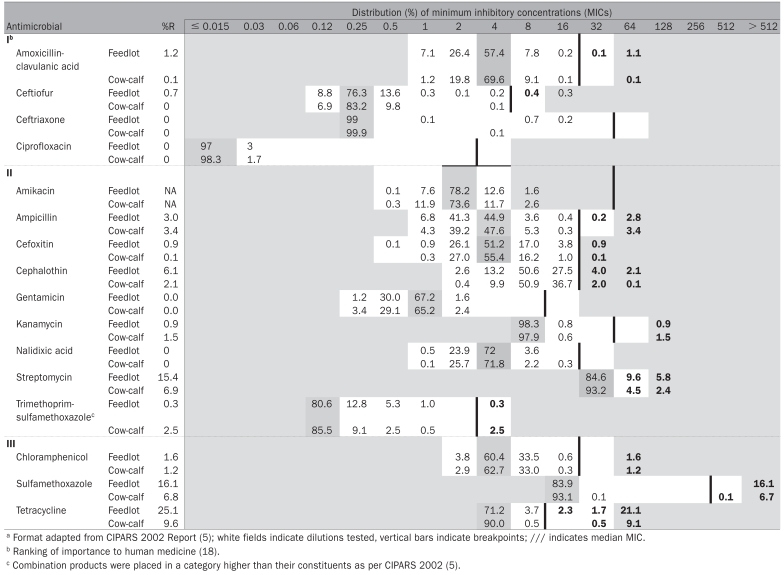 |
Table II.
Antimicrobial resistance patterns observed across all isolates
| Source of isolates | Number of isolates tested | Fully susceptible | Resistant to 1 antimicrobial | Resistant to 2 antimicrobials | Resistant to 3 antimicrobials | Resistant to > 3 antimicrobials | AC(K)SSuTa,b | ACSSuT + A3Ca,b,c |
|---|---|---|---|---|---|---|---|---|
| Feedlot (pooled); 15 farms | 993 | 688 | 114 | 44 | 121 | 22 | 5 | 7d |
| Cow (pooled) 13 farms | 602 | 538 | 24 | 7 | 18 | 15 | 3 | 0 |
| Calf (pooled); 13 farmse | 171 | 138 | 5 | 5 | 10 | 13 | 5 | 0 |
| Individually sampled feedlot animals; 2 farms | 1265 | 764 | 195 | 120 | 160 | 26 | 0 | 1 |
Isolates in this column are included in the column titled “Resistant to > 3 antimicrobials.”
Isolates in this column could also be resistant to additional antimicrobials.
Isolates in this column do not include isolates in the column titled “AC(K)SSuT.”
Two of these isolates also had reduced susceptibility to ceftriaxone.
The pooled samples included 34 isolates from other types of cattle on the cow-calf farms, including bulls, heifers, and steers. Of these isolates, only 2 were resistant, 1 to tetracycline and 1 to sulfamethoxazole-tetracycline.
For the pooled feedlot samples, 305/993 (31%) isolates were resistant to at least 1 antimicrobial, but there was no resistance observed to gentamicin or nalidixic acid. With respect to antimicrobials of highest importance to human health (18) all 7 of the pooled isolates resistant to ceftiofur were from feedlots. These 7 isolates had the ACSSuT + A3C pattern and 2 had reduced susceptibility to ceftriaxone. Six of these isolates came from the same farm, on the 1st visit only, and from 2 different pools. Overall, 25 different resistance patterns (including fully susceptible as a pattern) were observed and 15/993 (2%) isolates were resistant to ≥ 5 antimicrobials. Of these 15 isolates, 9 came from the same farm.
Of the pooled isolates from cows, 64/602 (11%) were resistant to ≥ 1 of the antimicrobials tested. Eighteen different resistance patterns were observed and 7/602 (1%) isolates were resistant to ≥ 5 antimicrobials. The isolates resistant to ≥ 5 antimicrobials were collected from 4 farms.
For the pooled isolates from calves, 33/171 (19%) isolates were resistant to ≥ 1 antimicrobials tested. Fifteen different resistance patterns were observed and 5/171 (3%) isolates were resistant to ≥ 5 antimicrobials. These 5 isolates were collected from 2 different farms. The AKSSuT and the AKSSuT-cephalothin patterns from both the cow and the calf isolates were from the same farm.
The pooled samples included 34 isolates from other types of cattle on the cow-calf farms, including bulls, heifers, and steers. Of these isolates, only 2 were resistant, 1 to tetracycline and 1 to sulfamethoxazole-tetracycline, and these 2 isolates came from the same farm.
Comparison of cow-calf farms to feedlots — pooled samples
Prevalences of resistance ≥ 5% were only observed to streptomycin, sulfamethoxazole, and tetracycline. With no other fixed effects in the model, farm type was significantly associated (P < 0.05) with prevalence of resistance to all 3 antimicrobials (conditional on the random effects). For the streptomycin model, the random effect farm was not significant at the 5% level and for the sulfamethoxazole model the random effect visit (farm) was not significant at the 5% level. For the tetracycline model, all the random effects were significant at the 5% level. The odds ratios indicated that cow-calf farms were, on average, at less risk than feedlots for having generic E. coli resistant to these drugs, conditional on the random effects (ORSTR = 0.24, 95% CISTR = 0.10 to 0.56; ORSMX = 0.25, 95% CISMX = 0.09 to 0.65; ORTCY = 0.21, 95% CITCY = 0.09 to 0.50). The models were repeated with all the random effects forced into the models and the results were the same, that is, farm type was significantly associated with the prevalence of resistance, the final odds ratios changed very slightly (for example, ORSTR = 0.26; 95% CISTR = 0.10 to 0.65).
In the absence of any analysis, the absolute value of the medians of the minimum inhibitory concentration (MIC) distributions did not differ between cow-calf farms and feedlots for any of these 3 antimicrobials (Table I); however, the medians were not corrected to account for the clustering of the data.
Comparison of cows to calves — pooled samples
For the cow-calf pooled data, with no other fixed effects in the model, animal type (cow versus calf) was significantly associated (P < 0.05) with prevalence of resistance to sulfamethoxazole. For this model the random effect farm was not significant at the 5% level. The odds ratios indicated that calves were, on average, at greater risk than cows for having generic E. coli resistant sulfamethoxazole, conditional on the random effects (ORSMX = 3.70, 95% CISMX = 1.11 to 12.34). There was no significant association between animal type and resistance to streptomycin or tetracycline. The models were repeated with all the random effects forced into the models, and the results were the same.
Prevalence of resistance — individual animal samples
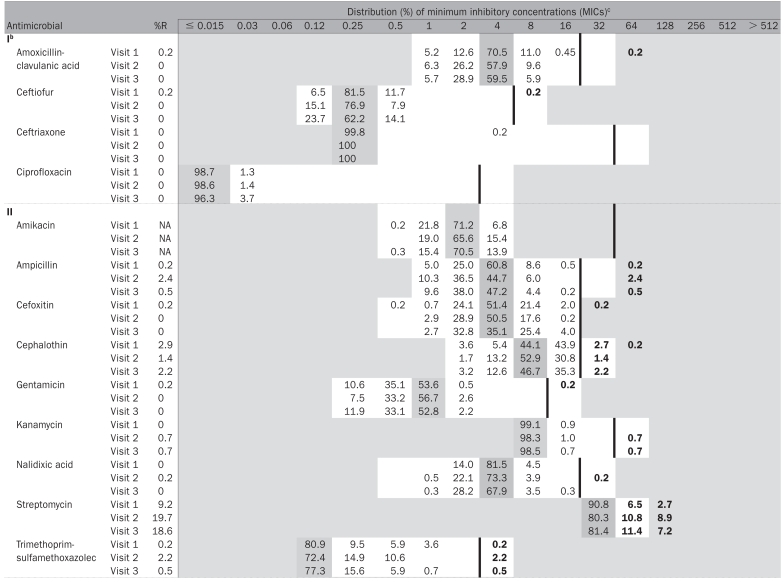
Antimicrobial susceptibility testing was conducted on 1265 isolates from individual animal fecal samples. The MICs and the prevalences of resistance are shown in Table III. Five hundred and one (40%) isolates were resistant to ≥ 1 antimicrobials. One isolate was resistant to ceftiofur and this isolate also was also resistant to 8 other antimicrobials (ACSSuT + A3C). One isolate was resistant to gentamicin (and also to 3 other antimicrobials) and another isolate was resistant to nalidixic acid and tetracycline. Overall, 23 different resistance patterns were observed and 11 isolates (< 1%) were resistant to ≥ 5 antimicrobials.
Table III.
Distribution of the minimum inhibitory concentrations (MICs) and the prevalence resistance in generic Escherichia coli recovered from individually sampled feedlot animals on two farms (visit 1 n = 444, visit 2 n = 416, visit 3 n = 405)a
|
Temporal changes — individual animal samples
Prevalences of resistance > 5% were only observed to streptomycin, sulfamethoxazole, and tetracycline. Interaction between “farm” and “visit” was tested for, and the interaction terms were significant so the results for changes in prevalence of resistance over time have farm specific interpretations. For streptomycin, there was no significant change in prevalence of resistance over time (all P > 0.09) for Farm 1. For streptomycin and for Farm 2, the prevalence of resistance significantly increased between the 1st and 2nd visits (P < 0.0001), but the prevalence of resistance was not significantly different between the 2nd and 3rd visits (P = 0.9837). For sulfamethoxazole and tetracycline, both farms had the same pattern: there was a significant increase in risk of resistance between visits 1 and 2 (all P < 0.05), and the risk of resistance between visits 2 and 3 was not significantly different (all P > 0.05).
Since the 2 farms had unequal days between the various visits, the data were re-evaluated using the actual number of days on feed (treated as a continuous variable) to determine whether days on feed was associated with risk of a resistant E. coli. In all the models, treating the variable “farm” as a fixed effect, the odds of having an E. coli resistant to streptomycin, sulfamethoxazole, or tetracycline significantly increased with increasing number of days on feed (P < 0.001 in all cases). There was a significant interaction between days on feed and farm for streptomycin and tetracycline.
The median MIC did not change over the 3 visits for any of the antimicrobials tested other than for tetracycline, for which the median MIC increased at each visit (1st visit ≤ 4 μg/mL, 2nd visit ≤ 8 μg/mL, 3rd visit ≤ 16 μg/mL), primarily because more isolates were in the > 32 μg/mL category. These median MICs, however, were not corrected to account for the clustering of the data.
Discussion
The intent of this study was to describe the prevalence and patterns of resistance in generic E. coli from beef cattle from southern Ontario. Antimicrobial use practices on these farms are published elsewhere (21) and relationships between use and resistance will be the subject of a future publication.
From a public health perspective and in relation to VDD’s categorization of antimicrobials with relation to human medicine, it was encouraging that for drugs in the category of highest importance to human medicine (Category I) there were no pooled isolates resistant to ceftriaxone, ciprofloxacin, and ≤ 1% of isolates were resistant to ceftiofur or amoxicillin-clavulanic acid. There were no pooled isolates resistant to gentamicin or nalidixic acid. Also, the prevalences of resistance were low (< 5%) for most drugs other than tetracycline, streptomycin, and sulfamethoxazole.
The study reports on A3C and AC(K)SSuT patterns as per CIPARS 2003, because the A3C pattern may be related to the presence of Amp-C beta-lactamases or extended spectrum beta-lactamases and the AC(K)SSuT pattern may be chromosomally encoded and has repeatedly been described in the past particularly for Salmonella Typhimurium DT104 (19). For the study, 8 isolates had the A3C pattern, and it was always accompanied by the ACSSuT pattern; 6 of these isolates were from the same farm.
The findings were consistent with respect to the Category I antimicrobials in comparison to a recent Canadian study in Alberta of 4 feedlots (9). For other antimicrobials in this Alberta study, 39% of generic E. coli isolates were resistant to tetracycline, 13% to streptomycin, and 13% to sulfamethoxazole (9), herein, the percentage of resistance was 25%, 15%, and 16%, respectively. It was found that 31% of isolates were resistant to ≥ 1 antimicrobials tested compared with 45% isolates (n = 500) found in the Alberta study (9). In another feedlot study in Saskatchewan published in 2006, AMR was investigated specifically in E. coli 0157 isolates recovered from 2 feedlots and 1 cow-calf operation (22). Among 131 isolates, no resistance was observed to ceftriaxone, ciprofloxacin, ceftiofur or amoxicillin-clavulanic acid; however, there was resistance to sulfisoxazole at a prevalence of 61% and tetracycline at 12% (22). Differing levels of resistance across the country may reflect differences in housing/management practices, E. coli strains/clones, seasons for sampling, as well as different AMU practices. For example, feedlot cattle are not housed in barns in western Canada, whereas in the Ontario farms studied feedlot cattle were housed in various combinations of barns and outdoor pens.
Several studies have reported AMR in generic E. coli recovered from beef cattle in the USA. In 1 study of a feedlot in Colorado (n = 2316 isolates) no resistance among generic E. coli was found to ceftriaxone, amikacin, apramycin, or gentamicin (23). One isolate was found to be resistant to ciprofloxacin, between 0.9 and 1.1% of isolates resistant to amoxicillin-clavulanic acid (depending on sampling protocol), and between 0 and 0.1% resistant to ceftiofur (depending on sampling protocol) (23). The highest levels of resistance in this study were also found to be to streptomycin, sulfamethoxazole, and tetracycline (23). In another feedlot study in Colorado, AMR was investigated in generic E. coli recovered from 95 pens of cattle from 2001 to 2003 (24). The investigators reported little resistance to antimicrobials that are of highest importance to human health, although the details of the prevalence and which drugs were included were not printed in the Web-report (24). They did comment, however, that resistance was most often seen to streptomycin, sulfamethoxazole, and tetracycline (24). In another study conducted in Michigan between 2002 and 2003, AMR of E. coli was investigated on 7 beef farms (type of farm not described) (25). This research group tested for slightly different antimicrobials and did not present their findings as prevalence of resistance, but they too did not find any isolates resistant to quinolones or fluoroquinolones (25). In a recent American clinical trial of feedlot animals reviewing the effects of ceftiofur crystalline free acid, the researchers noted that the most common prevalences of resistances in E. coli from the 61 steers were to sulfisoxazole (63%), tetracycline (41%), streptomycin (33%), chloramphenicol (23%), and ampicillin (21%) (26). For the control animals receiving no antimicrobials they noted no resistance to gentamicin, but found resistance to amoxicillin-clavulanic acid (4%), ceftiofur (4%), ceftriaxone (1%), ciprofloxacin (0.1%), and nalidixic acid (2%) (26). Generally, both Canadian and American researchers reported very low levels of resistance to antimicrobials that are included in Category I of the Canadian classification system, particularly to ciprofloxacin and ceftriaxone.
In another study conducted in western Canada (2002–2003), resistance among generic E. coli isolates was investigated from cow-calf pairs and the highest prevalences of resistance were observed to tetracycline, streptomycin, and sulfamethoxazole, similar to the findings from our study (13). Scottish researchers investigated resistance in E. coli from calves with and without diarrhea (27) and reported that 84% of all samples (1 sample per calf, multiple isolates per sample were tested) were resistant to ampicillin, 13% to apramycin, and 6% to nalidixic acid. Their conclusions were that antimicrobial resistant E. coli were more likely to be detected on farms with calf enteritis than on those without this condition (27). In another Scottish study, AMR of generic E. coli was investigated using a cohort of 48 newborn calves that were sampled weekly over a 4-month period, after which recovered isolates were tested for susceptibility to ampicillin, apramycin, and nalidixic acid. These researchers found that 67% of samples were resistant to at least 1 of these antimicrobials (28). They also found that the prevalence of nalidixic acid and ampicillin was initially high but declined significantly with age and that all calves had at one point E. coli isolates resistant to ampicillin or nalidixic acid (28). While there was no age specific information in the present study, no nalidixic acid resistance was seen and only 8% of the calf isolates were resistant to ampicillin (14/171 isolates). The difference in nalidixic acid resistance between Scotland and Canada might be explained by the fact that in the UK fluoroquinolones are licensed for use in calves. At the time of our data collection, fluoroquinolones were not licensed in Canada; however, they have since become licensed for use in treating bovine respiratory disease.
The observed significant differences in prevalence of streptomycin, sulfamethoxazole, and tetracycline resistance between cow-calf farms and feedlots may reflect differences in the types of antimicrobials used on the different farm types, age-related factors (although specific inferences cannot be made as there were no details on ages of calves sampled) or perhaps other farm-level factors. The antimicrobial resistance patterns may be serovar or strain specific, similar in nature to Salmonella serovars in which resistance is highly correlated with serovar (5); however, due to financial constraints, it was not possible to serotype the E. coli in this study. This study also showed that the prevalence of sulfamethoxazole resistant E. coli isolates was significantly higher on average in calves than in cows. The reason for this finding is unknown but may be related to the drug use practices in the calves.
Results from the 2 intensively sampled feedlots indicated that resistance to streptomycin, sulfamethoxazole, and tetracycline increased from the start of feeding until finishing, as related to number of days on feed. In this analysis the “days on feed” data were treated as a continuous variable; however, each feedlot was only sampled on 3 specific visits. When the visits were treated as categorical variables, streptomycin, sulfamethoxazole, and tetracycline resistance prevalences significantly increased from the 1st visit to the 2nd and that the 2nd and 3rd visits were not significantly different from one another (with the exception of streptomycin resistance on Farm 1, which did not significantly change over time). Results from this analysis, however, may be influenced by the fact the 2 feedlots had their cattle on feed for different lengths of time, and that the timing between visits differed between the farms. It could be hypothesized that the effects of drug usage and other factors in the short term after arrival at the feedlot promotes the selection for resistant bacteria that persist until the time of slaughter. Alternatively, the stress of mixing and shipping of animals to the feedlot may have promoted shedding and clonal spread of resistant bacteria that were detected only upon the 2nd visit. The lack of significant change of resistance prevalences between the 2nd and 3rd visits suggests that under the conditions of this study, resistance is stable within the population and persistent to slaughter. The timing of the rise of resistance is interesting particularly in relation to drug use practices; future studies could include more frequent sampling (every week) throughout the feeding period to study the association more closely. In other Canadian studies it was also noted that calves arrive at feedlots with resistant E. coli and that the prevalence of resistance was higher towards the end of the finishing stage (10,12,29). In a randomized clinical trial comparing isolates from cattle fed tetracycline, cattle given oxytetracycline metaphylactically, and control cattle receiving no antimicrobials, Checkley (10) conducted more frequent sampling during the feeding period and found that tetracycline resistance increased in fecal E. coli between arrival samples and preslaughter samples. However there was a significant difference between the cattle fed tetracycline versus the control cattle or the cattle given oxytetracycline metaphylactically, in that the group fed tetracycline had a much larger increase in tetracycline resistance that started to decline at roughly 13 days on feed but still remained significantly higher at preslaughter than at arrival (10). The unpublished results of a Colorado study indicated that resistance prevalences for chloramphenicol and nalidixic acid were lower at preslaughter but resistance to cephalothin and tetracycline were higher at the end of the feedlot duration (24).
Notably, some of our E. coli isolates were resistant to chloramphenicol, a drug that is no longer approved for use in beef cattle, which is similar to findings reported by CIPARS (5) and other Canadian researchers (9,22). Chloramphenicol resistance was always present in combination with resistance to tetracycline or sulfamethoxazole (+/− other antimicrobials) and its persistence, despite the banning of the drug, is likely related to co-selection with either tetracycline or sulfamethoxazole resistance. Alternatively, perhaps the use of florfenicol, a fluorinated derivative of chloramphenicol, is helping to maintain this resistance. White et al (30) noted increasing reports of cross-resistance to florfenicol and chloramphenicol attributable to a flo gene, which is a “homolog of the chloramphenicol resistance efflux gene, cmlA” (30).
It is important to recognize that the study herein was conducted on a small number of volunteer farms; therefore, caution is needed in extrapolating the results. In comparison to other Ontario beef farms, our study farms are generally larger than Ontario average farms [Ontario cow-calf farms have on average 20–25 cows and feedlots have on average 175 animals (31)]. The impact of these limitations on our overall findings is unknown, and for these reasons caution must be expressed regarding extending the conclusions of this study to the larger Ontario beef industry. Also, the combination farms, which had both cow-calf and feedlot operations (albeit for the most part on different sites), may have additional correlation that we did not adjust for, as they would be more likely to have similar isolates compared with other farms. The collection of pooled isolates occurred between January and August and seasonal patterns of disease and antimicrobial usage, and their effects on resistance patterns could not be assessed. Also, in describing the findings as overall prevalences of resistance and depicting them in the MIC tables, the clustering of the data is not taken into consideration. For example, though ceftiofur resistance was noted in 7 pooled feedlot isolates, 6 of these isolates came from the same farm on the same visit, although from 2 different pools. Since the isolates were from separate pools (different locations on the farm) it was not likely that all the isolates came from the same animal, but there might have been a ceftiofur-resistant clone populating the farm on that occasion.
While useful information can be derived from logistic modelling of the binary outcome (resistant or not), future explorations of associations between risk factors for AMR should also consider more advanced quantitative modelling options, such as modelling clustered data using a multinomial outcome to take advantage of the additional information present in the MIC distribution. Despite these limitations, on-farm investigation of AMR prevalence in generic E. coli provides important information at the beginning of the food chain. This assists in hypothesis generation for potential risk factors for AMR, and provides initial baseline information for potential development of on-farm AMR surveillance programs in beef cattle.
Acknowledgments
The authors sincerely thank the volunteer beef producers for participating in this study. We also thank Brent Avery, Graham Thatcher, Susan Willick, Adelle Adams, and Pat Pentney for assistance with the farm visits and/or data entry. The authors also thank Andrea Desruisseau, Tasnina Shireen, and Maria Popa for their assistance with the laboratory work. Significant thanks goes to Dave Leger for his assistance with Excel spreadsheets (and macros for AMR summaries) and to William Sears for his assistance with SAS coding.
Footnotes
This article is part of a PhD thesis, University of Guelph, Guelph, Ontario.
This research was supported by the Public Health Agency of Canada (formerly Health Canada), the Ontario Cattlemen’s Association, the Ontario Ministry of Agriculture, Food and Rural Affairs, and the Ontario Veterinary College PhD Fellowship Program.
References
- 1.Sunde M, Fossum K, Solberg A, Sørum H. Antibiotic resistance in Escherichia coli of the normal intestinal flora of swine. Microb Drug Resist. 1998;4:289–299. doi: 10.1089/mdr.1998.4.289. [DOI] [PubMed] [Google Scholar]
- 2.Kim S, Hu J, Gautom R, Kim J, Lee B, Boyle DS. CTX-M extended- spectrum beta-lactamases, Washington State. Emerg Infect Dis. 2007;13:513–514. doi: 10.3201/eid1303.060479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schroeder C, Zhao C, DebRoy C, et al. Antimicrobial resistance of Escherichia coli 0157 isolated from humans, cattle, swine, and food. Appl Environ Microbiol. 2002;68:576–581. doi: 10.1128/AEM.68.2.576-581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aarestrup FM, Wegener HC. The effects of antibiotic usage in food animals on the development of antimicrobial resistance of importance for humans in Campylobacter and Escherichia coli. Microbes Infect. 1999;1:639–644. doi: 10.1016/s1286-4579(99)80064-1. [DOI] [PubMed] [Google Scholar]
- 5.Government of Canada, Health Canada. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS)-2002. [Last accessed 20 February 2006]; [homepage on the Internet]. Available from http://www.phac-aspc.gc.ca/cipars-picra/index.html.
- 6.DANMAP. DANMAP 2006. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark. [Last accessed 13 September 2007]; [homepage on the Internet]. Available from http://www.danmap.org/pdfFiles/Danmap_2006.pdf.
- 7.National Veterinary Institute. Swedish Veterinary Antimicrobial Resistance Monitoring — 2002. [Last accessed 17 February 2006]; [homepage on the Internet]. Available from http://www.sva.se/pdf/svarm2002.pdf.
- 8.Read R. Antimicrobial resistance of human health importance in Alberta feedlots. Proceedings of the Agriculture’s Role in Managing Antimicrobial Resistance: The Road to Prudent Use Conference; October 23–26, 2005; Toronto, Ontario. [Google Scholar]
- 9.Checkley SL, Campbell JR, Renter DG, Dorin LC, McFall M. Microbial resistance in fecal E. coli isolates of Alberta. Proceedings of the 10th Symposium of the International Society for Veterinary Epidemiology and Economics (ISVEE); November 17–21, 2003; Vina del Mar, Chile. [Google Scholar]
- 10.Checkley S, Campbell J. Antimicrobial resistance of fecal E. coli isolates related to antimicrobial use in feedlot cattle. Proceedings of Agriculture’s Role in Managing Antimicrobial Resistance: The Road to Prudent Use; October 23–26, 2005; Toronto, Ontario. [Google Scholar]
- 11.Van Donkersgoed J. Baseline prevalence of AMR foodborne pathogens in Alberta feedlots. Proceedings of Agriculture’s Role in Managing Antimicrobial Resistance: The Road to Prudent Use; October 23–26, 2005; Toronto, Ontario. [Google Scholar]
- 12.McAllister T, Busz H, Stevenson S, Inglis D, Yanke L, Olson M, et al. Characterization of the ecology of antibiotic resistant E. coli in feedlot cattle and their environment. Proceedings of Agriculture’s Role in Managing Antimicrobial Resistance: The Road to Prudent Use; October 23–26, 2005; Toronto, Ontario. [Google Scholar]
- 13.Gow S. AMR in western Canadian cow-calf herds. Proceedings of Agriculture’s Role in Managing Antimicrobial Resistance: The Road to Prudent Use; October 23–26, 2005; Toronto, Ontario. [Google Scholar]
- 14.Bair C, Reid-Smith R, Irwin R, McEwen S. Antimicrobial Use in the Ontario Beef Industry. Proceedings of the Agriculture’s Role in Managing Antimicrobial Resistance Conference; Toronto, Ontario. October 24–26. [Google Scholar]
- 15.Clinical and Laboratory Standards Institute (CLSI) CLSI/NCCLS document M100-S15. Pennsylvania, USA: 2005. Performance Standards for Antimicrobial Susceptibility Testing: 15th Informational Supplement. [Google Scholar]
- 16.Clinical and Laboratory Standards Institute (CLSI) NCCLS/CLSI document M31-A2. 2. Pennsylvania, USA: 2002. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals: approved standard. [Google Scholar]
- 17.Centers for Disease Control and Prevention (CDC) National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) NARMS 2001 Annual Report. Atlanta, Georgia: U.S. Department of Health and Human Services; 2003. [Google Scholar]
- 18.Veterinary Drugs Directorate (VDD), Health Canada. Categorization of antimicrobial drugs based on importance in human medicine, (date modified Nov. 30, 2006) [Last accessed 19 March 2007]; [homepage on the Internet]. Available from http://www.hc-sc.gc.ca/dhp-mps/consultation/vet/consultations/amr_ram_hum-med_e.html.
- 19.Government of Canada, Public Health Agency of Canada. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS)-2003. [Last accessed 20 February 2006]; [homepage on the Internet]. Available from http://www.phac-aspc.gc.ca/cipars-picra/2003_e.html.
- 20.Dohoo I, Martin W, Stryhn H. Veterinary Epidemiologic Research. University of Prince Edward Island. Prince Edward Island, Canada: AVC Inc; 2003. [Google Scholar]
- 21.Carson CA, Reid-Smith R, Irwin RJ, Martin SW, McEwen S. Antimicrobial use on 24 beef farms in Ontario. Can J Vet Res. 2008;72:109–118. [PMC free article] [PubMed] [Google Scholar]
- 22.Vidovic S, Korber DR. Prevalence of Escherichia coli O157 in Saskatchewan cattle: Characterization of isolates by using random amplified polymorphic DNA PCR, antibiotic resistance profiles, and pathogenicity determinants. Appl Environ Microbiol. 2006;72:4347–4355. doi: 10.1128/AEM.02791-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner B, Dargatz D, Salman M, Morley P, Wittum T, Keefe T. Comparison of sampling techniques for measuring the antimicrobial susceptibility of enteric Escherichia coli recovered from feedlot cattle. Am J Vet Res. 2002;63:1662–1670. doi: 10.2460/ajvr.2002.63.1662. [DOI] [PubMed] [Google Scholar]
- 24.Morley P. Antimicrobial use and resistance in enteric bacteria. [Last accessed 29 August 2006]; Available from www.fda.gov/cvm/Morley_CompletedRpt.htm.
- 25.Sayah RS, Kaneene JB, Johnson Y, Miller R. Patterns of antimicrobial resistance observed in Escherichia coli isolates obtained from domestic- and wild-animal fecal samples, human septage, and surface water. Appl Environ Microbiol. 2005;71:1394–1404. doi: 10.1128/AEM.71.3.1394-1404.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowrance TC, Loneragan GH, Kunze DJ, et al. Changes in antimicrobial susceptibility in a population of Escherichia coli isolated from feedlot cattle administered ceftiofur crystalline-free acid. Am J Vet Res. 2007;68:501–507. doi: 10.2460/ajvr.68.5.501. [DOI] [PubMed] [Google Scholar]
- 27.Gunn GJ, Hall M, Low JC. Comparison of antibiotic resistance for Escherichia coli populations isolated from groups of diarrhoeic and control calves. Vet J. 2003;165:172–174. doi: 10.1016/s1090-0233(02)00244-7. [DOI] [PubMed] [Google Scholar]
- 28.Hoyle DV, Knight HI, Shaw DJ, et al. Acquisition and epidemiology of antibiotic-resistant Escherichia coli in a cohort of newborn calves. J Antimicrob Chemother. 2004;53:867–871. doi: 10.1093/jac/dkh177. [DOI] [PubMed] [Google Scholar]
- 29.Lefebvre B, Malouin F, Roy G, Giguere K, Diarra MS. Growth performance and shedding of some pathogenic bacteria in feedlot cattle treated with different growth-promoting agents. J Food Prot. 2006;69:1256–1264. doi: 10.4315/0362-028x-69.6.1256. [DOI] [PubMed] [Google Scholar]
- 30.White DG, Hudson C, Maurer JJ, et al. Characterization of chloramphenicol and florfenicol resistance in Escherichia coli associated with bovine diarrhea. J Clin Microbiol. 2000;38:4593–4598. doi: 10.1128/jcm.38.12.4593-4598.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farm Issues.com. Media Resource Centre. Housing: Where do beef cattle live? [Last accessed 21 February 2006]; [homepage on the Internet]. Available from http://www.farmissues.com/mediaPortal/beef/beef_basics.asp.


