Abstract
FOG-2 is a transcriptional co-regulator that is required for cardiac morphogenesis as mice deficient in this factor die during mid-gestation of cardiac malformations. FOG-2 interacts with GATA4 to attenuate GATA4-dependent gene expression. The first 12 amino acids of FOG-2 (the FOG Repression Motif) are necessary to mediate this repression. To determine the mechanism by which the FOG Repression Motif functions, we identified 7 polypeptides from rat cardiac nuclear extracts that co-purified with a GST-FOG-2 fusion protein. All proteins identified are members of the NuRD nucleosome-remodeling complex. Using in vitro binding and co-immunoprecipitation assays, we demonstrate that Metastasis-Associated proteins (MTA)-1, 2 and 3 and Retinoblastoma binding proteins RbAp46 and RbAp48 interact with FOG-2, but not with a mutant form of FOG-2 that is unable to repress transcription. Further, we define a novel domain located in the C-terminal portion of MTA-1 that mediates the FOG-2/MTA-1 interaction. We also demonstrate that knockdown of MTA protein expression dramatically impairs the ability of FOG-2 to repress GATA4 activity. Finally, we show that the zinc finger domain of MTA-1 is required for FOG-2 mediated transcriptional repression and that this domain interacts with RbAp46 and RbAp48 subunits of the NuRD complex. Together, these results demonstrate the importance of FOG-2/MTA/RbAp interactions for FOG-2 mediated transcriptional repression and further define the molecular interactions between the FOG Repression Motif and the NuRD complex.
INTRODUCTION
It is now well established that the modification of chromatin plays a crucial role in the regulation of transcription. The ability of regulatory factors to interact with their target elements requires that the tight repressive structure of chromatin be partially relaxed [1–3]. Two principal mechanisms by which chromatin state is regulated have been described. First, post-translational modifications of histones modulate the interactions of histone amino-terminal tails with DNA, other histones, and non-histone proteins. These modifications change higher order chromatin structure and alter the accessibility of DNA to regulatory factors [2, 4, 5]. Second, an ATP-dependent process carried out by large multi-subunit protein machines—collectively termed chromatin remodeling complexes—alters histone-DNA interactions and leads to the repositioning of histone octamers along the DNA to make critical cis-regulatory elements more accessible to transcription factors [1]. The three best characterized chromatin remodeling complexes, classified based on the identity of their ATP-dependent nucleosome remodeling subunit, are SWI/SNF-type complexes, the ISWI/SNF2L-containing machines, and NuRD complexes [1].
The Nucleosome Remodeling and Histone Deacetylase (NuRD) Complex is an approximately 2 MDa complex that was first identified by biochemical purification from both Xenopus laevis oocytes and human cell lines [6–8]. The NuRD complex comprises at least eight factors including the ATPase Mi2α/β, histone deacetylases HDAC1 and HDAC2, Retinoblastoma binding proteins RbAp46 and RbAp48, Metastasis Associated proteins (MTA)-1, 2 or 3, Methyl-CpG Binding Domain protein 3 (MBD3), and p66. The inclusion of Mi2α/β and HDACs in the NuRD complex physically combines the two major strategies for chromatin remodeling: histone modification and nucleosome remodeling. The association of these two activities supports the notion that chromatin remodeling mediated by the NuRD complex leads to repression of gene expression. Although it is well established that Mi2α/β lends ATP-dependent nucleosome remodeling activity and that HDAC1 and 2 lend histone deacetylase activity to the complex, the function of many of the other subunits is still unclear. MTA-1, for example, was first identified as a gene up-regulated in metastatic breast cancer cell lines [9], but its role in the NuRD complex has remained elusive. The MTA protein family has three members in mammals, all of which have been shown to be associated with the NuRD complex [10]. Some evidence indicates that MTA proteins confer functional specification to the NuRD complex [10, 11]. However, the interactions between MTA and the different members of the NuRD complex as well as interactions among all other subunits remain unexplored.
The Friend of GATA (FOG) family of proteins are transcriptional co-factors that have been shown to be critical for development of the hematopoietic as well as cardiovascular system in flies, frogs, and mice [12–15]. FOG proteins were first identified by their ability to physically interact with the N-terminal zinc fingers of members of the GATA family of transcriptional activators [16–19]. FOG-2 is required for proper cardiac morphogenesis as mice deficient in FOG-2 die of congestive heart failure at ED 13.5 due to multiple heart malformations [14, 15]. Interaction between FOG-2 and the N-terminal zinc finger of GATA3, 4, 5, and 6 results in repression of GATA-mediated transcriptional activation of target promoters [16, 17, 20]. Engineered mice in which the GATA4/FOG-2 interaction is disrupted display cardiac malformations similar to FOG-2 deficient embryos, suggesting that most of FOG-2’s functions in the heart are mediated by its interaction with GATA4 [21].
Our previous work demonstrated that FOG-2 is capable of repressing GATA4-mediated activation of target promoters and that the first twelve amino acids of FOG-2 are required for this repression [22, 23]. Furthermore, fusion of these 12 amino acids to the DNA binding domain of GAL4 demonstrated that this region is sufficient to mediate transcriptional repression even when recruited to a heterologous promoter [23]. Point mutation analysis defined a critical amino acid sequence, RRKQxxPxxI, which is present at the N-termini of FOG proteins from all vertebrate species examined to date as well as at the N-termini of several other partially characterized zinc finger transcriptional repressors. This region thus defines a novel transcriptional repression domain—the FOG Repression Motif—and a superfamily of zinc finger transcriptional repressors.
In this report, we show that the FOG Repression Motif of FOG-2 interacts with the MTA and RbAp proteins of the NuRD complex and that a novel domain in the C-terminal portion of MTA-1 mediates FOG-2/MTA-1 interaction. Consistent with this observation, we also demonstrate that MTA-1 expression is required for maximal FOG-2 repressive activity and that the zinc finger domain of MTA-1 is critical for this repression. Finally, we show that the zinc finger domain of MTA-1 physically interacts with the RbAp subunits of the NuRD complex, suggesting that the MTA/RbAp complex is a critical core necessary for FOG-2-mediated transcriptional repression.
MATERIALS AND METHODS
Plasmid Construction
The constructs p-638ANF GH, pVRβGal, pcDNA-GATA4, pcDNA-FOG-2, pcDNA-FOG-2(1–12), pcDNA-FOG-2(1–12K5A), pGEM7Zf[+]-rMTA1, pcDNA3CF-mMTA2, pcDNA3-Mi-2-β, pcDNA3-HDAC1, pBluescript-HDAC2, pcDNA3-RbAp46, pcDNA3-RbAp48, pcDNA3-p66, and pSPORT-MBD3 have been described previously [23, 24]. The Hey2 expression plasmid pcDNAHey2 was a generous gift of M. Chin [25]. MTA-1 truncations were generated by PCR from pcDNA3-rMTA1 and inserted into the BamHI and XhoI sites of pcDNA3 or pcDNA3-FlagA. For MTA-1 C1(1–450), C2(1–350), C3(1–250), and C4(1–148), the same forward primer was used: (5’-CGGGATCCCGGCATGGCCGCCAACATGTACAGGGTCG-3’). The reverse primers used were: MTA-1 C1(1–450) (5’-CCGCTCGAGCTAGCGTTTCTGCTGGACGTATCTGTCG-3’), MTA-1 C2(1–350) (5’-CCGCTCGAGCTACTGCCGCACGGAGCTGCTGCAATCC-3’), MTA-1 C3(1–250) (5’-CCGCTCGAGCTATTTATCAGCCAGGAGGGTCTTCTGC-3’), and MTA-1 (C4)1–148: (5’-CGGGATCCCGGCATGGCCGCCAACATGTACAGGGTCG-3’). For the generation of MTA-1 N1(148–703), N2(250–703), N3(350–703), and N4(450–703), N5(499–703), and N6 (533–703) the same reverse primer was used: (5’-CCGCTCGAGCTAGTCCTCAATAACAATGGGC-3’). The forward primers used were: MTA-1 N1(148–703) (5’-CGGGATCCCCTGGCTGATAAAGGGGAAATTCG-3’), MTA-1 N2(250–703) (5’-CGGGATCCCCGCGATGAGATGGAGGAGTGG-3’), MTA-1 N3(350–703) (5’-CGGGATCCCACTGTAGTGAATGGCACAGGGACACC-3’), MTA-1 N4(450–703) (5’-CGGGATCCCCATGGCATTCCAGCTCGGAGCAGTGG-3’), MTA-1 N5(499–703) (5’-CGGGATCCCCCCTACATGCCCATCAACAGTGC-3’), or MTA-1 N6 (533–703) (5’-CGGGATCCCCTGCCTCCAGTCAAGCGGCGGC-3’). pcDNA3-MTA-3 was generated from IMAGE clone ID#4038759 from American Type Culture Collection. cDNA was digested using EcoRV and NotI and inserted into the EcoRV and NotI sites of pcDNA3. To generate pGST-FOG-2(1–12) and pGST-FOG-2(1–12K5A), the following double stranded oligonucleotides were cloned into the BamHI-EcoRI site of pGEX-4T-1 (GE Healthcare): FOG-2(1–12), (5’GATCCAGCATGTCCCGGCGAAAGCAGAGTAAACCCCGGCAGATCG-3’ and 5’-AATTCGATCTGCCGGGGTTTACTCTGCTTTCGCCGGGACATGCTG-3’), and FOG-2(1–12K5A), (5’-GATCCAGCATGTCCCGGCGAGCGCAGAGTAAACCCCGGCAGATCG-3’ and 5’-AATTCGATCTGCCGGGGTTTACTCTGCGCTCGCCGGGACATGCTG-3’). pGST-MTA-1(Znf) was generated from pcDNA3-rMTA-1 using PCR and cloned into the BamHI-XhoI site of pGEX-4T-1. The primers used were: (5’-CGGGATCCCAGCAGAAACGTTTGAAAGCAGC-3’) and (5’ - CCGCTCGAGCTATACTACCTGCTTCAGCACCAGTGG-3’).
Affinity Purification
To express GST-fusion proteins, overnight cultures of BL21 cells containing pGEX-4T-1, pGST FOG-2(1–12), or pGST FOG-2(1–12K5A) were diluted 1:10 in LB broth and grown for 2 hours with shaking at 37°C. Isopropyl-β-D-thiogalactopyranoside was added to a final concentration of 1 mM and cultures were incubated at 30°C with agitation for three hours. Bacteria were pelleted, resuspended, and sonicated 3 × 10 seconds on ice. GST fusion proteins were then purified from bacterial extracts using glutathione-sepherose. Equivalent amounts of GST or GST fused to the first 12 amino acids of FOG-2 [GST-FOG2(1–12)] was incubated with 3.6 milligrams of rat neonatal cardiocyte nuclear extract overnight at 4°C in a buffer containing 150 mM NaCl, 50 mM Tris-HCl pH 7.5, 0.5% Igepal (Sigma), complete protease inhibitor cocktail tablet (Roche) and 1 mM DTT. Bound proteins were washed twice in the above buffer with 350 mM NaCl, three times with NaCl raised to 650 mM, followed by an additional wash in 350 mM NaCl. Complexes were purified using glutathione sepharose beads (GE Healthcare), resolved by SDS-PAGE, and visualized by staining with GelCode reagent (Pierce). Protein identification was performed by MALDI-TOF mass spectrometry by the University of Chicago Proteomics Core Facility on each visualized polypeptide. One NuRD subunit, Mi2β, was not identified by mass spectroscopy since this band possessed too little protein, but its presence was confirmed by Western blotting.
In vitro binding assays
NuRD subunits and MTA-1 truncations were in vitro translated in the presence of 35S-labeled Methionine using the TNT Quick Coupled Transcription/Translation Kit (Promega) according to manufacturer’s instructions. Purified GST, GST-FOG-2(1–12K5A), GST-FOG-2(1–12), or GST-MTA-1(Znf) fusion protein was incubated with the in vitro translated proteins. Resultant complexes were purified using glutathione sepharose beads, washed extensively in buffer containing 200mM NaCl, 50mM Tris pH7.5, 0.5% Igepal (Sigma), complete protease inhibitor cocktail tablet (Roche) and 1mM DTT. Complexes were resolved by SDS-PAGE and subject to autoradiography.
Immunoprecipitations
Five milligrams of rat neonatal cardiocyte nuclear extracts were incubated with a control IgG or a rabbit polyclonal antibody to FOG-2 (sc-10755, Santa Cruz) overnight at 4°C in a buffer containing 150 mM NaCl, 50 mM Tris-HCl pH 7.4, 1% Triton X-100 (Sigma), 1 mM EDTA, 1% Sodium deoxycholate, 0.1% SDS, and 1 mM PMSF. Immune complexes were precipitated using protein A sepharose (GE Healthcare) and resolved by SDS-PAGE along with 200 µg nuclear extract. Western analysis was performed using an antibody to MTA-1 (sc-9446, Santa Cruz) or MTA-2 (sc-9447, Santa Cruz).
Cell Culture and Transfections
Murine NIH 3T3 fibroblasts were cultured as previously described [23]. 1.1 × 105 cells were plated per well onto a 12-well plate and cultured overnight. Subsequently, 13.4 pmoles each of siRNAs designed against murine MTA-1, 2 and 3 (Dharmacon) were mixed with 2 ul of Lipofectamine 2000 (Invitrogen) and 50 ul Opti-MEM (Invitrogen) and added to cells with 0.5 ml growth medium lacking penicillin/streptomycin. Twenty-four hours after the siRNA treatment, cells were washed with phosphate-buffered saline (PBS). 300 ng of reporter plasmid (p638 ANF GH), 60 ng of pVRβGal, 200 ng of pcDNA-GATA4, 50 ng of pcDNA3-FOG-2 or 200ng pcDNAHey2, 0–200 ng pcDNA3-rMTA-1, and pcDNA3 to a total of 1 µg of DNA was mixed with 3 µl Superfect (Qiagen) and 75 µl of Opti-MEM and added to cells with 0.4 ml of growth medium. Three hours after the addition of the DNA/Superfect mixture, cells were washed with PBS, and 1 ml of growth medium was added. Forty-eight hours after transfection, medium and lysates were harvested and reporter assays were carried out as previously described [23].
Western Blot Analysis
siRNA-treated NIH 3T3 cells were harvested by trypsinization, washed with PBS, and lysed in SDS-PAGE Loading Buffer (Bio-Rad). Fifty µg whole cell lysates were then analyzed by Western analysis with specific antibodies against MTA-1 (sc-9446, Santa Cruz), or MTA-2 (sc-9447, Santa Cruz).
RESULTS
The FOG Repression Motif interacts with members of the NuRD complex
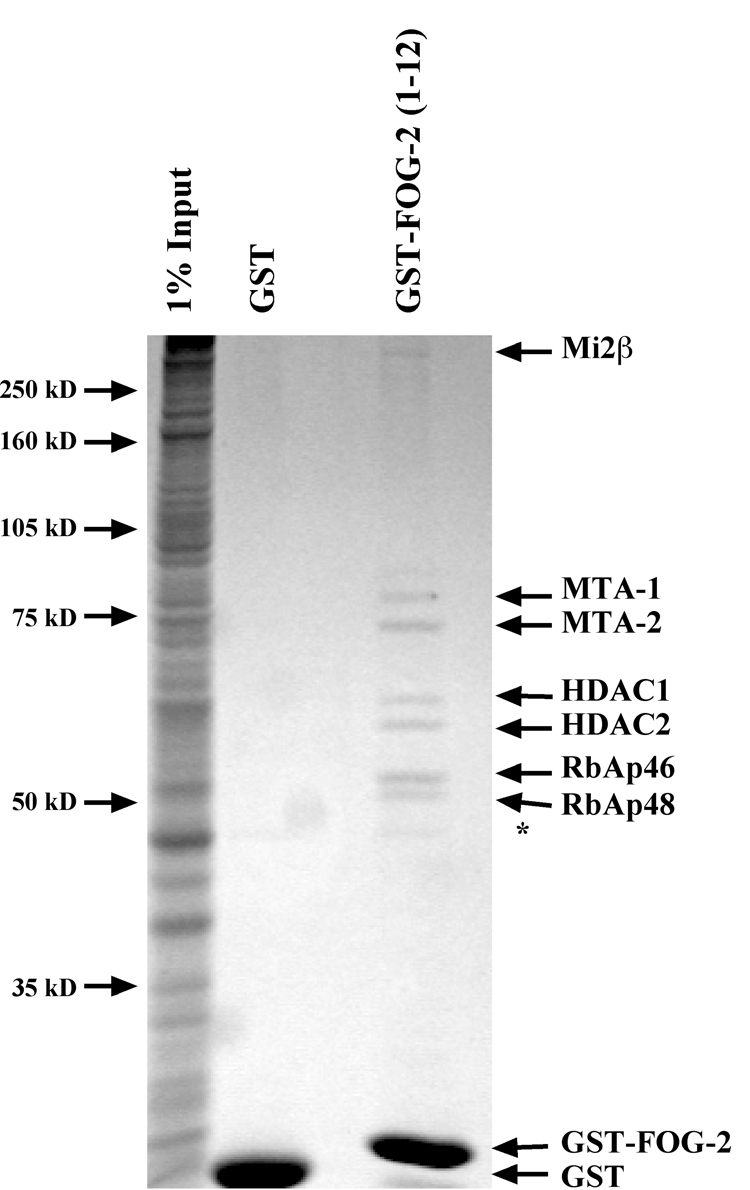
Given the requirement of the FOG Repression Motif for FOG-2 mediated transcriptional repression, we wanted to further elucidate the molecular mechanisms of transcriptional repression mediated by this domain. We reasoned that this motif might be involved in protein-protein interactions to recruit other transcriptional machinery to targeted promoters. To identify such proteins, we generated a bacterial expression plasmid that encoded the first 12 amino acids of FOG-2 fused to glutathione-S-transferase (GST). Purified GST-FOG-2(1–12) was bound to glutathione-sepharose beads and incubated with rat cardiocyte nuclear extract. As a negative control, a parallel sample of nuclear extract was incubated with GST alone. Resultant complexes were washed extensively, resolved by SDS-PAGE and visualized by Coomassie staining (Fig. 1). Seven polypeptides were observed to bind to GST-FOG-2(1–12), but not to GST alone. The polypeptides were identified by MALDI-TOF mass spectrometry or western analysis. All proteins identified correspond to members of the NuRD complex, including Mi2β, MTA-1, MTA-2, HDAC1, HDAC2, RbAp46, and RbAp48 [6–8]. Further, no other proteins were identified, suggesting that the NuRD complex is the critical partner for this repression domain. This result suggests that the FOG Repression Motif of FOG-2 functions as a transcriptional co-repressor by recruiting the NuRD complex to specific promoters.
Figure 1. Affinity Purification of FOG-2 Interacting Proteins.
Affinity purified glutathione-S-transferase (GST) or GST fused to the first 12 amino acids of FOG-2 (GST-FOG-2(1–12)) and was incubated with 3.6 milligrams of rat cardiocyte nuclear extract overnight at 4°C. Complexes were purified using glutathione sepharose beads, resolved by SDS-PAGE, and visualized by staining with GelCode reagent. Protein identification was performed by MALDI-TOF mass spectrometry on each visualized subunit. The * indicates a polypeptide isolated using both GST and GST-FOG-2(1–12).
MTA-1, 2, and 3 interact directly with the FOG Repression Motif
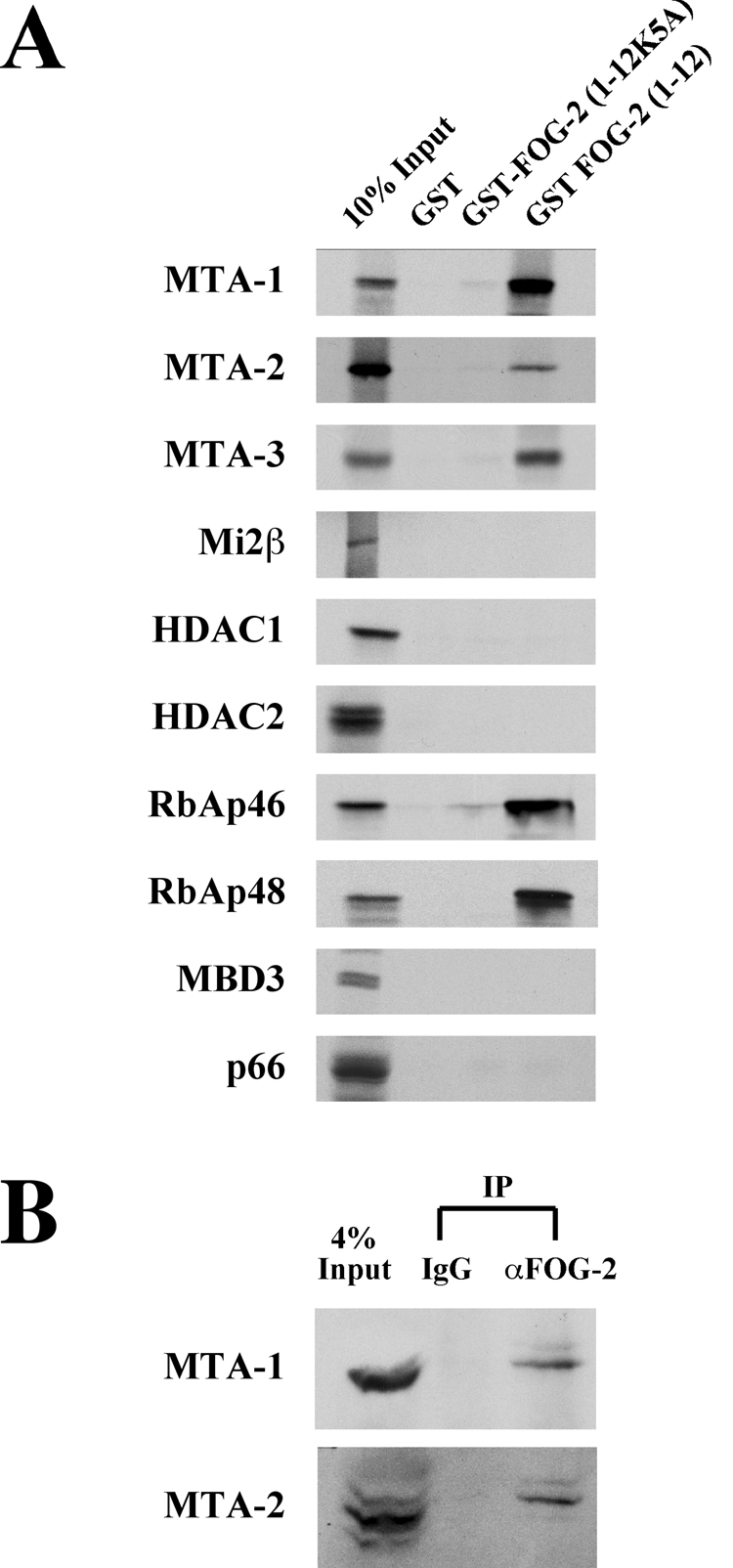
To determine which of the NuRD subunits associates with the FOG-2 repression motif, we performed in vitro binding assays between GST-FOG-2(1–12) and in vitro translated, 35S-labeled subunits of the NuRD complex. As an additional control, we also used a GST-FOG-2 fusion protein containing amino acids 1–12 of FOG-2 with an alanine substitution in amino acid 5 (K5A) that we have previously shown abrogates repression mediated by this motif as well as by the highly similar repression motif found in FOG-1 [23, 24]. The first twelve amino acids of FOG-2 bound MTA-1, -2 and -3, RbAp46, and RbAp48, but not HDAC1, HDAC2, MBD3, p66 or Mi2β (Fig. 2A). The binding of GST-FOG-2 to MTA and RbAp proteins was nearly abolished by the K5A mutation in the FOG Repression Motif, correlating binding of MTA and RbAp proteins with the repressive activity of the FOG Repression Motif.
Figure 2. MTA-1, 2, and 3 Interact with the FOG Repression Motif.
A. Purified GST-FOG-2(1–12) fusion protein was incubated with 35S-labeled, in vitro translated subunits of the NuRD complex. Resultant complexes were washed extensively, resolved by SDS-PAGE, and subject to autoradiography. B. Five milligrams of rat neonatal cardiocyte nuclear extract was incubated with a control IgG or a rabbit polyclonal antibody to FOG-2 overnight at 4°C. Immune complexes were precipitated using protein A sepharose and resolved by SDS-PAGE. Western analysis was performed using an antibody to MTA-1 or MTA-2 as indicated.
To investigate the interaction between FOG-2 and MTA proteins further, we performed co-immunoprecipitation experiments using anti-FOG-2 antibodies and nuclear extracts from rat neonatal cardiocytes. Resultant protein complexes were analyzed by western blot with antibodies to MTA-1 or MTA-2 (Fig. 2B). Both MTA-1 and 2 co-immunoprecipitated with the anti-FOG-2 antibody but not a control antibody. This observation suggests that FOG-2 and the MTA proteins physically interact in cardiocytes when expressed at endogenous levels and is consistent with FOG-2 physically associating with the NuRD complex in vivo.
The FOG interaction domain localizes to the C-terminal portion of MTA-1
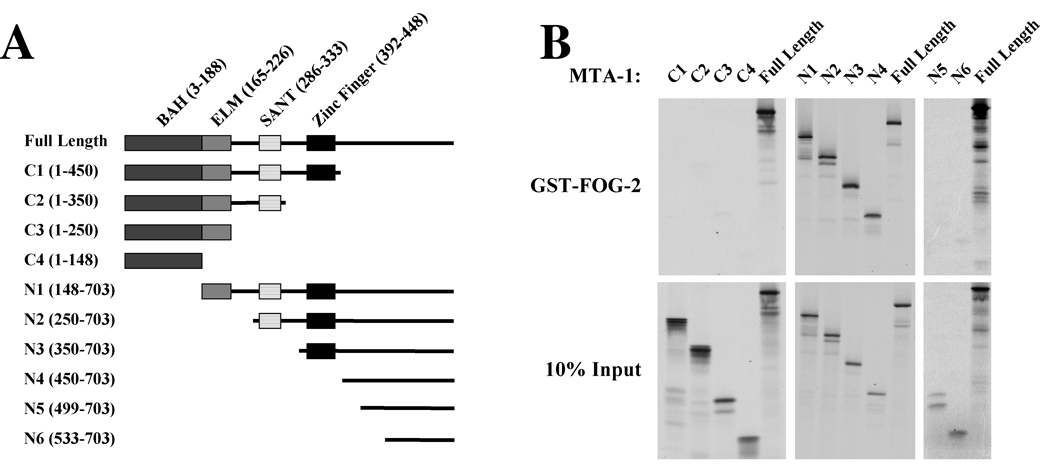
There are three known members of the MTA protein family in mammals. The primary amino acid sequence of MTA-1 is 59% and 58% identical to MTA-2 and MTA-3, respectively. All MTA proteins contain a number of conserved domains in the N-terminal half of the protein [10, 11] (Fig. 3A). These domains include the BAH (Bromo-Adjacent Homology) domain, an ELM2 domain, a SANT domain, and a zinc finger domain [10, 11]. The BAH and ELM2 domains have been proposed to mediate protein-protein interactions [11, 26, 27], while SANT domains are thought to bind histone tails [28–30]. To identify the domain of MTA proteins critical for mediating FOG-2 interactions, we generated a series of expression constructs for truncations of rat MTA-1 (Fig. 3A). These constructs were used to generate in vitro translated, 35S-labeled protein for binding reactions with GST-FOG-2. To our surprise, none of the previously identified domains of MTA-1 were required for binding to FOG-2 (Fig. 3B). Instead, a 253 amino acid domain at the C-terminus of MTA-1 (aa 450–703) was sufficient to mediate interaction with the FOG Repression Motif (Fig. 3B, N4 truncation). However, further truncations containing only the last 204 (N5 truncation, aa 499–703) or 170 (N6 truncation, aa 533–703) amino acids of the C-terminus were unable to bind to FOG-2 (Fig. 3B), suggesting that the region from residue 450 to residue 499 is required for FOG-2/MTA-1 interactions. To examine whether the FOG-2 and MTA-1 interaction modules are conserved, we studied the binding between the N-terminus of FOG-1 (amino acids 1–45) and a set of human MTA-1 truncations. Similar to the above results, we found that MTA-1 binding by FOG-1 required amino acids 449 to 524 (Supplemental Figure 1). An NCBI BLAST search revealed that this region is highly conserved among human, mouse and rat MTA-1 but shows a more moderate conservation among MTA-1, MTA-2 and MTA3 proteins. However, surprisingly, this search did not identify a similar region in any other proteins. Taken together, these observations define a novel domain in the C-terminal portion of MTA-1 that is required for FOG/MTA-1 interactions.
Figure 3. The C-terminal Portion of MTA-1 Interacts with FOG-2.
A. A schematic of the known protein domains of MTA-1 and N- and C-terminal truncations of MTA-1 used in the binding assays. B. In vitro binding assays between GST-FOG-2(1–12) and N- and C-terminal truncations of MTA1. The binding assays were performed as described in Figure 2. The top panels show the results with GST-Fog-2(1–12) fusion protein and the truncation indicated above. The bottom panels show the input MTA truncations.
MTA proteins are required for maximal FOG-2-mediated transcriptional repression
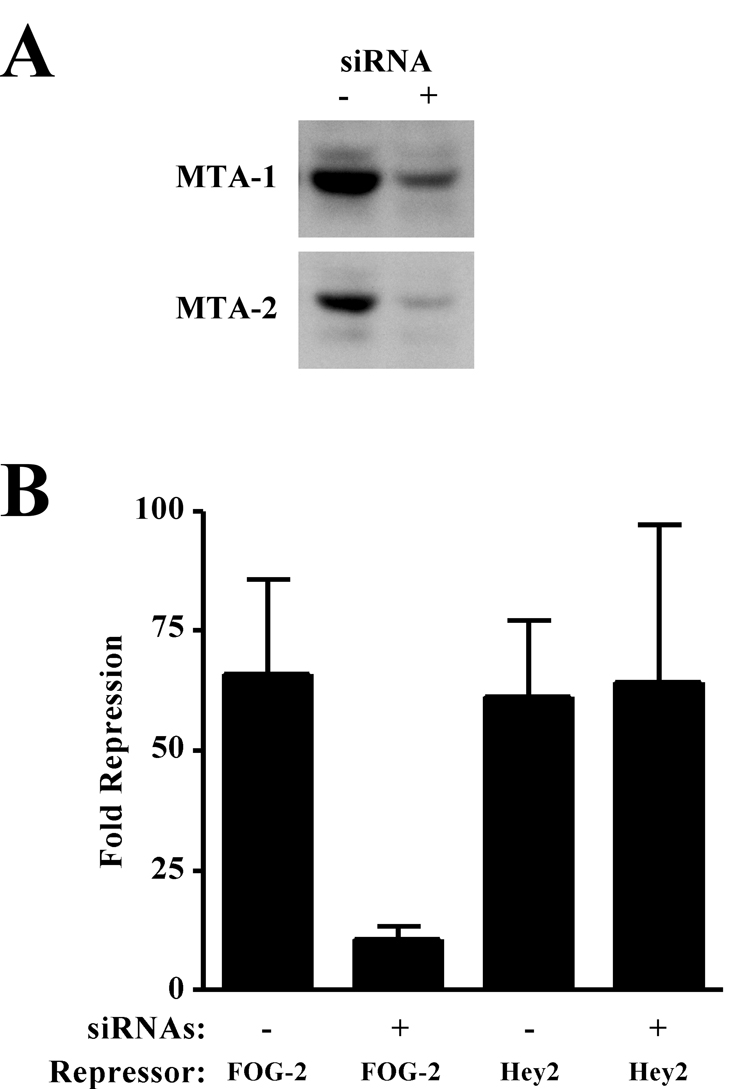
We have previously shown that FOG-2 can repress GATA4 activation of the atrial natriuretic factor (ANF) promoter using a transient transfection assay in NIH 3T3 fibroblasts [23]. Consistent with the notion that this repression is mediated by FOG-2/MTA-1 interactions, this cell line is known to express components of the NuRD complex, including MTA-1 [31]. Therefore, to determine the functional significance of FOG-2/MTA interactions, we endeavored to knock down the levels of MTA in these cells using a siRNA approach. We transfected 3T3 fibroblasts with a mixture of siRNAs directed against MTA-1, 2, and 3. Western analysis of siRNA treated fibroblasts revealed a substantial reduction in MTA-1 and 2 protein levels (Fig. 4A). In both untransfected and transfected 3T3 fibroblasts, we were unable to detect MTA-3 protein expression with three independent MTA-3-specific antibodies, although northern analysis of untransfected fibroblasts revealed the expression of MTA-3 mRNA (data not shown). To test the ability of FOG-2 to repress GATA4 transactivation of the ANF promoter, we then transfected these fibroblasts with a reporter construct containing the ANF promoter driving human growth hormone (hGH) expression and expression constructs for GATA4 and FOG-2. Consistent with our previous results, FOG-2 repressed GATA4-mediated transactivation of the ANF promoter 66±19 fold in the presence of endogenous MTA proteins (Fig. 4B, column 1). However, knockdown of MTA expression by siRNA pretreatment resulted in a dramatic reduction in the ability of FOG-2 to repress transactivation of the ANF promoter, with only a 10±3 fold repression observed (Fig. 4B, column 2). The difference in FOG-2-mediated repression was statistically significant (p=0.018). To rule out the possibility that knockdown of MTA expression globally affects all repressors of GATA-mediated activation, we tested the effect of siRNA treatment on Hey2-mediated repression. Hey2 (also known as Hrt2, Hesr2, Herp1, and CHF1) is a regulator of cardiac development that has been shown to inhibit GATA4-driven ANF promoter activity by more than 90% in vitro [32, 33]. As shown, Hey2 is able to repress GATA4 transactivation of the ANF promoter by 61±16 fold, and this inhibition is not significantly affected by knock down of MTA protein levels. Thus, these results demonstrate that MTA proteins are required specifically for maximal FOG-2-mediated transcriptional repression.
Figure 4. MTA Proteins are Necessary for Maximal FOG-2 Mediated Repression.
A. NIH 3T3 fibroblasts were transfected with a mixture of siRNAs directed against MTA-1, 2, and 3. Twenty-four hours post transfection, total cell lysates were prepared and western analysis was performed using an antibody to MTA-1 (top panel) or MTA-2 (bottom panel). B. NIH 3T3 fibroblasts were transfected with siRNAs directed against MTA-1, 2, and 3. Twenty-four hours later, cells were transfected with a reporter plasmid containing 638 bp of the ANF promoter driving human growth hormone (hGH) and expression plasmids for murine GATA4 and FOG-2 or Hey2. Forty-eight hours later, the media was assayed for hGH and normalized for transfection efficiency. Fold repression is calculated as the ratio of the GATA-mediated transactivation of the ANF promoter in the absence vs. presence of FOG-2 or Hey2. Results represent the mean ± S.E.M. (n=6).
The zinc finger domain of MTA-1 is required for FOG-2 mediated Transcriptional Repression
As a next step in the characterization of FOG-2/MTA interactions, we sought to develop a functional assay for MTA-1 activity in FOG-mediated transcriptional repression. As above, we knocked down endogenous MTA levels in 3T3 fibroblasts using siRNA, then transfected these fibroblasts with a reporter construct containing the ANF promoter driving hGH expression and expression constructs for GATA4 and FOG-2. As above, the addition of anti-MTA siRNA greatly attenuated FOG-2’s ability to repress the ANF promoter (Fig. 5B, columns 1 and 2, 64±20 fold versus 15±2 fold, p=0.025). Co-transfection of an expression construct encoding rat MTA-1, whose expression should not be affected by siRNAs directed against their mouse homologues, restored FOG-2 mediated repression to 42±7 fold (Fig. 5B, column 3), providing further support for the notion that maximal FOG-mediated repression requires MTA expression.
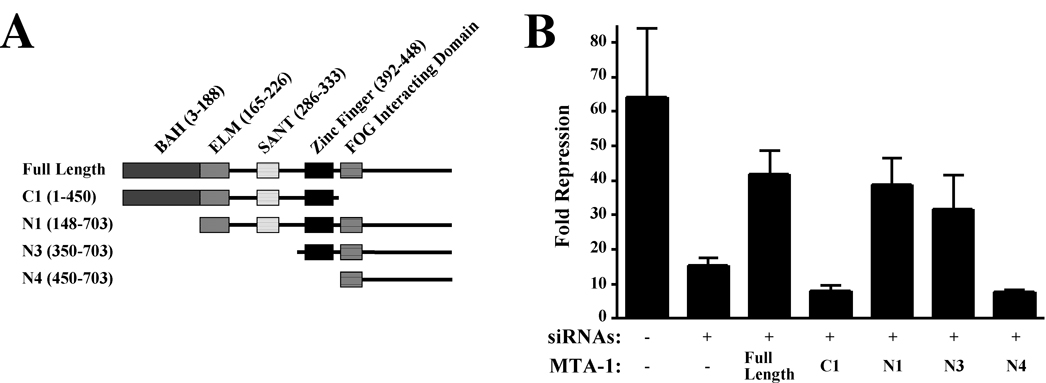
Figure 5. The Zinc Finger of MTA-1 is Required for Maximal FOG-2-Dependent Transcriptional Repression.
A. A schematic of the protein domains of MTA-1 and MTA truncations. B. NIH 3T3 cells were transfected with siRNAs directed against murine MTA family members. Twenty-four hours later, cells were transfected with a reporter hGH plasmid and expression plasmids for murine GATA4, FOG-2 and full length or truncated MTA-1 as indicated. Forty-eight hours later, the media was assayed for hGH and normalized for transfection efficiency. Fold repression is calculated as the ratio of the GATA-mediated transactivation of the ANF promoter in the absence versus presence of FOG-2. Results represent the mean ± S.E.M. (n=9).
To identify domains of MTA-1 that are essential for FOG-2-mediated repression, we generated expression plasmids encoding N- and C-terminal truncations of MTA-1 and tested their ability to rescue FOG-2-dependent repression in the transient transfection assay described above (Fig. 5A). A construct lacking the C-terminal 253 amino acids of MTA-1 (C1), the region encoding the FOG interaction domain, was unable to restore FOG-2-mediated repression (Fig. 5B, column 4), consistent with the requirement of this domain for interaction with the FOG Repression Motif. However, a construct lacking the BAH domain (N1) restored repression, suggesting that this domain is dispensable for FOG-2’s ability to repress transcription (Fig. 5B, column 5). While a construct encoding solely the C-terminal 353 amino acids of MTA-1 (N3) was capable of restoring FOG-2 mediated repression (Fig. 5B, column 6), a construct encoding the C-terminal 253 amino acids of MTA-1 (the FOG interaction domain, N4) failed to do so (Fig. 5B, column 7). Taken together, these results suggest that the BAH, ELM2, and SANT domains of MTA-1 are not required for FOG-mediated repression in this assay. Consistent with the results of our in vitro binding assays, the C-terminal region of MTA-1 is required, but not sufficient for FOG-2- mediated repression. In addition, the MTA-1 zinc finger domain is required, suggesting that it mediates interaction with other members of the NuRD complex that are critical for FOG-2-dependent repression.
The zinc finger domain of MTA-1 interacts specifically with the RbAp subunits of the NuRD complex
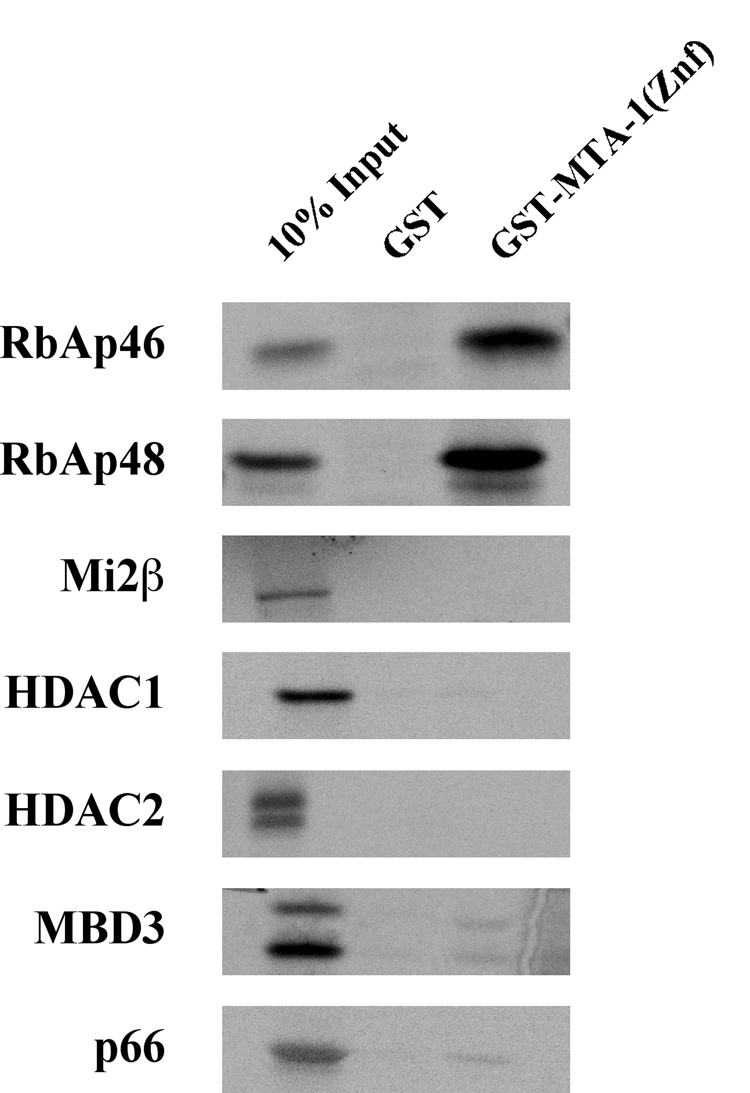
To identify which NuRD subunit(s) MTA-1 is recruiting via its zinc finger domain, in vitro binding assays were performed using a fusion protein containing GST and the zinc finger domain of MTA-1 (amino acids 392–448). This fusion protein was incubated with 35S-labeled, in vitro translated subunits of the NuRD complex, washed extensively, and resultant complexes resolved by SDS-PAGE. As shown in Figure 6, the MTA-1 zinc finger domain was able to interact specifically with RbAp46 and RbAp48, but none of the other NuRD complex subunits. This suggests that interaction between MTA-1 and RbAp46/48 is required for FOG-mediated transcriptional repression.
Figure 6. The Zinc Finger Domain of MTA-1 Interacts with NuRD Complex Subunits RbAp46 and RbAp48.
A fusion protein containing GST and the zinc finger domain of MTA-1 (aa 392–448) was purified from bacteria and incubated with 35S-labeled, in vitro translated NuRD subunits as indicated. Resultant complexes were washed extensively, resolved by SDS-PAGE, and subjected to autoradiography.
DISCUSSION
This work provides further insights into the mechanism by which the NuRD complex mediates transcriptional repression through its association with the FOG Repression Motif. We have shown that the FOG Repression Motif of FOG-2 associates with the NuRD complex through its interaction with MTA and RbAp proteins. We have mapped the FOG-2/MTA interaction to a domain within the C-terminal portion of MTA-1. In addition, we have demonstrated that this interaction is functionally significant as MTA expression is required for maximal FOG-2 mediated transcriptional repression. Further, FOG-2’s ability to repress transcription appears to require both the C-terminal portion as well as the zinc finger domain of MTA-1. The zinc finger domain of MTA-1 interacts with RbAp46/48, suggesting that FOG-2, MTA and RbAp proteins all associate with one another to form a complex that is required for FOG-2 mediated transcriptional repression.
Our work is consistent with studies of other proteins that possess the FOG Repression Motif. FOG-1 has been shown to bind the NuRD complex in vitro and in vivo via the FOG repression motif. Similar to FOG-2, FOG-1 was found to interact with MTA-1 and MTA-2 as well as RbAp46/48 [24]. Sall1 is a multi-zinc finger transcription factor that contains the FOG Repression Motif and is critical for kidney development [34]. Sall1 has been shown to bind the NuRD complex via this motif, although the exact component(s) of the NuRD complex that Sall1 interacts with was not determined [35]. Finally, BCL11B is another transcriptional repressor that we have previously identified to contain the FOG Repression Motif at its N-terminus [23, 24]. This protein has also been shown to bind the NuRD complex via an interaction with MTA-1 and MTA-2 [36]. Taken together, these observations support the notion that the FOG Repression Motif functions as a repressor domain by the recruitment of the NuRD complex to targeted promoters. The results reported here go beyond these previous reports by (a) defining a novel domain in MTA-1 required for interaction with the FOG Repression Motif, (b) demonstrating the importance of MTA expression for function of the FOG Repression Motif, (c) demonstrating the requirement of the zinc finger domain of MTA-1 for FOG-mediated repression, and (d) demonstrating MTA/RbAp interactions mediated by the zinc finger of MTA-1.
MTA proteins directly interact with other transcriptional repressors that lack a FOG Repression Motif. The transcriptional repressor Bcl-6 associates with the NuRD complex via binding to MTA-3 [37] and the tumor suppressor p53 has been shown to bind MTA-2 [38]. Interestingly, these interactions are specific for a particular MTA family member. In contrast, the FOG Repression Motif interacts with all MTA proteins (Fig. 2). Consistent with this observation, the critical region of MTA-1 identified for FOG interaction (aa 450–499) is 58% identical at the amino acid level to sequences within both MTA-2 and MTA-3.
It is likely that MTA proteins serve other functions within the NuRD complex. The BAH, ELM2, and SANT domains have all been implicated in protein-protein interactions [11, 26, 28–30]. Thus, the presence of these domains within the MTA proteins suggest that they are capable of forming multiple protein interactions and that these regions may, in addition to the zinc finger domain, be facilitating the recruitment and assembly of the NuRD complex on target DNA. Interestingly, the BAH, ELM2, and SANT domains were not required for rescue of FOG-mediated repression in our siRNA-based functional assay for MTA-1 (Fig. 5). This suggests that either these domains are not necessary for the function of MTA-1 in this system, or that the small amount of full-length MTA proteins present in siRNA treated fibroblasts is sufficient to compensate for the functions mediated by these domains.
It is interesting that both MTA proteins and RbAp proteins associate with the 12 amino acid FOG Repression Motif as demonstrated by our in vitro binding assays. The binding of both protein families is disrupted by the K5A mutation within the FOG Repression Motif, suggesting that both of these protein families require this residue for binding. Similar results were seen with the binding of FOG-1 to each of these proteins [24]. It is currently unclear if both MTA and RbAp proteins can bind to FOG-2 at the same time or if their binding is mutually exclusive. Complicating such an analysis is our observation that MTA-1 and RbAp proteins also directly interact with one another through the zinc finger domain of MTA-1, independent of binding to FOG-2. Despite the shared ability to bind to proteins containing the FOG Repression Motif, MTA and RbAp proteins do not share any obvious structural domains. In addition, we have been unable to identify any significant amino acid homology between the FOG interacting domain of MTA-1 and any domain within either RbAP46 or RbAp48. Further work will be required to identify the FOG-interacting domain in the RbAp proteins and to determine the functional significance of RbAp/FOG-2 interactions.
This work suggests a model for FOG-2 mediated transcriptional repression in which FOG-2 is targeted to specific promoters through its interaction with GATA4, and then recruits the NuRD complex via interactions between the FOG Repression Motif and MTA proteins and/or RbAp proteins. Further in vivo evidence for the functional significance of the FOG Repression Motif will await the generation of mice with specific point mutations in the FOG Repression Motif of FOG-1 and FOG-2.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (HL071063 to E.C.S., DK058044 to G. A. B., and A.E. R. by T32 GM007197). We gratefully thank Michael Chin for providing the pcDNAHey2 expression plasmid.
The abbreviations used are
- FOG
Friend of GATA
- NuRD
nucleosome remodeling and deacetylase complex
- MTA
metastasis associated protein
- HDAC
histone deacetylase
- RbAp
retinoblastoma associated protein
- MBD3
methyl-CpG-binding domain protein
- ANF
atrial naturetic factor
- GST
glutathione-S-transferase
- aa
amino acid.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD, LaPointe JW, Lorch Y. Preparation of nucleosomes and chromatin. Methods Enzymol. 1989;170:3–14. doi: 10.1016/0076-6879(89)70039-2. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Grunstein M. 25 years after the nucleosome model: chromatin modifications. Trends Biochem Sci. 2000 Dec;25(12):619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 4.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000 Jan 6;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 5.Imhof A, Becker PB. Modifications of the histone N-terminal domains. Evidence for an "epigenetic code"? Mol Biotechnol. 2001 Jan;17(1):1–13. doi: 10.1385/MB:17:1:01. [DOI] [PubMed] [Google Scholar]
- 6.Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998 Jul 2;8(14):843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998 Oct 16;95(2):279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 8.Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998 Oct 29;395(6705):917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 9.Toh Y, Pencil SD, Nicolson GL. A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression, and protein analyses. The Journal of biological chemistry. 1994 Sep 16;269(37):22958–22963. [PubMed] [Google Scholar]
- 10.Bowen NJ, Fujita N, Kajita M, Wade PA. Mi-2/NuRD: multiple complexes for many purposes. Biochimica et biophysica acta. 2004 Mar 15;1677(1–3):52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Yao YL, Yang WM. The metastasis-associated proteins 1 and 2 form distinct protein complexes with histone deacetylase activity. The Journal of biological chemistry. 2003 Oct 24;278(43):42560–42568. doi: 10.1074/jbc.M302955200. [DOI] [PubMed] [Google Scholar]
- 12.Deconinck AE, Mead PE, Tevosian SG, Crispino JD, Katz SG, Zon LI, et al. FOG acts as a repressor of red blood cell development in Xenopus. Development (Cambridge, England) 2000 May;127(10):2031–2040. doi: 10.1242/dev.127.10.2031. [DOI] [PubMed] [Google Scholar]
- 13.Fossett N, Tevosian SG, Gajewski K, Zhang Q, Orkin SH, Schulz RA. The Friend of GATA proteins U-shaped, FOG-1, and FOG-2 function as negative regulators of blood, heart, and eye development in Drosophila. Proc Natl Acad Sci U S A. 2001 Jun 19;98(13):7342–7347. doi: 10.1073/pnas.131215798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svensson EC, Huggins GS, Lin H, Clendenin C, Jiang F, Tufts R, et al. A syndrome of tricuspid atresia in mice with a targeted mutation of the gene encoding Fog-2. Nat Genet. 2000 Jul;25(3):353–356. doi: 10.1038/77146. [DOI] [PubMed] [Google Scholar]
- 15.Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, et al. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell. 2000 Jun 23;101(7):729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- 16.Lu JR, McKinsey TA, Xu H, Wang DZ, Richardson JA, Olson EN. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol Cell Biol. 1999 Jun;19(6):4495–4502. doi: 10.1128/mcb.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svensson EC, Tufts RL, Polk CE, Leiden JM. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc Natl Acad Sci U S A. 1999 Feb 2;96(3):956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tevosian SG, Deconinck AE, Cantor AB, Rieff HI, Fujiwara Y, Corfas G, et al. FOG-2: A novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc Natl Acad Sci U S A. 1999 Feb 2;96(3):950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, et al. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997 Jul 11;90(1):109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 20.Anttonen M, Ketola I, Parviainen H, Pusa AK, Heikinheimo M. FOG-2 and GATA-4 Are coexpressed in the mouse ovary and can modulate mullerian-inhibiting substance expression. Biol Reprod. 2003 Apr;68(4):1333–1340. doi: 10.1095/biolreprod.102.008599. [DOI] [PubMed] [Google Scholar]
- 21.Crispino JD, Lodish MB, Thurberg BL, Litovsky SH, Collins T, Molkentin JD, et al. Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG cofactors. Genes Dev. 2001 Apr 1;15(7):839–844. doi: 10.1101/gad.875201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svensson EC, Huggins GS, Dardik FB, Polk CE, Leiden JM. A functionally conserved N-terminal domain of the friend of GATA-2 (FOG-2) protein represses GATA4-dependent transcription. The Journal of biological chemistry. 2000 Jul 7;275(27):20762–20769. doi: 10.1074/jbc.M001522200. [DOI] [PubMed] [Google Scholar]
- 23.Lin AC, Roche AE, Wilk J, Svensson EC. The N termini of Friend of GATA (FOG) proteins define a novel transcriptional repression motif and a superfamily of transcriptional repressors. The Journal of biological chemistry. 2004 Dec 31;279(53):55017–55023. doi: 10.1074/jbc.M411240200. [DOI] [PubMed] [Google Scholar]
- 24.Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, et al. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. Embo J. 2005 Jul 6;24(13):2367–2378. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin MT, Maemura K, Fukumoto S, Jain MK, Layne MD, Watanabe M, et al. Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. The Journal of biological chemistry. 2000 Mar 3;275(9):6381–6387. doi: 10.1074/jbc.275.9.6381. [DOI] [PubMed] [Google Scholar]
- 26.Oliver AW, Jones SA, Roe SM, Matthews S, Goodwin GH, Pearl LH. Crystal structure of the proximal BAH domain of the polybromo protein. Biochem J. 2005 Aug 1;389(Pt 3):657–664. doi: 10.1042/BJ20050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding Z, Gillespie LL, Paterno GD. Human MI-ER1 alpha and beta function as transcriptional repressors by recruitment of histone deacetylase 1 to their conserved ELM2 domain. Mol Cell Biol. 2003 Jan;23(1):250–258. doi: 10.1128/MCB.23.1.250-258.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grune T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, et al. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003 Aug;12(2):449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 29.Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell. 2002 Oct;10(4):935–942. doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- 30.Boyer LA, Latek RR, Peterson CL. The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol. 2004 Feb;5(2):158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 31.Yaguchi M, Wada Y, Toh Y, Iguchi H, Kono A, Matsusue K, et al. Identification and characterization of the variants of metastasis-associated protein 1 generated following alternative splicing. Biochimica et biophysica acta. 2005 Dec 30;1732(1–3):8–14. doi: 10.1016/j.bbaexp.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Fischer A, Klattig J, Kneitz B, Diez H, Maier M, Holtmann B, et al. Hey basic helix-loop-helix transcription factors are repressors of GATA4 and GATA6 and restrict expression of the GATA target gene ANF in fetal hearts. Mol Cell Biol. 2005 Oct;25(20):8960–8970. doi: 10.1128/MCB.25.20.8960-8970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang F, Sakata Y, Cui L, Youngblood JM, Nakagami H, Liao JK, et al. Transcription factor CHF1/Hey2 suppresses cardiac hypertrophy through an inhibitory interaction with GATA4. American journal of physiology. 2006 May;290(5):H1997–H2006. doi: 10.1152/ajpheart.01106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishinakamura R, Matsumoto Y, Nakao K, Nakamura K, Sato A, Copeland NG, et al. Murine homolog of SALL1 is essential for ureteric bud invasion in kidney development. Development (Cambridge, England) 2001 Aug;128(16):3105–3115. doi: 10.1242/dev.128.16.3105. [DOI] [PubMed] [Google Scholar]
- 35.Lauberth SM, Rauchman M. A conserved 12-amino acid motif in Sall1 recruits the nucleosome remodeling and deacetylase corepressor complex. The Journal of biological chemistry. 2006 Aug 18;281(33):23922–23931. doi: 10.1074/jbc.M513461200. [DOI] [PubMed] [Google Scholar]
- 36.Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 2005 Oct 13;24(45):6753–6764. doi: 10.1038/sj.onc.1208904. [DOI] [PubMed] [Google Scholar]
- 37.Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, et al. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004 Oct 1;119(1):75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 38.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000 Nov 16;408(6810):377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.