Abstract
Recent evidence suggests that probiotic bacteria may stabilize gut barrier function via induction of anti-microbial peptides such as defensins. This study aimed to elucidate the induction mechanism of the human beta defensin-2 (hBD-2) gene by different probiotic lactobacillus strains. The expression of hBD-2 mRNA peaked at 6 h of incubation upon treatment of Caco-2 cells and increased with higher dosage of various probiotic bacteria. Deletion of nuclear factor (NF)-κB and activator protein-1 (AP-1) binding sites on the hBD-2 promoter resulted in a complete abrogation of promoter activation by probiotics. As revealed by the use of specific mitogen-activated protein kinase (MAPK) inhibitors the hBD-2 induction was dependent on the MAPK extracellular regulated kinase (ERK 1/2), p38 and c-Jun N-terminal kinase (JNK), although to varying degrees. Several Lactobacillus strains and VSL#3, a probiotic cocktail of four lactobacilli, three bifidum and one streptococcus species, induced the secretion of the hBD-2 peptide into the culture media as shown by enzyme-linked immunosorbent assay (ELISA). Thus, the present study suggests that lactobacilli and the VSL#3 bacterial mixture strengthen intestinal barrier functions through the up-regulation of hBD-2 via induction of proinflammatory pathways including NF-κB and AP-1 as well as MAPKs.
Keywords: anti-microbial activity, defensins, intestinal epithelial cells, probiotics
Introduction
Probiotics, defined as ‘live microbial food supplements which improve the health of the host’[1], have obtained increasing medical relevance. In the intestine they may prevent the overgrowth of pathogenic bacteria, increase the resistance of the gut to invasion by pathogens and ameliorate disease processes. For example, infant formula with Bifidobacterium bifidum and Streptococcus thermophilus diminished the incidence of acute diarrhoea and rotavirus shedding in infants admitted to hospital [2]. The administration of a synbiotic formula containing Pediococcus pentosaceus, Leuconostoc mesenteroides, Lactobacillus paracasei ssp. paracasei and L. plantarum reduced the infection and sepsis rates in multiple trauma patients [3].
A local dysbiosis with a relative lack of the mucosa-associated lactobacilli seems to be typical for ulcerative colitis [4]. Accordingly, lactobacilli [L. rhamnosus (LGG)] were administered to patients with ulcerative colitis and indeed promoted remission maintenance in ulcerative colitis [5]. However, LGG failed to induce or extend remission in Crohn's disease patients [6]. In contrast to this approach with a single bacterial strain, the administration of multiple probiotic organisms might expand their capacity of immunological modulation. This aspect was followed with VSL#3, a cocktail of eight live bacterial species such as lactobacilli, bifidobacteria and a streptococcus strain. Indeed, several clinical trials report efficient application of this probiotic mixture. For instance, VSL#3 has been shown to prevent the occurrence of pouchitis, an unspecific inflammation of the ileoanal pouch, after restorative proctocolectomy for ulcerative colitis [7]. Moreover, VSL#3 is able to maintain remission in refractory pouchitis following antibiotic therapy [8].
Many mechanisms of action are proposed by which probiotics might enhance mucosal protection against gastrointestinal infections and reduce idiopathic inflammation. The concept of a competition between probiotic and pathogenic bacteria for specific binding sites on intestinal epithelial cells is well established [9, 10]. It is postulated that Saccharomyces boulardii protects against Clostridium difficile-induced enteritis through degradation of toxin molecules and their corresponding receptor [11–13]. Several studies have demonstrated probiotic effects on the barrier function, e.g. enhanced phosphorylation of actinin and occludin in the tight junction region of epithelial cells which inhibited the invasion of enteroinvasive Escherichia coli[14]. Furthermore, probiotics may provoke an increased IgA response, activate leucocytes [15–19] and regulate the cytokine homeostasis [10, 20, 21]. Bacterial DNA of probiotics has been shown to modulate the nuclear factor (NF)-κB pathway in response to tumour necrosis factor (TNF)-α[22]. VSL#3 might maintain remission in pouchitis by inducing an increase of interleukin (IL)-10 and suppressing proinflammatory cytokine tissue levels of patients [23]. In children with acute diarrhoea an enhancement of cytotoxic T suppressor (CD8+) and T helper cells (CD4+) was determined after probiotic bifidobacterium and streptococcus treatment [2]. An improved tissue repair, enhanced phagocytosis of Pseudomonas aeruginosa and a decrease in apoptosis in a burned-mouse model were reported in another study as responses following probiotic lactobacillus treatment [24].
Previously, we demonstrated that specific probiotic strains including lactobacilli up-regulate expression of the anti-microbial peptide human-beta defensin 2 (hBD-2) in Caco-2 cells [25]. The mechanisms of the probiotic-mediated hBD-2 induction have been investigated only for E. coli Nissle 1917, but not for lactobacilli or others. Thus the aim of the present study was to examine by which mechanisms lactobacillus probiotics regulate hBD-2 gene transcription.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains used in this study are shown in Table 1. VSL#3 was obtained as a lyophilized mixture consisting of eight different Gram-positive organisms (B. longum, B. infantis, B. breve, L. acidophilus, L. casei, L. delbrueckii ssp. bulgaricus, L. plantarum and Streptococcus salivarius ssp. thermophilus). Each bag contained 450 billion bacteria which were washed with phosphate-buffered saline (PBS), reconstituted in fetal calf serum (FCS)- and antibiotic-free Dulbecco's modified Eagle's medium (DMEM) and adjusted to the required cell concentration. E. coli Nissle 1917 was grown overnight at 37°C under gentle agitation at 200 r.p.m. in trypticase soy broth (TSB). To obtain bacteria in a linear growth phase, 100 μl of the bacterial suspension were added to 10 ml fresh TSB medium and grown under permanent shaking for 5 h. Heat inactivation was carried out in a water bath at 65°C for 1 h. Bacteria were concentrated by centrifugation and the pellet was washed with PBS and adjusted to a density of 3 × 108 cells/ml with FCS- and antibiotic-free culture medium. Lactobacilli and Pediococcus were grown anaerobically according to De Man, Rogosa and Sharpe (MRS) in MRS broth for 18–24 h in a CO2-enriched atmosphere in an anaerobic jar; 300–500 μl bacteria were transferred into fresh media and grown for another 5–7 h depending on the individual growth rate. The culture supernatant of each bacterial strain was used after dilution with the same dilution factor as the pellet in DMEM. For testing of dose-dependency, bacteria were used at concentrations between 106 and 109 bacteria/ml.
Table 1.
Bacterial strains used in this study.
| Species of subspecies (isolate) | Strain designation/serotype | Type of isolate | Source and/or reference |
|---|---|---|---|
| Esherichia coli Nissle 1917 (EcN) | O6:K5:H1 | Pharmaceutical | †, [25] |
| Lactobacillus acidophilus | PZ 1138 | Industrial | † (‡) |
| L. fermentum | PZ 1162 | Intestinal isolate | † |
| Pediococcus pentosaceus (16:1) ATCC25745 | LMG P-20608 | Intestinal isolate | § |
| L. paracasei ssp. paracasei (F19) | LMG P-17806 | Growing rye | § |
| VSL#3 | ¶ |
Ardeypharm Collection of Strains, Pharma-Zentrale GmbH, Herdecke, Germany.
Catalogue of DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany.
BCCM, Belgian Coordinated Collection of Microorganisms.
Sigma-tau Arzneimittel GmbH, Düsseldorf, Germany.
Cell culture
Caco-2 cells [German Collection of Microorganisms and Cell cultures (DSMZ) ACC (169)] were cultivated in DMEM containing 25 mM HEPES, 2 mM glutamine, 10% FCS, 50 μg/ml gentamicin and 1% non-essential amino acids. The cells were grown as monolayers in 75 cm2 flasks (Greiner, Frickenhausen, Germany) at 37°C in a 5% CO2−95% air atmosphere with 90% humidity. For stimulation experiments, undifferentiated cells were seeded at a density of 2·8 × 105 cells/well in 12-well culture plates (Becton Dickinson GmbH, Heidelberg, Germany). Cells grown to ∼70% confluence in culture wells were incubated with serum- and antibiotic-free medium for at least 12 h. Our intention was to eliminate serum-induced hBD-2 expression and prevent any influence of antibiotics on the immune response. To determine hBD-2 mRNA expression, Caco-2 cells were incubated with the bacteria for 6 h. For signalling pathway studies the mitogen-activated protein kinase (MAPK) inhibitors AG126 (Calbiochem, Darmstadt, Germany), SB203580 and SP600125 (Tocris, Ellisville, MO, USA) were resuspended in dimethyl sulphoxide (DMSO). Cells were pretreated with the specific ERK 1/2, p38 and c-Jun N-terminal kinase (JNK) inhibitors 1 h prior to stimulation with bacteria. Cell viability after inhibitor and DMSO treatment was assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell proliferation assay (ATCC, Manassas, USA), according to the manufacturer's protocol.
Luciferase reporter gene assay
To assess hBD-2 promoter activity, Caco-2 cells were seeded into 12-well culture plates and transfected upon 70% confluence. The luciferase reporter constructs for hBD-2, hBD-2 mut activator protein-1 (AP-1), hBD-2 mutN1+2, and hBD-2 mutN1+2+AP-1 have been described recently [25]. Cells were transfected with 0·5 μg hBD-2 reporter plasmid and 0·05 μg of an internal control Renilla luciferase expression plasmid (phRG-TK; Promega, Madison, WI, USA) by using 1 μl of the transfection reagent FuGENE 6 (Roche Diagnostics GmbH, Mannheim, Germany), according to the manufacturer's protocol. Twenty-four hours after transfection, cells were stimulated for 4·5 h with bacteria. Cells were harvested using 250 μl of passive lysis buffer (Promega) per well. Firefly luciferase activity from the hBD-2-pGL3 reporter vector and Renilla luciferase activity were analysed with the Dual-Luciferase® Reporter Assay System (Promega) using a luminometer (Berthold). Promoter activity was normalized to the activity of the internal Renilla luciferase control.
RNA isolation and cDNA synthesis
At the end of the stimulation experiment, cells were washed with PBS and harvested with TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the supplier's protocol. Subsequently, 1 μg of total RNA was reverse-transcribed into cDNA with oligo(dT) primers and 15 U/μg avian myeloblastosis virus (AMV) reverse transcriptase (Promega), according to standard procedures.
Real-time reverse transcription–polymerase chain reaction (RT–PCR)
Real-time RT–PCR analyses were performed in a fluorescence temperature cycler (LightCycler; Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instructions. cDNA corresponding to 10 ng of RNA served as a template in a 10 μl reaction mixture containing 3 mM MgCl2, 0·5 μM of each primer and 1 × lightCycler-FastStart DNA Master SYBR Green I mix (Roche Diagnostics GmbH). Initial denaturation at 95°C for 10 min was followed by 45 cycles, each cycle consisting of 95°C for 15 s, the primer-specific annealing temperature for 5 s and elongation at 72°C for 15 s. For hBD-2 (sense 5′-ATCAGCCATGAGGGTCTTGT-3′; anti-sense 5′-GAGACCACAGGTGCCAATTT-3′) the annealing temperature was set at 62°C. Amplification using these primers resulted in a 172 base pair (bp) fragment. As an internal control gene we used glyceraldehyde-3-phosphate dehydrogenase (GAPDH), one of the most commonly used housekeeping genes. For GAPDH (sense 5′-CCAGCCGAGCCACATCGCTC-3′; anti-sense 5′-ATGAGCCCCAGCCTTCTCCAT-3′) we used a touchdown protocol with a primary temperature of 66°C and a target temperature of 60°C. At the end of each run melting curve profiles were achieved by cooling the sample to 65°C for 15 s and then heating slowly at 0·20°C/s up to 95°C with continuous measurement of fluorescence to confirm amplification of specific transcripts. Cycle-to-cycle fluorescence emission readings were monitored and analysed using LightCycler software (Roche Diagnostics GmbH). Melting curves were generated after each run to confirm amplification of specific transcripts. The specificity of the amplification products was verified by subjecting the amplification products to electrophoresis on a 2% agarose gel and visualization by ethidium bromide staining. For the quantitative evaluation plasmids served as an external homologous DNA standard of known number of copies. To create standard curves, the plasmids were serially diluted (1:10) covering the appropriate concentration range. The mRNA expression is given as a ratio between the target gene and GAPDH gene expression.
hBD-2 ELISA
To determine the secretion of hBD-2 peptide by Caco-2 cells upon stimulation with probiotic bacteria, cell culture supernatants were collected at the end of the experiment, 1% bovine serum albumin (BSA) was added as a protein carrier and samples were stored at −20°C. Cationic proteins were extracted overnight at 4°C under constant gentle agitation with a weak cation exchange matrix (Macro Prep CM®; Bio-Rad Laboratories, Hercules, CA, USA), added at a 1:40 ratio of matrix to cell culture supernatant including a protease inhibitor cocktail (PIC) [phenylmethylsulphonyl fluoride (PMSF) 10 μM, pepstatin 10 μg/ml, leupeptin 10 μg/ml]. Subsequently the beads were washed with ammonium acetate and the absorbed cations were eluted twice with 5% acetic acid [26]. The eluate was lyophilized under vacuum and resuspended in 0·01% acetic acid (+PIC). This solution was finally tested for hBD-2 by ELISA as described by Wehkamp et al. [25].
Statistics
Data were analysed using Excel (Microsoft) and GraphPadInstat (version 3·1 for Windows; GraphPad Software, San Diego, CA, USA). For the description of random samples, the arithmetic mean ± standard deviation (s.d.) or median ± minimum or maximum values was used. Comparisons with Gaussian distribution were made using the Kolmogorov–Smirnov test. When data were not normally distributed, non-parametric analysis of variance (anova) was performed using the Kruskal–Wallis test with Dunn's post-test.
Results
Time- and dose-dependence of hBD-2 induction by probiotic lactobacilli
First, the time- and dose–response of hBD-2 gene expression upon treatment of Caco-2 cells with defined probiotic lactobacilli was examined. In previous studies we had compared the induction of hBD-2 mRNA expression by the strains L. fermentum, P. pentosaceus and L. acidophilus PZ 1129 with the strong stimulant E. coli Nissle 1917 under standard conditions [25]. In the current studies several lactobacilli strains and a combination with bifidum bacteria (VSL#3) promoted hBD-2 mRNA transcription in a time-dependent manner. The maximal amount of hBD-2 mRNA was expressed after 6 h of incubation decreased remarkably at 12 h and reached basal levels again after 24 h of incubation (Fig. 1a). The highest induction of hBD-2 was elicited by the probiotic cocktail VSL#3, followed by L. fermentum and P. pentosaceus, whereas L. acidophilus PZ 1129 and L. paracasei had only a weak effect. The housekeeping gene expression (GAPDH) remained stable at all time-points.
Fig. 1.
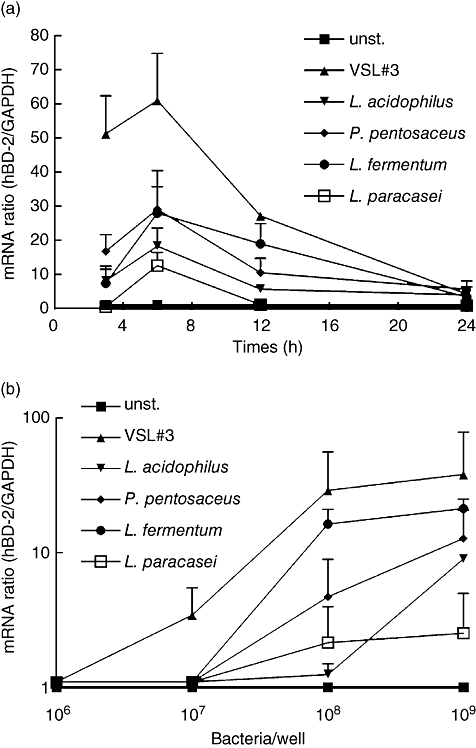
Probiotics induce human-beta defensin 2 (hBD-2) mRNA expression in Caco-2 cells in a time- and dose-dependent manner. (a) Caco-2 cells were treated with five different heat-inactivated bacterial preparations at a concentration of 3 × 108 bacteria per ml for 3, 6, 12 and 24 h. The RNA was isolated, reverse-transcribed into cDNA and the amount of hBD-2 copies was determined by real-time polymerase chain reaction (PCR). The data represent means ± standard deviation (s.d.) of three to four independent experiments performed in duplicate. (b) Caco-2 cells were stimulated for 6 h with 106−109 heat-killed bacteria per ml. hBD-2 mRNA expression was determined by real-time PCR. The data shown represent the means ± s.d. of three independent experiments performed in duplicate.
The induction of hBD-2 was also shown to be dose-dependent (Fig. 1b). At the lowest concentration of 1 × 106 bacteria per ml, hBD-2 expression remained unaffected. Only the VSL#3 combination exerted an inducing effect at the concentration of 1 × 107 cells/ml. P. pentosaceus, L. fermentum and VSL#3 induced hBD-2 mRNA significantly at concentrations of 1 × 108 cells/ml, which was enhanced further at the highest bacteria cell number tested. L. acidophilus induced hBD-2 slightly at 1 × 109 cells/ml but L. paracasei exhibited only a weak influence on hBD-2 expression in Caco-2 cells. Next, we compared the bacterial pellet with the supernatant of different lactic acid bacteria with respect to the hBD-2 expression. Consistently, the supernatant was slightly more active than the pellet (L. fermentum 1·3-fold, L. acidophilus 2·2-fold and P. pentosaceus 3·5-fold), although the difference was not statistically significant.
Effect of lactic acid bacteria on hBD-2 promoter activity in Caco-2 cells
To test whether the probiotic induced hBD-2 mRNA expression is reflected by hBD-2 promoter activation, Caco-2 cells were transfected with hBD-2 luciferase reporter constructs (Fig. 2a). The hBD-2 promoter was activated by E. coli Nissle 1917 (6·8-fold), P. pentosaceus (2·5-fold), L. acidophilus PZ 1129 (2·7-fold), L. fermentum (4·5-fold) and VSL#3 (2·4-fold).
Fig. 2.
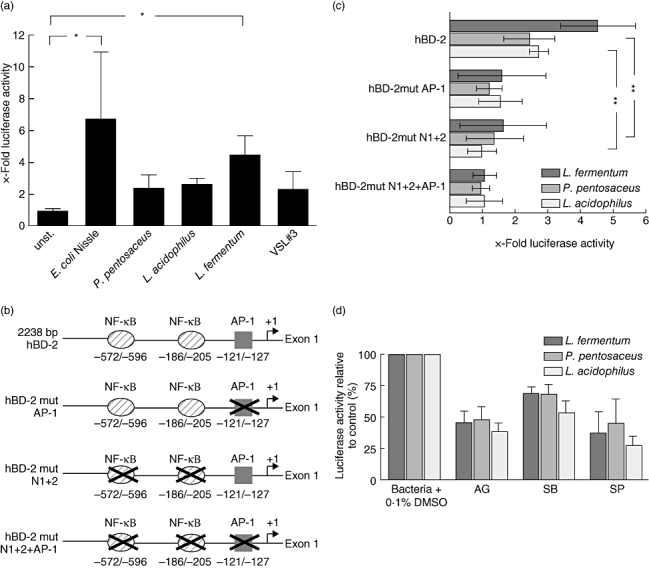
Probiotics activate the human-beta defensin 2 (hBD-2) promoter. (a) Caco-2 cells were transiently transfected with a luciferase gene reporter vector containing the hBD-2 promoter and an internal control Renilla luciferase plasmid as described previously [25]; 24 h after transfection the cells were stimulated for 4·5 h with 3 × 108 bacteria per ml. hBD-2 promoter activity was determined as a ratio of firefly and Renilla luciferase activities. The data represent means ± standard deviation (s.d.) of three experiments performed in duplicate. The means were normalized to basal unstimulated luminescence of controls, set at 1. P < 0·05 was considered significant. Relevance of the nuclear factor (NF)-κB and activator protein-1 (AP-1) binding sites for hBD-2 promoter activation by the strains Lactobacillus fermentum, Pediococcus pentosaceus and L. acidophilus PZ 1138. (b) The hBD-2 promoter constructs used are diagrammed. Nucleotide positions are marked relative to the hBD-2 transcription start. Two NF-κB sites and one AP-1 site in the hBD-2 promoter (base pairs −2338 to −1), linked to the luciferase gene, were mutated in different combinations. (c) Caco-2 cells were transfected with the wild-type (−2338-luc) or mutated hBD-2 promoter luciferase plasmids together with the internal control Renilla plasmid. After transfection, cells were incubated with 3 × 108 bacteria per ml. The ratio of firefly and Renilla luciferase activity was determined via luminescence measurement. The data are means ± s.d. of three independent experiments performed in duplicate. P < 0·01 was considered very significant. (d) Inhibition of the mitogen-activated protein kinases (MAPK) extracellular regulated kinase (ERK) 1/2, p38 and c-Jun N-terminal kinase (JNK) partially blocks L. fermentum-, Pediococcus- and L. acidophilus PZ 1138-mediated hBD-2 promoter activation in Caco-2 cells; 50 μM AG126 (ERK 1/2), 10 μM SB203580 (p38) and 10 μM SP600125 (JNK) were added to hBD-2 transfected Caco-2 cells 1 h prior to stimulation with 3 × 108 bacteria per ml. Data are expressed as a percentage of the control response (bacteria with dimethylsulphoxide 0·1%) and are represented as the mean ± s.d. of duplicate values from three to four independent experiments.
Involvement of transcription factors NF-κB and AP-1 in hBD-2 promoter activation
We then asked whether the binding of NF-κB and AP-1 to their respective binding sites on the hBD-2 promoter is essential for the probiotic-induced human β-defensin-2 gene transcription (Fig. 2b and c). The mutation of two NF-κB recognition motifs reduced luciferase expression after stimulation with L. fermentum by 64% and P. pentosaceus by 44%, and with L. acidophilus PZ 1129 it was abolished completely. After deletion of the binding site for the transcription factor AP-1, the promoter activation by L. fermentum, P. pentosaceus and L. acidophilus was diminished by 65%, 50% or 43%, respectively. Thus AP-1 binding to the hBD-2 promoter might be as important as NF-κB binding for hBD-2 promoter activation initiated by L. fermentum and P. pentosaceus. Mutation of the three transcription factor binding sites completely abrogated any promoter activation by the bacterial strains.
Involvement of ERK 1/2, JNK and p38 in hBD-2 promoter activation
To gain more insight into the signalling pathways of probiotic strains, we blocked three main MAPK pathways with specific inhibitors: p38 using SB203580, JNK by SP600125 and ERK 1/2 through AG126. For all experiments, IL-1β stimulation was used simultaneously as a positive control (data not shown). Inhibition of ERK 1/2 reduced hBD-2 promoter activation through L. fermentum, P. pentosaceus and L. acidophilus by up to 60% (Fig. 2d). The inhibition of p38 decreased promoter activation by L. fermentum or P. pentosaceus by 31%, and that by L. acidophilus by 41% compared to the bacterial treatment with DMSO as control. Luciferase induction by L. fermentum was inhibited by 63%, by P. pentosaceus by 55% and by L. acidophilus by 75%, respectively, when cells were preincubated with the JNK inhibitor. Thus, JNK and ERK 1/2 definitely play a role in the signalling pathway mediated by these three probiotic strains, whereas p38 seems to be of minor importance. Because VSL#3 contains eight different probiotic strains which probably differ in their mechanisms we did not include this bacterial cocktail in the signalling studies.
The concentrations of MAPK inhibitors AG126, SB203580 and SP600125 used for the cell culture experiments had no influence on cell viability.
Secretion of hBD-2 peptide after stimulation with probiotics
Finally, we tested whether probiotics also promote the secretion of hBD-2 peptide. Culture supernatants from stimulated Caco-2 cells were collected, concentrated, and the hBD-2 peptide was detected by ELISA as described previously [25]. Unlike controls, treatment of Caco-2 cells with VSL#3, L. fermentum, P. pentosaceus and L. acidophilus also resulted in detectable amounts of hBD-2 in the culture supernatant (Fig. 3). Compared to the control E. coli Nissle 1917, these results indicate that other probiotics also have a capacity to induce the synthesis of the anti-microbial hBD-2 peptide.
Fig. 3.
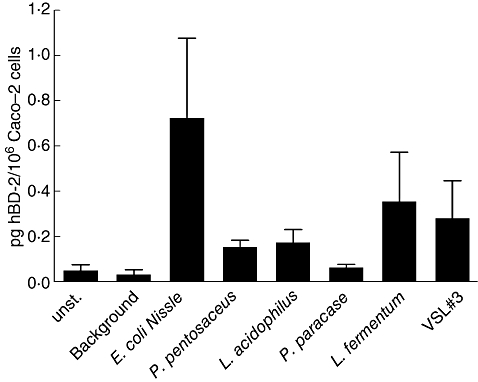
Detection of the human-beta defensin 2 (hBD-2) peptide in culture supernatants from Caco-2 cells treated with probiotics. Caco-2 epithelial cells were incubated for 6 h with 3 × 108 bacteria per ml and hBD-2 released in the culture supernatant was collected. Cationic protein was extracted by MacroPrep CM® cation exchange matrix as described in Methods and hBD-2 was detected by hBD-2 enzyme-linked immunosorbent assay. Data are represented as the mean ± standard deviation of triplicate values from two independent experiments.
Discussion
The present and a previous [25] report suggest that several apathogenic probiotic bacteria including lactobacilli and others induce innate immunity through defensin induction. Notably, the pathogenic strains Salmonella ssp. and Helicobacter pylori have also been reported to induce hBD-2 expression [27, 28]. In contrast, lactobacilli and other probiotics appear to induce the intestinal barrier defence system without provoking inflammatory events in patients. The results detailed above suggest that, nevertheless, this induction is mediated by classical proinflammatory pathways.
The time-dependence experiments showed a similar pattern as already described for E. coli Nissle 1917 [25], with a maximum of hBD-2 induction after 6 h of incubation. At the highest bacteria concentration of 1 × 109 cells/ml the effect of VSL#3 and L. fermentum reached a plateau, in contrast to P. pentosaceus and L. acidophilus. Our results reveal that the potential to induce hBD-2 depends on the lactobacilli tested, merely no class effect. This observation is in accordance with the varying magnitude of dendritic cell activation exerted by different lactobacilli strains [29]. When taking the induction of hBD-2 promoter activation and protein secretion into consideration, L. fermentum was the most potent of the lactobacilli tested and second only to E. coli Nissle 1917. Although VSL#3 induced the highest amount of hBD-2 mRNA compared to the other probiotic strains used, it exerted a weaker capacity of hBD-2 peptide induction than L. fermentum. The differences between promoter activation, mRNA expression and peptide secretion with respect to the order of strains are minor and constitute no evidence for post-transcriptional regulation.
Similar to E. coli Nissle 1917 [25], the hBD-2 gene transcription induced by lactic acid bacteria depended on the transcription factors NF-κB and AP-1, both of which also regulate the expression of proinflammatory cytokines, immune receptors and cell surface adhesion molecules [30, 31]. Activated NF-κB is also found in intestinal epithelial cells of patients with Crohn's disease and ulcerative colitis, which variably induce epithelial defensin formation [32]. Invasive and non-invasive enteric pathogens typically trigger inflammatory responses through NF-κB activation [33–35], whereas many commensal bacteria including various E. coli are inactive [25]. Interestingly, we observed an individually different contribution of NF-κB or AP-1 for hBD-2 activation for each probiotic lactobacillus strain. L. acidophilus stimulated hBD-2 promoter activity mainly through NF-κB, whereas the mutation of the AP-1 binding site alone also reduced L. acidophilus-mediated luciferase gene expression, implicating a role for both transcription factors as synergistically acting partners. P. pentosaceus and L. fermentum seem to induce hBD-2 promoter activity equally by NF-κB and by AP-1 binding. These strains most probably exhibit a differing variety of pathogen-associated molecular pattern molecules, thus leading to subtle distinctions of transcriptional hBD-2 regulation.
Recent investigations confirm the induction of proinflammatory responses by probiotics because B. lactis and L. casei were reported to trigger both the phosphorylation of the p65 subunit of NF-κB and the MAPK p38 [36, 37]. The limited activation of the transcription factors NF-κB and AP-1 may be favourable, as the activation below the inflammation threshold might render the immune system more alert versus hostile confrontations. However, different probiotics may have opposite effects on the NF-κB system [36–39]. In contrast to the above-mentioned transcription factors, the MAPKs ERK 1/2, JNK and p38 were apparently activated by all lactobacillus strains in a similar fashion and ERK 1/2 and JNK were consistently more important than p38.
In previous investigations we found a 13-fold higher hBD-2-inducing capacity by the bacterial culture supernatant of E. coli Nissle 1917 than by the pelleted bacteria indicating a soluble factor as the main responsible inducer [40]. We also therefore compared whether supernatants of L. fermentum, P. pentosaceus and L. acidophilus exerted a stronger activity than the bacterial pellet. The lactobacillus pellet actually displayed no significantly higher hBD-2-inducing capacity than the culture supernatant showing that these strains might rather induce hBD-2 by a bacterial cell wall component. Despite the heat treatment of the bacteria, this component seems not to be largely secreted or shed into the supernatant, as reported for the flagellin-mediated hBD-2 induction by E. coli Nissle 1917 [40]. However, secreted bacterial glycoproteins of probiotics seem to act as important mediators to promote epithelial cell survival, differentiation and growth [41]. The culture supernatant of probiotics has also been shown to exert bacterial killing activity and therefore possesses a potent immunostimulatory capacity [42].
We used heat-killed bacteria to prevent bacterial overgrowth during the experiment and to avoid adverse effects by an eventual alteration of the media composition. Viable bacteria might induce a higher variety of immune responses, but heat-killed L. acidophilus and L. casei were also shown to exhibit the ability to suppress candidiasis in immunodeficient mice [43]. Therefore, probiotics do not necessarily have to be viable for efficient use. It remains to be investigated which bacterial components of probiotic lactobacilli induce the defensin expression and thus have impact on the anti-bacterial activity of the gut.
In conclusion, we believe that the induction of defensins by probiotics including lactobacilli might be an interesting new therapeutic strategy to strengthen innate defence mechanisms.
Acknowledgments
We thank Stig Bengmark and Corinne Enders for the supply of bacteria and Sabine Nuding for the helpful discussions. We gratefully acknowledge the excellent technical support of Kathleen Siegel. This work was supported by the Robert Bosch Foundationand the Deutsche Forschungsgemeinschaft (SFB 617).
References
- 1.Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989;66:365–78. [PubMed] [Google Scholar]
- 2.Saavedra JM, Bauman NA, Oung I, Perman JA, Yolken RH. Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. Lancet. 1994;344:1046–9. doi: 10.1016/s0140-6736(94)91708-6. [DOI] [PubMed] [Google Scholar]
- 3.Kotzampassi K, Giamarellos-Bourboulis EJ, Voudouris A, Kazamias P, Eleftheriadis E. Benefits of a synbiotic formula (Synbiotic 2000Forte (R)) in critically ill trauma patients: early results a randomized controlled trial. World J Surg. 2006;30:1848–55. doi: 10.1007/s00268-005-0653-1. [DOI] [PubMed] [Google Scholar]
- 4.Zhang M, Liu B, Zhang Y, Wei H, Lei Y, Zhao L. Structural shifts of mucosa-associated lactobacilli and Clostridium leptum subgroup in patients with ulcerative colitis. J Clin Microbiol. 2007;45:496–500. doi: 10.1128/JCM.01720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zocco MA, dal Verme LZ, Cremonini F, et al. Efficacy of Lactobacillus GG in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther. 2006;23:1567–74. doi: 10.1111/j.1365-2036.2006.02927.x. [DOI] [PubMed] [Google Scholar]
- 6.Schultz M, Timmer A, Herfarth HH, Sartor RB, Vanderhoof JA, Rath HC. Lactobacillus GG in inducing and maintaining remission of Crohn's disease. BMC Gastroenterol. 2004;4:5. doi: 10.1186/1471-230X-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gionchetti P, Rizzello F, Helwig U, et al. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124:1202–9. doi: 10.1016/s0016-5085(03)00171-9. [DOI] [PubMed] [Google Scholar]
- 8.Mimura T, Rizzello F, Helwig U, et al. Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut. 2004;53:108–14. doi: 10.1136/gut.53.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YK, Puong KY. Competition for adhesion between probiotics and human gastrointestinal pathogens in the presence of carbohydrate. Br J Nutr. 2002;88(Suppl. 1):S101–8. doi: 10.1079/BJN2002635. [DOI] [PubMed] [Google Scholar]
- 10.Otte JM, Podolsky DK. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am J Physiol Gastrointest Liver Physiol. 2004;286:G613–26. doi: 10.1152/ajpgi.00341.2003. [DOI] [PubMed] [Google Scholar]
- 11.Castagliuolo I, LaMont JT, Nikulasson ST, Pothoulakis C. Saccharomyces boulardii protease inhibits Clostridium difficile toxin A effects in the rat ileum. Infect Immun. 1996;64:5225–32. doi: 10.1128/iai.64.12.5225-5232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castagliuolo I, Riegler MF, Valenick L, LaMont JT, Pothoulakis C. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect Immun. 1999;67:302–7. doi: 10.1128/iai.67.1.302-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pothoulakis C, Kelly CP, Joshi MA, et al. Saccharomyces boulardii inhibits Clostridium difficile toxin A binding and enterotoxicity in rat ileum. Gastroenterology. 1993;104:1108–15. doi: 10.1016/0016-5085(93)90280-p. [DOI] [PubMed] [Google Scholar]
- 14.Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC) Gut. 2003;52:988–97. doi: 10.1136/gut.52.7.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaila M, Isolauri E, Soppi E, Virtanen E, Laine S, Arvilommi H. Enhancement of the circulating antibody secreting cell response in human diarrhea by a human Lactobacillus strain. Pediatr Res. 1992;32:141–4. doi: 10.1203/00006450-199208000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Link-Amster H, Rochat F, Saudan KY, Mignot O, Aeschlimann JM. Modulation of a specific humoral immune response and changes in intestinal flora mediated through fermented milk intake. FEMS Immunol Med Microbiol. 1994;10:55–63. doi: 10.1111/j.1574-695X.1994.tb00011.x. [DOI] [PubMed] [Google Scholar]
- 17.Chiang BL, Sheih YH, Wang LH, Liao CK, Gill HS. Enhancing immunity by dietary consumption of a probiotic lactic acid bacterium (Bifidobacterium lactis HN019): optimization and definition of cellular immune responses. Eur J Clin Nutr. 2000;54:849–55. doi: 10.1038/sj.ejcn.1601093. [DOI] [PubMed] [Google Scholar]
- 18.Dogi CA, Perdigon G. Importance of the host specificity in the selection of probiotic bacteria. J Dairy Res. 2006;73:357–66. doi: 10.1017/S0022029906001993. [DOI] [PubMed] [Google Scholar]
- 19.Malin M, Suomalainen H, Saxelin M, Isolauri E. Promotion of IgA immune response in patients with Crohn's disease by oral bacteriotherapy with Lactobacillus GG. Ann Nutr Metab. 1996;40:137–45. doi: 10.1159/000177907. [DOI] [PubMed] [Google Scholar]
- 20.Cukrowska B, Lodinova-Zadnikova R, Enders C, Sonnenborn U, Schulze J, Tlaskalova-Hogenova H. Specific proliferative and antibody responses of premature infants to intestinal colonization with nonpathogenic probiotic E. coli strain Nissle 1917. Scand J Immunol. 2002;55:204–9. doi: 10.1046/j.1365-3083.2002.01005.x. [DOI] [PubMed] [Google Scholar]
- 21.Madsen K, Cornish A, Soper P, et al. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121:580–91. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 22.Jijon H, Backer J, Diaz H, et al. DNA from probiotic bacteria modulates murine and human epithelial and immune function. Gastroenterology. 2004;126:1358–73. doi: 10.1053/j.gastro.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Ulisse S, Gionchetti P, D'Alo S, et al. Expression of cytokines, inducible nitric oxide synthase, and matrix metalloproteinases in pouchitis: effects of probiotic treatment. Am J Gastroenterol. 2001;96:2691–9. doi: 10.1111/j.1572-0241.2001.04139.x. [DOI] [PubMed] [Google Scholar]
- 24.Valdez JC, Peral MC, Rachid M, Santana M, Perdigon G. Interference of Lactobacillus plantarum with Pseudomonas aeruginosa in vitro and in infected burns: the potential use of probiotics in wound treatment. Clin Microbiol Infect. 2005;11:472–9. doi: 10.1111/j.1469-0691.2005.01142.x. [DOI] [PubMed] [Google Scholar]
- 25.Wehkamp J, Harder J, Wehkamp K, et al. NF-kappaB- and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: a novel effect of a probiotic bacterium. Infect Immun. 2004;72:5750–8. doi: 10.1128/IAI.72.10.5750-5758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porter EM, Poles MA, Lee JS, Naitoh J, Bevins CL, Ganz T. Isolation of human intestinal defensins from ileal neobladder urine. FEBS Lett. 1998;434:272–6. doi: 10.1016/s0014-5793(98)00994-6. [DOI] [PubMed] [Google Scholar]
- 27.Ogushi KF, Wada AF, Niidome TF, et al. Salmonella enteritidis FliC (flagella filament protein) induces hu beta-defensin-2 mRNA production by Caco-2 cells. J Biol Chem. 2001;276:30521–6. doi: 10.1074/jbc.M011618200. [DOI] [PubMed] [Google Scholar]
- 28.Wehkamp J, Schmidt K, Herrlinger KR, et al. Defensin pattern in chronic gastritis: HBD-2 is differentially expressed with respect to Helicobacter pylori status. J Clin Pathol. 2003;56:352–7. doi: 10.1136/jcp.56.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen HR, Larsen CN, Kaestel P, et al. Immunomodulating potential of supplementation with probiotics: a dose–response study in healthy young adults. FEMS Immunol Med Microbiol. 2006;47:380–90. doi: 10.1111/j.1574-695X.2006.00109.x. [DOI] [PubMed] [Google Scholar]
- 30.Neish AS. Molecular aspects of intestinal epithelial cell–bacterial interactions that determine the development of intestinal inflammation. Inflamm Bowel Dis. 2004;10:159–68. doi: 10.1097/00054725-200403000-00015. [DOI] [PubMed] [Google Scholar]
- 31.Philpott DJ, Girardin SE, Sansonetti PJ. Innate immune responses of epithelial cells following infection with bacterial pathogens. Curr Opin Immunol. 2001;13:410–16. doi: 10.1016/s0952-7915(00)00235-1. [DOI] [PubMed] [Google Scholar]
- 32.Rogler G, Brand K, Vogl D, et al. Nuclear factor κB is activated in macrophages and epithelial cells of inflamed intestinal mucosa. Gastroenterology. 1998;115:357–69. doi: 10.1016/s0016-5085(98)70202-1. [DOI] [PubMed] [Google Scholar]
- 33.Berin MC, Darfeuille-Michaud A, Egan LJ, Miyamoto Y, Kagnoff MF. Role of EHEC O157:H7 virulence factors in the activation of intestinal epithelial cell NF-kappaB and MAP kinase pathways and the upregulated expression of interleukin 8. Cell Microbiol. 2002;4:635–48. doi: 10.1046/j.1462-5822.2002.00218.x. [DOI] [PubMed] [Google Scholar]
- 34.Gewirtz AT, Rao AS, Simon PO, et al. Salmonella typhimurium induces epithelial IL-8 expression via Ca(2+)-mediated activation of the NF-kappaB pathway. J Clin Invest. 2000;105:79–92. doi: 10.1172/JCI8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Philpott DJ, Yamaoka S, Israel A, Sansonetti PJ. Invasive Shigella flexneri activates NF-kappa B through a lipopolysaccharide-dependent innate intracellular response and leads to IL-8 expression in epithelial cells. J Immunol. 2000;165:903–14. doi: 10.4049/jimmunol.165.2.903. [DOI] [PubMed] [Google Scholar]
- 36.Kim YG, Ohta T, Takahashi T, et al. Probiotic Lactobacillus casei activates innate immunity via NF-kappaB and p38 MAP kinase signaling pathways. Microbes Infect. 2006;8:994–1005. doi: 10.1016/j.micinf.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 37.Ruiz PA, Hoffmann M, Szcesny S, Blaut M, Haller D. Innate mechanisms for Bifidobacterium lactis to activate transient pro-inflammatory host responses in intestinal epithelial cells after the colonization of germ-free rats. Immunology. 2005;115:441–50. doi: 10.1111/j.1365-2567.2005.02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsen CN, Nielsen S, Kaestel P, et al. Dose–response study of probiotic bacteria Bifidobacterium animalis subsp lactis BB-12 and Lactobacillus paracasei subsp paracasei CRL-341 in healthy young adults. Eur J Clin Nutr. 2006;60:1284–93. doi: 10.1038/sj.ejcn.1602450. [DOI] [PubMed] [Google Scholar]
- 39.Petrof EO, Kojima K, Ropeleski MJ, et al. Probiotics inhibit nuclear factor-kappaB and induce heat shock proteins in colonic epithelial cells through proteasome inhibition. Gastroenterology. 2004;127:1474–87. doi: 10.1053/j.gastro.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Schlee M, Wehkamp J, Altenhoefer A, Oelschlaeger TA, Stange EF, Fellermann K. The induction of human beta-defensin-2 by the probiotic Escherichia coli Nissle 1917 is mediated through flagellin. Infect Immun. 2007;75:2399–407. doi: 10.1128/IAI.01563-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132:562–75. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coconnier-Polter MH, Lievin-Le MV, Servin AL. A Lactobacillus acidophilus strain of human gastrointestinal microbiota origin elicits killing of enterovirulent Salmonella enterica Serovar Typhimurium by triggering lethal bacterial membrane damage. Appl Environ Microbiol. 2005;71:6115–20. doi: 10.1128/AEM.71.10.6115-6120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner RD, Pierson C, Warner T, et al. Biotherapeutic effects of probiotic bacteria on candidiasis in immunodeficient mice. Infect Immun. 1997;65:4165–72. doi: 10.1128/iai.65.10.4165-4172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


