Abstract
Tumour necrosis factor (TNF)-α, an important proinflammatory cytokine, has been implicated in the pathogenesis of sarcoidosis, a multi-systemic granulomatous disorder of unknown aetiology. Here, we report for the first time the association of TNF haplotypes and genotypes with sarcoidosis and its prognosis in the Indian population. Five potentially functional promoter polymorphisms in the TNFA gene and a LTA_NcoI polymorphism (+252 position) of the LTA gene were genotyped in a clinically well-defined cohort of North-Indian patients with sarcoidosis (n = 96) and their regional controls (n = 155). Serum TNF-α (sTNF-α) and serum angiotensin converting enzyme (SACE) levels were measured and correlated with genotypes and haplotypes. The TNFA_-1031 and TNFA_-863 polymorphisms were identified as markers for disease onset (FET P = 0·006 and 0·042 for TNFA_-1031 and TNFA_-863, respectively). Additionally, the allele A of LTA_NcoI polymorphism was shown to be prevalent in the ‘no treatment’ group (FET P = 0·005), while the G allele was associated with frequent relapses on drug withdrawal (P = 0·057). Furthermore, the TNFA-308G>A and the TNFA-238G>A polymorphisms were found to influence sTNF-α (P = 0·054 and 0·0005, respectively) and SACE levels (P = 0·0017 and 0·056, respectively). The haplotype frequencies were significantly different in the patients and the controls (P = 0·0067). The haplotype GTCCGG was identified as the major risk/susceptibility haplotype (P = 0·003) and was associated with increased SACE levels in the patient population. In conclusion, our study suggests an association of TNF polymorphisms with sarcoidosis.
Keywords: Indian population, LTA, polymorphism, sarcoidosis, TNF-α
Introduction
Sarcoidosis, a multi-systemic granulomatous disorder of unknown aetiology, occurs in almost all populations, with prevalence differing in each population on the basis of geographical location and racial differences. Its manifestations are localized essentially to lung and skin, but the involvement of other sites such as eyes, lymph nodes, parotid glands, heart, liver, spleen, cranial nerves and central nervous system can also occur [1,2]. It affects predominantly the young and middle-aged adults in both sexes [3]. Clinical manifestations are characterized by the presence of non-caseating granulomas and numerous immunological abnormalities. It has been recognized that granulomas are formed as a result of exaggerated cellular immune response to one or more external or self-antigens and subsequent accumulation of hyperactive macrophages and CD4+ T cells at the inflammatory site. Although no evidence for the presence of pathogenic organism(s) has been found at the centre of granulomas, the immunological pattern of cells at the site suggests the role of an exogenous antigen for the cause of disease [4,5]. Additionally, the characteristic features of the cells in sarcoid granulomas share a common immunological pathway, as noticed in other granulomatous disorders such as mycobacterial and fungal infections. The disease is characterized frequently by remissions and relapses. The diagnosis of sarcoidosis is always obscure among Indian patients, particularly because of the prevailing high incidence of tuberculosis in the subcontinent. However, in recent years several studies have reported on the extent of disease severity and the prevalence of sarcoidosis in India [6–8].
Several polymorphisms within the major histocompatibility complex (MHC) class I and class II genes have been implicated in susceptibility to sarcoidosis as well as with disease prognosis [9–12]. The genes for tumour necrosis factor α (TNF-α; TNFA) and lymphotoxin-α (LT-α; LTA) are located on the short arm of chromosome 6 and are linked closely to the region between the class I human leucocyte antigen-B (HLA-B) and class II (HLA D-related; HLA-DR) loci [13]. Both these genes encode proteins that are involved in the immune response and may, therefore, be relevant to the inflammatory reaction in the sarcoid lung [14,15]. In particular, TNF-α is thought to be a principal cytokine in sarcoidosis as it regulates granuloma formation and its sustenance [16]. In addition, TNF-α levels are known to fluctuate with disease activity [17–19].
Although the reported studies on sarcoidosis have indicated heterogeneity in the TNF allelic association in different populations (Table 1) [14,15,20–24], no such study is available from the Indian subcontinent. The aim of the present study was to investigate the potentially functional TNFA and LTA gene polymorphism(s) in Indian patients with sarcoidosis and to determine the association between these polymorphisms and sarcoidosis. In addition, we have attempted to explore whether any of these polymorphisms may be related to the clinical manifestations, onset and the associated features of sarcoidosis. We also attempt to correlate these polymorphisms with serum TNF-α (sTNF-α) and serum angiotensin converting enzyme (SACE) levels.
Table 1.
Association studies of the TNF genes with sarcoidosis.
| S. No. | Reference | Polymorphisms investigated | Location | Population | Results |
|---|---|---|---|---|---|
| 1. | Grutters et al. [20] | -1031, -863, -857, -308, -238 | Promoter | British (UK) and Dutch | •An increase in the TNF_-857T allele in the sarcoid patients was observed (P = 0·003) •Subgroup analysis showed a significant increase in the TNF_308A allele in patients with Lofgren's syndrome |
| 2. | Yamaguchi et al. [24] | -308, -244, -238; TNFB_NcoI (LTA_NcoI) | Promoter; Intron 1 | 110 Japanese patients and 161 control subjects | •No association with sarcoidosis •Identified TNFB*1 (LTA_NcoI_G) allele as the marker for prolonged clinical course in patients |
| 3. | Seitzer et al. [22] | -308; TNFB_NcoI (LTA_NcoI) | Promoter; Intron 1 | 101 pulmonary sarcoidosis patients and 216 controls from Germany | •No association with sarcoidosis •Increased frequency of the TNFA2 (TNF_308 A) in patients with Lofgren's syndrome when compared to the control group (P = 0·0078) and the non-Lofgren's syndrome patient group (P = 0·0212) |
| 4. | Pandey et al. [21] | -308, -238 | Promoter | 278 Caucasian and 219 African American patients and equal number of matched controls | •No association with sarcoidosis in both populations |
| 5. | Swider et al. [37] | -308 | Promoter | 78 sarcoidosis patients and 50 controls from Germany | •No association with sarcoidosis •Increased frequency of the TNFA2 (TNF_308A) in patients with Lofgren's syndrome when compared to the control group |
| 6. | Rybicki et al. [38] | -308 | Promoter | 225 African American nuclear families with a history of sarcoidosis | •No association with sarcoidosis |
| 7. | Takashige et al. [36] | -308; -26 (LTA_NcoI) | Promoter; Intron 1 | 26 cardiac sarcoidosis Japanese patients and 125 normal controls | •The TNFA2 (TNF_308 A) was associated significantly with sarcoidosis (P = 0·001) |
| 8. | Mrazek et al. [15] | -308; LTA+252 (LTA_NcoI) | Promoter; Intron 1 | 114 Czech patients and 425 controls | •LTA + 252*G (LTA_NcoI_G) were more frequent in the patients (P < 0·01) •LTA + 252*G (LTA_NcoI_G) and TNF-308*A were over-represented in patients with Lofgren's syndrome when compared with non-Lofgren's syndrome patients (P = 0·038 and P = 0·04) |
LTA: lymphotoxin-α; TNF: tumour necrosis factor.
Methods
Subjects
Ninety-six unrelated patients with sarcoidosis (mean age 43·5 ± 9 years), who presented to the out-patient department of internal medicine, the chest clinic or admitted to the tertiary care centre, All India Institute of Medical Sciences Hospital, New Delhi during the period 1996–2005 were included in the study. All patients were diagnosed based on the presence of the clinical symptoms, radiographic features compatible with sarcoidosis and biopsy evidence of non-caseating epithelioid cell granuloma. The clinical characteristics and results of the laboratory investigations, including complete haemogram, serum biochemistry and Mantoux test (5TU), were recorded carefully during the initial presentation as well as in the subsequent follow-up visits. The clinical stage of the disease was classified based on the chest radiographic findings at the initial presentation. The study cohort thus consisted of three patients in stage 0 (had no pulmonary involvement), 35 in stage I (had bilateral hilar lymphadenopathy without parenchymal infiltrate), 57 patients in stage II (with bilateral hilar lymphadenopathy along with parenchymal infiltrate) and one patient had stage III that had parenchymal infiltrate without hilar lymphadenopathy. As there was only one patient in stage III, he was therefore grouped with stage II for analysis. Definitive histopathological evidence for disease was obtained frequently from biopsies of liver (n = 3), lymph nodes (n = 24), skin (n = 23) and lungs (n = 45). Biopsies were performed, albeit rarely, in other sites such as nerve (n = 2), parotid gland and muscle. Multiple biopsies were positive in 89 patients. Patients with evidence of mycobacterial, fungal and/or parasitic infections, and those with a history of exposure to organic/inorganic material known to cause granulomatous lung diseases, were excluded from the study. The institutional ethics committee approved the study proposal, and informed written consent was obtained from each study subject.
Healthy volunteers (referred to as normal controls, n = 155) were recruited from the general population with the same socio-economic status and ethnic background as that of the patients. All individuals were screened negative for a family history of tuberculosis or any other related disease. In addition, these individuals had a negative tuberculin test and their chest radiographs and peripheral blood counts were normal.
Pulmonary functions
The lung volumes and their subdivisions were measured using a constant volume variable body pressure plethysmograph (P. K. Morgan, Chatham, Kent, UK), as described previously [25]. Pulmonary diffusing capacity (DLco) was measured by the steady state technique using Rahn and Otis end-tidal sampler for obtaining alvelolar air [9].
Serum TNF-α and SACE activity measurement
TNF-α levels in serum were determined by the OPTEIAtm enzyme-linked immunosorbent assay (ELISA) kit (BD Biosciences, Franklin Lakes, NJ, USA), as per the manufacturer's instructions. SACE activity was measured in 72 patients with sarcoidosis and 141 normal controls, as described previously [26,27]. Running reference ACE activity in our laboratory during study period was 12–35 U/l.
Genotyping
Genomic DNA was extracted from peripheral blood leucocytes using the modified salting-out procedure [28] and genotyped for TNFA-1031T>C, TNFA-863C>A, TNFA-857T>C, TNFA-308G>A, TNFA-238G>A and LTA_NcoI polymorphisms, as described elsewhere [29]. The accuracy of genotyping was confirmed by direct sequencing of the DNA samples (n = 5) for all three respective genotypes for all the loci investigated.
Statistical analysis
The allele frequencies were calculated and agreement with Hardy–Weinberg equilibrium was tested using a χ2 goodness-of-fit test for each locus. The association between two categorical variables was evaluated by likelihood ratio (LR) χ2 and Fisher's exact tests. All statistical tests performed were two-tailed. Odds ratios (ORs) were calculated with their 95% confidence intervals (CIs). Analysis of variance (anova) was performed to test the effect of the polymorphisms and haplotypes on sTNF-α and SACE levels. Haplotypes for each individual were inferred using phase statistical software [30]. To determine the primary difference in the haplotype frequencies, clump22 software was used with 1 000 000 Monte Carlo simulations [31].
Results
Association of TNF polymorphisms with sarcoidosis
The allele frequencies of the investigated TNF polymorphisms in the study population are summarized in Table 2. The distribution of genotype frequencies for all the polymorphisms investigated was consistent with Hardy–Weinberg expectations in both control and patient groups (P > 0·05). A trend for association was observed for the LTA_NcoI[LR χ2 = 2·95, d.f. = 1, P = 0·083, OR = 1·45, 95% CI = (0·95, 2·19)] and TNFA_-1031[LR χ2 = 3·01, d.f. = 1, P = 0·086, OR = 0·71, 95% CI = (0·48, 1·05)] polymorphisms; however, this significance was lost on applying correction for multiple testing (raw P-values are multiplied by 6; pc = 0·50 for LTA_NcoI and 0·52 for TNFA_-1031 polymorphisms).
Table 2.
Allele frequencies of the polymorphisms investigated in the TNFA and LTA genes. Data are given in absolute numbers with percentages in parentheses.
| Polymorphism | Allele | Sarcoidosis patients (2n = 192) | Control subjects (2n = 310) | FET P-value |
|---|---|---|---|---|
| NcoI | A | 138 (71·88) | 244 (78·72) | 0·086 |
| G | 54 (28·13) | 66 (21·29) | ||
| -1031 | C | 54 (28·13) | 110 (35·48) | 0·096 |
| T | 138 (71·88) | 200 (64·52) | ||
| -863 | A | 54 (28·13) | 100 (32·26) | 0·370 |
| C | 138 (71·88) | 210 (67·74) | ||
| -857 | C | 171 (89·06) | 276 (89·03) | 1·00 |
| T | 21 (10·94) | 34 (10·97) | ||
| -308 | A | 11 (5·73) | 27 (8·71) | 0·297 |
| G | 181 (94·27) | 283 (91·29) | ||
| -238 | A | 7 (3·65) | 14 (4·52) | 0·819 |
| G | 185 (96·35) | 296 (95·48) |
Association of TNF polymorphisms in sarcoidosis subgroups
To determine the role of specific TNF alleles with disease prognosis, patients were subgrouped into different categories based on disease onset, radiographic severity and extrapulmonary manifestations such as joint, skin and ocular involvement. Individually, the TNFA_-1031 polymorphism and TNFA_-863 polymorphisms showed a significant association with the disease onset (FET P = 0·006 and 0·042 for TNFA_-1031 and TNFA_-863, respectively) with a high frequency of the C allele at -1031 and A allele at -863 loci in patients with insidious onset and vice versa (Fig. 1). None of the other polymorphisms showed a significant association with the other subgroups.
Fig. 1.
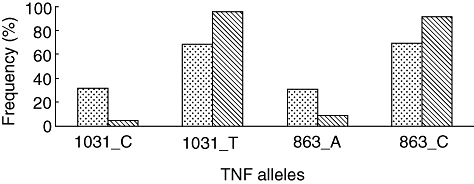
Selected TNF allele frequencies with the clinical category of onset − insidious (dotted bars) and acute (hatched bars) in patients with sarcoidosis.
Association of TNF polymorphisms with treatment and follow-up
The course of the disease was monitored and the patients were subgrouped on the basis of chronic disease, duration beyond 2 years and analysed for the distribution of the polymorphisms. None of the studied polymorphisms revealed a significant association with the course of the disease. Furthermore, of the total patients with sarcoidosis, 86 were followed-up for a period of 7 years and the laboratory tests and pulmonary functions were measured before and during the treatment. Of these, 12 patients required no treatment (no treatment group), and in 11 patients vital capacity (VC) and DLco showed a significant improvement for the duration of treatment with corticosteroids. However, these patients showed deterioration in pulmonary functions after stopping the treatment and required frequent courses of steroids. On the other hand, 63 patients showed more than 10% improvement in VC and DLco with prednisolone treatment and their pulmonary function remained stable for the period of follow-up. When analysis was performed with respect to treatment and response (no treatment group versus average and good combined), the allele A of LTA_NcoI was found to be prevalent in the ‘no treatment’ group (FET P = 0·005). Further analysis in patients who had frequent relapses of symptoms (24 of 74) on tapering off the dosages of prednisolone showed an over-representation of the G allele of LTA_NcoI (FET P = 0·057) and a trend for the C allele of the TNFA_-863 (FET P = 0·094), compared with the patients with no relapses.
Association of TNF haplotypes with sarcoidosis
Statistical software, phase, showed the presence of 13 haplotypes in the 96 patients and 155 healthy controls used for the study. However, only five haplotypes were present at frequencies above 5% in the study population (Fig. 2). Because the counts per haplotypes were very low, Clump22 was used with 1 000 000 Monte Carlo simulations. Haplotype frequencies showed highly significant differences between the cases and controls [normal T1 χ2 = 27·48, d.f. = 12, P = 0·0067, maximum (T4) χ2 = 0·0075].
Fig. 2.
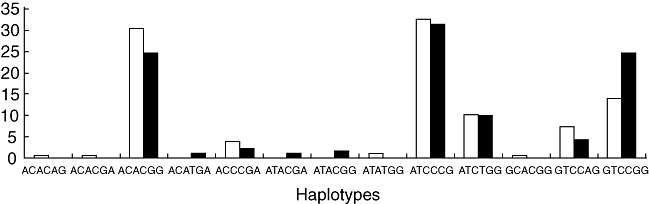
The frequency distribution of the haplotypes of the TNFA and LTA genes in sarcoidosis patients (dark bars) and unrelated controls (light bars). The haplotypes were plotted on the x-axis and their respective relative frequencies (%) on the y-axis.
Individually, the haplotype GTCCGG was the most frequent haplotype in the patient population (LR χ2 = 8·85, d.f. = 1, P = 0·003). The odds of patients rather than controls having the haplotype GTCCGG was 2·01, with 95% CI = 1·27, 3·19. This significance was retained on applying correction for multiple testing (raw P-values multipled by 13; Pc = 0·039). Hence, the haplotype GTCCGG is an important risk/susceptibility haplotype and is associated positively with sarcoidosis in the Indian population.
In addition to this, the distribution of the risk/susceptibility haplotype GTCCGG was significantly different when the patients with acute disease activity (i.e. disease lasts < 2 years) were compared with patients with a chronic disease activity (i.e. disease lasts > 2 years) (FET P = 0·048). None of the other subgroups showed a significant difference in distribution with respect to this haplotype (P > 0·05; data not shown).
Functional correlation of TNF polymorphisms with its level in the serum
To find any functional correlation of the investigated TNF polymorphisms with its level in the serum, sTNF-α levels were measured in 83 unrelated sarcoidosis patients and 112 normal controls. In the patients, the TNFA-308G>A and the TNFA-238G>A polymorphisms were found to influence sTNF-α levels (F-ratio = 3·83, d.f. = 1, P = 0·054; and F-ratio = 13·32, d.f. = 1, P = 0·0005, respectively). The TNFA-308G allele and the TNFA-238A allele were associated with higher sTNF-α (Table 3). However, in the control population no such effect of the polymorphisms on the sTNF-α levels was observed.
Table 3.
Serum tumour necrosis factor (TNF)-α levels in context of different TNF genotypes. Numbers in parentheses indicate [± standard deviation (s.d.)].
| Patients | Controls | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Number | Log TNF-α levels (±s.d.) | F-ratio | d.f. | P-value | Number | Log TNF-α levels (±s.d.) | F-ratio | d.f. | P-value |
| TNFA-308G>A | 3·83 | 1 | 0·054 | 0·1 | 2 | 0·91 | ||||
| AA | – | – | 12 | 1·94 (± 0·08) | ||||||
| GA | 79 | 0·96 (± 0·6) | 18 | 1·64 (± 0·95) | ||||||
| GG | 74 | 1·65 (± 1·02) | 92 | 1·67 (± 0·91) | ||||||
| TNFA-238G>A | 13·32 | 1 | 0·0005 | 0·128 | 1 | 0·72 | ||||
| GA | 7 | 2·82 (± 1·07) | 12 | 1·58 (± 0·71) | ||||||
| GG | 76 | 1·46 (± 0·93) | 100 | 1·68 (± 0·93) | ||||||
Furthermore, when haplotypes (counts ≥ 2) were analysed for sTNF-α levels in the patients, a highly significant difference was observed (F-ratio = 3·05, d.f. = 8, P = 0·003). However, sTNF-α levels for the individuals homozygous for the major haplotype GTCCGG were not significantly different from the individuals lacking it (P > 0·05).
Additionally, when the patient subgroups were analysed with respect to sTNF-α levels, it was observed that the patients with insidious onset had higher sTNF-α (1·64 ± 1·05) when compared with patients with acute onset (0·96 ± 0·25) (F-ratio = 2·84, d.f. = 1, P = 0·096). Similarly, higher sTNF- α (F-ratio = 3·32, d.f. = 1, P = 0·07) was detected in the patients with chronic manifestations (log 1·46 ± 1·03) when compared with those patients with acute manifestations (log 1·92 ± 0·96).
Effect of TNF polymorphisms on SACE levels
Because SACE levels have been found to be affected by sarcoidosis and have been used as markers in corroborating the diagnosis, assessing the likelihood of spontaneous remission, confirming the clinical status of the patient and determining the adequacy of steroid therapy for the disease [32–35], SACE levels were measured from 72 unrelated sarcoidosis patients and 141 normal controls. Although no significant difference was observed in the patients with respect to the disease onset, stage, course and treatment and with the tendency of the disease to relapse (P > 0·05), a few interesting observations were made. The only stage III patient had the highest log SACE level (2·32) compared to the stage 0 (1·88 ± 0·15), stage I (1·66 ± 0·24) and stage II (1·71 ± 0·25) patients. In addition to this, the patients in the ‘no-treatment group’ had the lowest log SACE levels (1·59 ± 0·16) compared to the patients who showed an average (1·73 ± 0·22) or good response (1·72 ± 0·28) to the administered drugs. However, none of these differences were statistically significant (P > 0·05). Furthermore, the genetic effects of the polymorphisms in the TNF genes were also investigated on the SACE levels and it was found that the TNFA-308G>A and the TNFA-238G>A polymorphisms influenced ACE levels in the patients' sera samples (F-ratio = 10·622, d.f. = 1, P = 0·0017 and F-ratio = 3·79, d.f. = 1, P = 0·056, respectively). The presence of the GG genotype for TNFA-308 and GA genotype for TNFA-238 polymorphism was associated with higher ACE levels (Table 4). No such effect was observed in the control population (Table 4).
Table 4.
Serum angiotensin converting enzyme (ACE) levels in context of different tumour necrosis factor (TNF) genotypes and haplotypes. Numbers in parenthesis indicate [± standard deviation (s.d.)].
| Patients | Controls | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Number | Log ACE levels (± s.d.) | F-ratio | d.f. | P-value | Number | Log ACE levels (± s.d.) | F-ratio | d.f. | P-value |
| TNFA-308G>A | 10·62 | 1 | 0·0017 | 0·08 | 2 | 0·93 | ||||
| AA | – | – | 2 | 1·51 (± 0·16) | ||||||
| GA | 8 | 1·44 (± 0·09) | 20 | 1·51 (± 0·11) | ||||||
| GG | 64 | 1·74 (± 0·25) | 119 | 1·52 (± 0·11) | ||||||
| TNFA-238G>A | 3·79 | 1 | 0·056 | 1·05 | 1 | 0·31 | ||||
| GA | 5 | 1·91 (± 0·24) | 14 | 1·55 (± 0·13) | ||||||
| GG | 67 | 1·69 (± 0·11) | 127 | 1·52 (± 0·11) | ||||||
| Haplotype | ||||||||||
| GTCCGG/GTCCGG | 6 | 1·91 (± 0·43) | 5·06 | 2 | 0·0082 | 3 | 1·50 (± 0·17) | 1·30 | 2 | 0·274 |
| GTCCGG/Non-GTCCGG | 24 | 1·74 (± 0·24) | 30 | 1·49 (± 0·12) | ||||||
| Non-GTCCGG/Non GTCCGG | 42 | 1·66 (± 0·22) | 108 | 1·53 (± 0·11) | ||||||
Further, when haplotypes (counts ≥ 2) were analysed with respect to SACE levels in the patients, a significant difference was observed (F-ratio = 2·09, d.f. = 8, P = 0·04). Individuals homozygous for the risk haplotype GTCCGG had higher SACE levels than the others (F-ratio = 2·74, d.f. = 6, P = 0·07, Table 4).
Discussion
This is the first comprehensive study on the association of TNF alleles with sarcoidosis from the Indian subcontinent. Here, we have concentrated on five potentially functional promoter polymorphisms in the TNFA gene and also on a polymorphism present in intron 1 (+252) of the LTA gene (LTA_NcoI) which has been associated earlier with the variation in TNF-α production and sarcoidosis.
Our study, performed in a cohort consisting of 96 sarcoidosis patients (with varying degrees of severity) and 155 normal controls, suggests that the polymorphisms and haplotypes in the TNF genes are associated with sarcoidosis and its subphenotypes in the North Indian population. Even though the patients recruited for the study had substantial diagnostic classifications and had undergone rigorous testing and follow-up during the course of the study, none of the polymorphic markers investigated in the study showed any significant association with sarcoidosis individually. A marginal association was observed for the LTA_NcoI and TNFA_-1031 polymorphisms with sarcoidosis, while highly significant differences in the distribution of both TNFA_-1031 and TNFA_-863 polymorphisms were observed when the sarcoidosis patients were subdivided into those with acute and chronic disease onset, thereby suggesting their roles as promising markers of sarcoidosis onset in the Indian population. Additionally, the allele A of LTA_NcoI polymorphism was shown to be prevalent in the ‘no treatment’ group (FET P = 0·005), while the G allele was associated with frequent relapses on drug withdrawal (P = 0·057). An earlier study identified that the G allele (allele 1) for this polymorphism was associated with a clinically prolonged clinical course and a reduced probability of remission in the Japanese population [24]. The differences in the results obtained could be due to ethnic variations that persist in different populations, study design and patient selection criteria. In contrast to our study, their study was limited to patients with a relatively benign course who did not require corticosteroid therapy [24].
It is interesting to note here that although the TNFA-308G>A and the TNFA-238G>A polymorphisms failed to show any significant association with sarcoidosis, these polymorphisms were found to influence the sTNF-α (Table 3) and SACE levels (Table 4). Also, no significant differences were observed when these sTNF-α and SACE levels were compared in the control population (Tables 3 and 4, respectively). Thus, the differences observed in the sTNF-α and SACE levels for the different genotypes for these polymorphisms could therefore be attributed to other factors, such as clinical disease severity, onset, etc. Additionally, earlier studies carried out in populations of diverse origins failed to find the association of these polymorphisms with sarcoidosis [20,21,24].
As shown in Table 1, several case–control [15,20–22,24,36,37] as well as family-based studies [38] from a range of ethnic backgrounds were conducted for the TNFA-308G>A polymorphism for investigating its association with sarcoidosis. Most of these studies report an association of the TNFA-308 A allele (allele 2) with a clinical phenotype, Lofgren's syndrome, while no significance was observed when the sarcoidosis patients were compared with the controls [15,20,21,24,37]. Consistent with these findings, our study also failed to detect any significant association for this polymorphism with sarcoidosis. Additionally, the incidence of the Lofgren's syndrome is very rare in the Indian population [2]; the analysis for this polymorphism could not be performed with this phenotype.
Previous in vivo and in vitro studies conducted for the TNFA-308G>A polymorphism have been somewhat confusing. Whereas there are studies which report the minor TNFA-308 A allele (allele 2) to be associated with higher inducible levels of gene transcription and TNF-α protein production [39,40], a study measuring the spontaneous and lipopolysaccharide-induced TNF-α release from bronchoalveolar lavage (BAL) cells and peripheral blood mononuclear cells from sarcoidosis patients failed to show the effect of this polymorphism on these cells during the course of active sarcoid inflammation [23]. In our study, the patients homozygous for the TNFA-308G allele had higher sTNF-α when compared with GA heterozygotes (Table 3). Additionally, the SACE levels for these individuals were also significantly higher than the GA heterozygotes (Table 4). This is not surprising, as ACE converts angiotensin I to angiotensin II [41], which has been known to induce TNF-α production from fibroblasts and nuclear factor-κB production from monocytes [42]. Additionally, ACE inhibitors have also shown to inhibit TNF-α production from mononuclear cells [43]. Thus, it might be possible that the two genes TNFA and ACE interact and lead to phenotypes, as observed here. In future, it would be interesting to investigate further the effect of ACE gene polymorphisms on sTNF-α.
It is important to note that the TNFA-238G allele also showed a significant association with lower sTNF-α and SACE levels. It is therefore likely that the TNFA-238 polymorphism plays an important role in modulating the levels of this important cytokine. Additionally, analysis with the software tfsearch (http://www.cbrc.jp/research/db/TFSEARCH.html) predicted that the substitution of G to A at this position resulted in abrogation of the binding of transcription factors such as ADR1 to this site. However, this prediction needs to be verified experimentally in the future. Alternatively, the possibility that other mutations in and around the TNFA gene, linked to the TNFA-238 polymorphism, may influence the expression of TNF-α cannot be excluded.
To supplement further our understanding of the contribution of these genetic variants to sarcoidosis and its associated phenotypes, six-locus haplotypes were constructed and their distribution was compared in the patients and the control population. The haplotype GTCCGG was identified as the major risk/susceptibility haplotype. In addition, the risk haplotype was associated with increased SACE levels in the patient population.
An earlier study carried out in the British and the Dutch populations identified the TNF_-857T allele as the major risk/susceptibility marker for sarcoidosis. In addition to this, they identified a major five-locus haplotype (haplotype 4) to be over-represented in the sarcoidosis patients [20]. Our study failed to detect any significant difference in the distribution of the C and T alleles for this locus in the studied population (P < 0·05; Table 2). However, it would be important to note that the risk haplotype identified in our study (GTCCGG) is an extended haplotype in comparison to their haplotypes 1 and 4 (TNFA-1031T_-863C_-308G_-238G) [20], which showed increased prevalence in the sarcoidosis patients compared to controls.
Our earlier study reported the association of several HLA-DR and HLA-DQ alleles, known to exist in tight LD with TNF genes [20], with sarcoidosis in the Indian population [9], thereby making the determination of the relative roles of each of these genes in the immunogenetics of sarcoidosis extremely difficult. Further high-resolution HLA typing data in addition to fine mapping of the TNF genes would be of help in resolving these problems and, hence, in the elucidation of the genetic basis of sarcoidosis.
In summary, we report here the risk haplotype in the TNF gene cluster, which is associated with sarcoidosis in the Indian population. In addition, we report the association of TNFA_-1031 and TNFA_-863 polymorphisms with disease onset and LTA_NcoI polymorphism with drug response. Also, the TNFA-308G>A and TNFA-238G>A polymorphisms were identified as predictors of sTNF-α and SACE levels. However, further studies on the functionality of these polymorphisms and fine mapping across the MHC region is required to clarify the eventual role of the TNF genes in the immunogenetics of sarcoidosis.
Acknowledgments
We thank all patients and healthy volunteers for participating in this study. The authors acknowledge the help extended by Ms Yogita Dixit, Ms Shweta Tamta and Mr Hemant Misra during the course of this work. The work was supported by the Department of Biotechnology, Government of India on Program Support for Immunoproteomics-based Diagnosis of Infectious Disease (BT/PR5100/MED/12/2005/2004). Shilpy Sharma acknowledges CSIR for her fellowship.
References
- 1.Sharma SK, Mohan A. Sarcoidosis: global scenario and Indian perspective. Indian J Med Res. 2002;116:221–47. [PubMed] [Google Scholar]
- 2.Sharma SK, Mohan A. Uncommon manifestations of sarcoidosis. J Assoc Physicians India. 2004;52:210–14. [PubMed] [Google Scholar]
- 3.Hart LA, Conron M, du Bois RM. Sarcoidosis. Int J Tuberc Lung Dis. 2001;5:791–806. [PubMed] [Google Scholar]
- 4.Hunninghake GW, Costabel U, Ando M, et al. ATS/ERS/WASOG statement on sarcoidosis. Sarcoidosis Vasc Diffuse Lung Dis. 1999;16:149–73. American Thoracic Society/European Respiratory Society/World Association of Sarcoidosis and other Granulomatous Disorders. [PubMed] [Google Scholar]
- 5.Muller-Quernheim J. Sarcoidosis: immunopathogenetic concepts and their clinical application. Eur Respir J. 1998;12:716–38. doi: 10.1183/09031936.98.12030716. [DOI] [PubMed] [Google Scholar]
- 6.Bambery P, Behera D, Gupta AK, et al. Sarcoidosis in north India: the clinical profile of 40 patients. Sarcoidosis. 1987;4:155–8. [PubMed] [Google Scholar]
- 7.Gupta SK, Mitra K, Chatterjee S, Chakravarty SC. Sarcoidosis in India. Br J Dis Chest. 1985;79:275–83. [PubMed] [Google Scholar]
- 8.Sharma SK, Mohan A, Guleria JS. Clinical characteristics, pulmonary function abnormalities and outcome of prednisolone treatment in 106 patients with sarcoidosis. J Assoc Physicians India. 2001;49:697–704. [PubMed] [Google Scholar]
- 9.Sharma SK, Balamurugan A, Pandey RM, Saha PK, Mehra NK. Human leukocyte antigen-DR alleles influence the clinical course of pulmonary sarcoidosis in Asian Indians. Am J Respir Cell Mol Biol. 2003;29:225–31. doi: 10.1165/rcmb.2003-0007OC. [DOI] [PubMed] [Google Scholar]
- 10.Seitzer U, Gerdes J, Muller-Quernheim J. Genotyping in the MHC locus: potential for defining predictive markers in sarcoidosis. Respir Res. 2002;3:6. doi: 10.1186/rr178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinetti M, Luisetti M, Cuccia M. HLA and sarcoidosis: new pathogenetic insights. Sarcoidosis Vasc Diffuse Lung Dis. 2002;19:83–95. [PubMed] [Google Scholar]
- 12.Foley PJ, Lympany PA, Puscinska E, Zielinski J, Welsh KI, du Bois RM. Analysis of MHC encoded antigen-processing genes TAP1 and TAP2 polymorphisms in sarcoidosis. Am J Respir Crit Care Med. 1999;160:1009–14. doi: 10.1164/ajrccm.160.3.9810032. [DOI] [PubMed] [Google Scholar]
- 13.Nedwin GE, Naylor SL, Sakaguchi AY, et al. Human lymphotoxin and tumor necrosis factor genes: structure, homology and chromosomal localization. Nucleic Acids Res. 1985;13:6361–73. doi: 10.1093/nar/13.17.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma OP. Tumor necrosis factor polymorphism in sarcoidosis. Chest. 2001;119:678–9. doi: 10.1378/chest.119.3.678. [DOI] [PubMed] [Google Scholar]
- 15.Mrazek F, Holla LI, Hutyrova B, et al. Association of tumour necrosis factor-alpha, lymphotoxin-alpha and HLA-DRB1 gene polymorphisms with Lofgren's syndrome in Czech patients with sarcoidosis. Tissue Antigens. 2005;65:163–71. doi: 10.1111/j.1399-0039.2005.00370.x. [DOI] [PubMed] [Google Scholar]
- 16.Baughman RP, Lower EE, du Bois RM. Sarcoidosis. Lancet. 2003;361:1111–18. doi: 10.1016/S0140-6736(03)12888-7. [DOI] [PubMed] [Google Scholar]
- 17.Ziegenhagen MW, Benner UK, Zissel G, Zabel P, Schlaak M, Muller-Quernheim J. Sarcoidosis: TNF-alpha release from alveolar macrophages and serum level of sIL-2R are prognostic markers. Am J Respir Crit Care Med. 1997;156:1586–92. doi: 10.1164/ajrccm.156.5.97-02050. [DOI] [PubMed] [Google Scholar]
- 18.Zheng L, Teschler H, Guzman J, Hubner K, Striz I, Costabel U. Alveolar macrophage TNF-alpha release and BAL cell phenotypes in sarcoidosis. Am J Respir Crit Care Med. 1995;152:1061–6. doi: 10.1164/ajrccm.152.3.7663784. [DOI] [PubMed] [Google Scholar]
- 19.Prasse A, Georges CG, Biller H, et al. Th1 cytokine pattern in sarcoidosis is expressed by bronchoalveolar CD4+ and CD8+ T cells. Clin Exp Immunol. 2000;122:241–8. doi: 10.1046/j.1365-2249.2000.01365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grutters JC, Sato H, Pantelidis P, et al. Increased frequency of the uncommon tumor necrosis factor -857T allele in British and Dutch patients with sarcoidosis. Am J Respir Crit Care Med. 2002;165:1119–24. doi: 10.1164/ajrccm.165.8.200110-0320. [DOI] [PubMed] [Google Scholar]
- 21.Pandey JP, Frederick M. TNF-alpha, IL1-beta, and immunoglobulin (GM and KM) gene polymorphisms in sarcoidosis. Hum Immunol. 2002;63:485–91. doi: 10.1016/s0198-8859(02)00399-3. [DOI] [PubMed] [Google Scholar]
- 22.Seitzer U, Swider C, Stuber F, et al. Tumour necrosis factor alpha promoter gene polymorphism in sarcoidosis. Cytokine. 1997;9:787–90. doi: 10.1006/cyto.1997.0224. [DOI] [PubMed] [Google Scholar]
- 23.Somoskovi A, Zissel G, Seitzer U, Gerdes J, Schlaak M, Muller Q. Polymorphisms at position -308 in the promoter region of the TNF-alpha and in the first intron of the TNF-beta genes and spontaneous and lipopolysaccharide-induced TNF-alpha release in sarcoidosis. Cytokine. 1999;11:882–7. doi: 10.1006/cyto.1999.0498. [DOI] [PubMed] [Google Scholar]
- 24.Yamaguchi E, Itoh A, Hizawa N, Kawakami Y. The gene polymorphism of tumor necrosis factor-beta, but not that of tumor necrosis factor-alpha, is associated with the prognosis of sarcoidosis. Chest. 2001;119:753–61. doi: 10.1378/chest.119.3.753. [DOI] [PubMed] [Google Scholar]
- 25.Sharma SK, Pande JN, Singh YN, et al. Pulmonary function and immunologic abnormalities in miliary tuberculosis. Am Rev Respir Dis. 1992;145:1167–71. doi: 10.1164/ajrccm/145.5.1167. [DOI] [PubMed] [Google Scholar]
- 26.Cushman DW, Cheung HS. Spectrophotometric assay and properties of the angiotensin-converting enzyme of rabbit lung. Biochem Pharmacol. 1971;20:1637–48. doi: 10.1016/0006-2952(71)90292-9. [DOI] [PubMed] [Google Scholar]
- 27.Sharma SK, Rao DN, Pande JN, Guleria JS. Serum angiotensin-converting enzyme activity in sarcoidosis. Indian J Med Res. 1987;85:638–44. [PubMed] [Google Scholar]
- 28.Nagpal K, Sharma S, Rao C, et al. TGFbeta1 haplotypes and asthma in Indian populations. J Allergy Clin Immunol. 2005;115:527–33. doi: 10.1016/j.jaci.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 29.Sharma S, Sharma A, Kumar S, Sharma SK, Ghosh B. Association of TNF haplotypes with asthma, serum IgE levels, and correlation with serum TNF-alpha levels. Am J Respir Cell Mol Biol. 2006;35:488–95. doi: 10.1165/rcmb.2006-0084OC. [DOI] [PubMed] [Google Scholar]
- 30.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sham PC, Curtis D. Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet. 1995;59:97–105. doi: 10.1111/j.1469-1809.1995.tb01608.x. [DOI] [PubMed] [Google Scholar]
- 32.DeRemee RA, Rohrbach MS. Serum angiotensin-converting enzyme activity in evaluating the clinical course of sarcoidosis. Ann Intern Med. 1980;92:361–5. doi: 10.7326/0003-4819-92-3-361. [DOI] [PubMed] [Google Scholar]
- 33.Khan AH, Ghani F, Khan A, Khan MA, Khurshid M. Role of serum angiotensin converting enzyme in sarcoidosis. J Pak Med. 1998;48:131–3. [PubMed] [Google Scholar]
- 34.Rodriguez GE, Shin BC, Abernathy RS, Kendig EL., Jr. Serum angiotensin-converting enzyme activity in normal children and in those with sarcoidosis. J Pediatr. 1981;99:68–72. doi: 10.1016/s0022-3476(81)80959-6. [DOI] [PubMed] [Google Scholar]
- 35.Sainani GS, Mahbubani V, Trikannad V. Serum angiotensin converting enzyme activity in sarcoidosis and pulmonary tuberculosis. J Assoc Physicians India. 1996;44:29–30. [PubMed] [Google Scholar]
- 36.Takashige N, Naruse TK, Matsumori A, et al. Genetic polymorphisms at the tumour necrosis factor loci (TNFA and TNFB) in cardiac sarcoidosis. Tissue Antigens. 1999;54:191–3. doi: 10.1034/j.1399-0039.1999.540211.x. [DOI] [PubMed] [Google Scholar]
- 37.Swider C, Schnittger L, Bogunia-Kubik K, et al. TNF-alpha and HLA-DR genotyping as potential prognostic markers in pulmonary sarcoidosis. Eur Cytokine Netw. 1999;10:143–6. [PubMed] [Google Scholar]
- 38.Rybicki BA, Maliarik MJ, Poisson LM, Iannuzzi MC. Sarcoidosis and granuloma genes: a family-based study in African-Americans. Eur Respir J. 2004;24:251–7. doi: 10.1183/09031936.04.00005904. [DOI] [PubMed] [Google Scholar]
- 39.Wilson AG, Symons JA, McDowell TL, McDevitt HO, Duff GW. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci USA. 1997;94:3195–9. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouma G, Crusius JB, Oudkerk PM, et al. Secretion of tumour necrosis factor alpha and lymphotoxin alpha in relation to polymorphisms in the TNF genes and HLA-DR alleles. Relevance for inflammatory bowel disease. Scand J Immunol. 1996;43:456–63. doi: 10.1046/j.1365-3083.1996.d01-65.x. [DOI] [PubMed] [Google Scholar]
- 41.Chagani T, Pare PD, Zhu S, et al. Prevalence of tumor necrosis factor-alpha and angiotensin converting enzyme polymorphisms in mild/moderate and fatal/near-fatal asthma. Am J Respir Crit Care Med. 1999;160:278–82. doi: 10.1164/ajrccm.160.1.9808032. [DOI] [PubMed] [Google Scholar]
- 42.Klahr S, Morrissey J. Angiotensin II and gene expression in the kidney. Am J Kidney Dis. 1998;31:171–6. doi: 10.1053/ajkd.1998.v31.pm9428470. [DOI] [PubMed] [Google Scholar]
- 43.Genctoy G, Altun B, Kiykim AA, et al. TNF alpha-308 genotype and renin–angiotensin system in hemodialysis patients: an effect on inflammatory cytokine levels? Artif Organs. 2005;29:174–8. doi: 10.1111/j.1525-1594.2005.29029.x. [DOI] [PubMed] [Google Scholar]


