Abstract
ETR1 represents a prototypical ethylene receptor. Homologues of ETR1 have been identified in Arabidopsis as well as in other plant species, indicating that ethylene perception involves a family of receptors and that the mechanism of ethylene perception is conserved in plants. The amino-terminal half of ETR1 contains a hydrophobic domain responsible for ethylene binding and membrane localization. The carboxyl-terminal half of the polypeptide contains domains with homology to histidine kinases and response regulators, signaling motifs originally identified in bacteria. The putative histidine kinase domain of ETR1 was expressed in yeast as a fusion protein with glutathione S-transferase and affinity purified. Autophosphorylation of the purified fusion protein was observed on incubation with radiolabeled ATP. The incorporated phosphate was resistant to treatment with 3 M NaOH, but was sensitive to 1 M HCl, consistent with phosphorylation of histidine. Autophosphorylation was abolished by mutations that eliminated either the presumptive site of phosphorylation (His-353) or putative catalytic residues within the kinase domain. Truncations were used to delineate the region required for histidine kinase activity. An examination of cation requirements indicated that ETR1 requires Mn2+ for autophosphorylation. These results demonstrate that higher plants contain proteins with histidine kinase activity. Furthermore, these results indicate that aspects of ethylene signaling may be regulated by changes in histidine kinase activity of the receptor.
Histidine protein kinases have taken on increasing importance of late, proportionate with our growing knowledge of the signaling pathways and the organisms in which they function. The prototypical histidine kinases were identified in bacteria as belonging to “two-component” signaling systems (1–3). In these systems, histidine kinases autophosphorylate a conserved histidine residue, often in response to an environmental stimulus; this phosphate then is transferred to an aspartic acid residue on a second component referred to as the response regulator. Recently, proteins with homology to the bacterial histidine kinases have been identified in eukaryotes, including animals, fungi, and plants (2). Interestingly, whereas the SLN1 osmosensor of yeast was demonstrated to possess histidine kinase activity (4), homologues from animals were found to be serine/threonine kinases (5, 6). Thus, from sequence alone, the biochemical nature of the kinase cannot be inferred.
In plants, proteins with homology to histidine kinases have been implicated in signal transduction by the plant hormone ethylene (7). Ethylene is a simple two-carbon gas important at many stages of the plant’s life, including seed germination, seedling growth, leaf and petal abscission, organ senescence, and pathogen responses (8). Several components of the ethylene signal transduction pathway have been identified by a mutational approach with Arabidopsis thaliana (9). One of these components is the ETR1 gene product (10, 11), which functions as an ethylene receptor in Arabidopsis (12). The ETR1 protein contains an ethylene-binding site in its amino-terminal half (12) and regions with homology to histidine kinases and response regulators in its carboxyl-terminal half (11). This modular design is similar to that of many bacterial histidine kinases, which contain a sensory domain in the amino-terminal region and a histidine kinase domain in the carboxyl-terminal region (1). Based on this similarity in design, it has been proposed that the ETR1 ethylene receptor could function analogously to these bacterial sensor proteins, with ethylene binding regulating activity of the proposed histidine kinase domain (12).
ETR1 is not the only putative histidine kinase implicated in plant signal transduction. In Arabidopsis, other proteins closely related to ETR1 also function in ethylene signal transduction. For example, the ERS1 protein contains sequence similarity to ETR1 throughout its entire length, including the ethylene binding and histidine kinase domains, but lacks a response regulator domain (13). The presence of multiple isoforms indicates that a family of ethylene receptors may function in Arabidopsis. Additional proteins related to ETR1 also have been identified in tomato (14–16), indicating that the mechanism of ethylene perception is conserved in other plant species. Histidine kinases may be involved in other plant signaling pathways as well. CKI1, a putative histidine kinase involved in cytokinin signaling, recently has been identified in Arabidopsis (17).
The finding that ETR1 and CKI1 have homology to the histidine kinases and response regulators of bacteria provides evidence that the two-component signal transduction systems of bacteria are present in plants. However, questions remain as to how this evolutionarily ancient system has been adapted for signal transduction in eukaryotes. In this paper, we demonstrate that the ETR1 gene encodes a functional histidine kinase with biochemical features similar to those of its bacterial counterparts. This finding supports a role for histidine phosphorylation as a central regulatory mechanism in plants and has specific implications for ethylene signal transduction.
MATERIALS AND METHODS
Plasmid Constructions.
For expression of glutathione S-transferase (GST) fusions in yeast, the vector pEG(KT) was used (18). This vector contains the GST domain under control of a galactose-inducible promoter and allows for URA selection in yeast. One GST fusion was designed to express that portion of ETR1 corresponding to amino acids 164–738, and for this a DraI–HindIII fragment of cETR1–5.2 (19) was cloned between the SmaI and HindIII sites of pEG(KT). To express that portion of ETR1 corresponding to amino acids 164–609, PCR was performed using cETR1–5.2 as the template, with 5′ primer (CTGGATCCAAGACTACACTTGTTGAG) and 3′ primer (ACAAGCTTCCAGTGAAATTTGAATG). The PCR product was digested with BamHI and HindIII for subsequent cloning. To express that portion of ETR1 corresponding to amino acids 333–609, PCR was performed with 5′ primer (TTGGATCCGCTAGACGAGAAGCAGAA) and the same 3′ primer as above, then digested with BamHI and HindIII for subsequent cloning. Site-directed mutants of ETR1 were made by using the Altered Sites Mutagenesis System (Promega) according to the manufacturer. Mutations were made in the same PCR products used for expression of wild-type ETR1, and the presence of desired mutations was confirmed by sequencing.
Yeast Expression.
Constructs were transformed (20) into yeast Saccharomyces cerevisiae, strain sc295 (MATa GAL4 GAL80 ura3–52 leu2–3,112 reg1–501 gal1 pep4–3), containing the pMTL4c plasmid (LEU selection) for production of GAL4 protein. The sc295 strain has the reg1 mutation, so GAL promoters are not glucose-repressed, and the pep4–3 mutation for protease deficiency. Standard media and procedures were used for growth (21). For induction of the GST-fusion proteins, 0.5% galactose was used.
Isolation of GST-Fusion Proteins.
After growth to midlog phase, yeast cells were harvested, washed, and resuspended in extraction buffer [50 mM Tris⋅HCl, pH 7.6/100 mM NaCl/1 mM EDTA, 10% (vol/vol) glycerol, with 1 mM phenylmethylsulfonyl fluoride as a protease inhibitor] at 2 ml buffer/g of cells. Cells were broken open by vortexing with chilled glass beads (21), and the homogenate was centrifuged at 3,000 × g for 5 min to remove debris. This supernatant was centrifuged at 100,000 × g for 30 min to separate the soluble protein fraction from the membrane fraction. The soluble fraction was incubated with glutathione-agarose beads, which allowed the GST-fusion protein to bind. The beads then were washed with 30 mM Tris, pH 7.3, 150 mM NaCl, 0.1% (vol/vol) Tween20.
In Vitro Kinase Assay.
Assays for kinase activity were performed on GST fusions bound to glutathione-agarose beads. Unless specified otherwise, beads (50 μl volume) were resuspended in 30 μl assay buffer (50 mM Tris⋅HCl, pH 7.6/50 mM KCl/2 mM DTT/5 mM MnCl2/10% glycerol) containing 500 μM [γ32P]ATP (2 Ci/mmol). Reactions were incubated at 22°C for 45 min, then terminated by the addition of 500 μl of 30 mM Tris, pH 7.3, 150 mM NaCl, 10 mM EDTA. With a 45-min assay, approximately 50% of the maximal incorporation of 32P was observed for the wild-type protein (data not shown). Beads were washed twice with termination buffer, then resuspended in SDS/PAGE loading buffer. Samples were subjected to SDS/PAGE (22) using 8% polyacrylamide gels. After electrophoresis, proteins were electrotransferred to Immobilon nylon membrane (Millipore). Stability of incorporated phosphate was determined by treatment of the membrane with either alkali solution (3 M NaOH) or acid solution (1 M HCl) for 2 h at 22°C (4). Membranes were rinsed with water, dried, and incorporated phosphate visualized by autoradiography. Western blotting was performed by using either an anti-ETR1 (HRR) antibody (19) or a polyclonal anti-GST antibody (Santa Cruz Biotechnology). Immunodecorated proteins were visualized by enhanced chemiluminescence detection according to the manufacturer (Pierce). Densitometric analysis was performed by using NIH Image after first scanning the exposed film using Adobe Photoshop and a LaCie scanner. Scans were quantified by comparison to a standard dilution series.
RESULTS
Expression and Purification of ETR1 Domains.
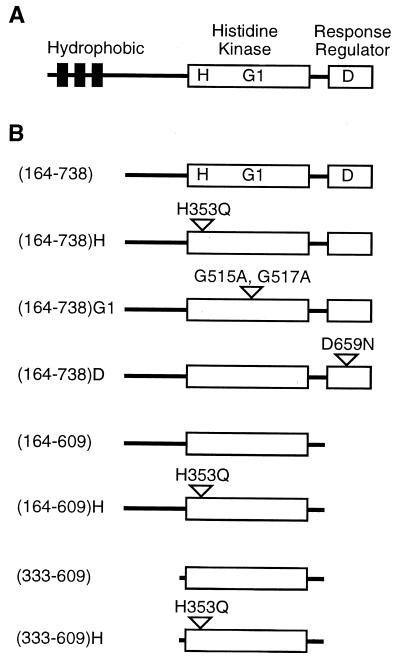
The ETR1 protein has a modular structure similar to that of many bacterial histidine kinases (Fig. 1). The amino-terminal region contains a hydrophobic domain that is responsible for both membrane localization and the binding of ethylene (12, 19). The carboxyl-terminal half of the protein contains separate domains with homology to bacterial histidine kinases and bacterial response regulators (11). The putative histidine kinase and response regulator domains of ETR1 contain all of the conserved residues considered essential for enzymatic activity. Thus, ETR1 contains an input domain coupled to two modules that use phosphorylation as a means of information transfer.
Figure 1.
(A) Domains of the ETR1 protein. Hydrophobic, histidine kinase, and response-regulator domains are indicated. H indicates histidine-353, the putative phosphorylation site. G1 indicates position of the G1 box within the putative kinase domain. D indicates aspartate-659, a potential phosphorylation site. (B) Versions of ETR1 expressed as GST fusions in yeast. For truncations, the first and last amino acids of the expressed region are indicated, the full-length ETR1 protein being 738 amino acids long. For site-directed mutations, single letter abbreviations for amino acids Ala (A), Asn (N), Asp (D), Gln (Q), Gly (G), and His (H) are used.
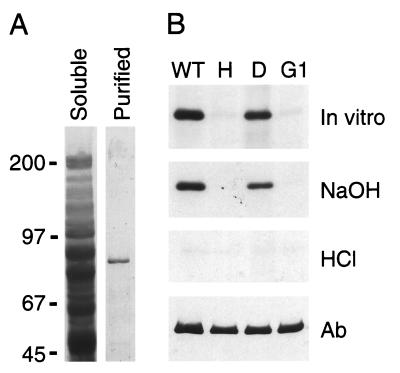
It has been found that the input domain of some bacterial histidine kinases exerts negative regulation over histidine kinase activity, such that the histidine kinase domain is constitutively activated when expressed without the input domain (23). We hypothesized that expression of the soluble domain of ETR1, without the amino-terminal ethylene-binding domain, could allow us to assess the enzymatic activity of the putative histidine kinase domain. To this end, constructs were made in which soluble portions of ETR1 were expressed as fusions with GST (Fig. 1), using the yeast Saccharomyces cerevisiae as an expression system. Yeast was used for two reasons. First, initial experiments using fusion proteins expressed in E. coli did not yield any kinase activity (results not shown), possibly because of misfolding of the protein. Second, previous work had demonstrated that full-length ETR1 expressed in yeast had similar biochemical properties to the native protein found in Arabidopsis (19), adopting a functional structure that was capable of binding ethylene (12). GST fusions with truncated versions of ETR1 were soluble and were purified from yeast by binding to glutathione-agarose beads. Fusion proteins were analyzed by SDS/PAGE to determine purity and to confirm that each construct produced an ETR1 species of the appropriate size (Fig. 2A).
Figure 2.
(A) Purification of GST-ETR1(164–738) from yeast. SDS/PAGE profiles of proteins are shown from the soluble fraction, and after affinity purification of GST-ETR1(164–738) by binding to glutathione-agarose beads. Proteins are stained with Coomassie blue. Migration positions of molecular mass markers are indicated in kDa. (B) In vitro phosphorylation of GST-ETR1(164–738). Wild-type (WT) and mutant versions of the fusion protein were examined for the ability to autophosphorylate. The site-directed mutations were His-353–Gln (H), Asp-659–Asn (D), and in the G1 box (G1) of ETR1. Affinity- purified protein was incubated with 32P-ATP, subjected to SDS/PAGE, then transferred to nylon membrane (in vitro). Proteins were sequentially treated with alkali (NaOH) and acid (HCl). After each treatment, incorporated phosphate was visualized by autoradiography. Finally, protein was visualized by Western blot using a polyclonal antibody against ETR1 (Ab).
Histidine Kinase Activity of ETR1.
A GST fusion with the soluble domain of ETR1 (amino acids 164–738), encompassing both histidine kinase and response regulator domains, was purified from yeast. The purified polypeptide displayed an apparent molecular mass of 89 kDa when analyzed by SDS/PAGE (Fig. 2A), consistent with the predicted molecular mass of 91 kDa for the fusion protein [GST = 27 kDa; ETR1(164–738) = 64 kDa]. When purified GST-ETR1(164–738) was incubated in the presence of [γ-32P]ATP, 32P was incorporated into the fusion protein (Fig. 2B).
The stability of the incorporated phosphate under alkali and acidic conditions was tested as a means of assessing the type of phosphate linkage (24). Phosphoramidates, such as phosphohistidine, are stable under basic conditions but sensitive to acidic conditions. Acylphosphates, such as phosphoaspartic acid are sensitive to both acidic and basic conditions. On the other hand, the O-phosphate linkages of phosphoserine, phosphothreonine, and phosphotyrosine are acid-stable. The incorporated phosphate was resistant to treatment with 3 M NaOH, but was sensitive to 1 M HCl (Fig. 2B), suggesting that the fusion protein contained phosphohistidine. We found no evidence for the formation of phosphoaspartic acid, even when pulse–chase experiments were performed (results not shown).
To further assess phosphorylation of ETR1, the conserved histidine residue predicted to be the site of phosphorylation was mutated (H353Q). No phosphate was incorporated into the mutant fusion protein (Fig. 2B), indicating that His-353 is required for autophosphorylation. On the other hand, mutation of the aspartic acid residue within the response regulator domain (D659N) did not affect the ability of ETR1 to autophosphorylate at the histidine residue (Fig. 2B). This is consistent with the properties of bacterial two-component systems, where the initial site of phosphorylation is histidine; only subsequent to histidine phosphorylation is the phosphate transferred to the response regulator (1). In addition to mutating potential sites of phosphorylation, we mutated the G1 box (G515A, G517A) within the kinase domain of ETR1. The G1-box mutation is predicted to abolish histidine kinase activity (25) as it disrupts the region implicated in ATP binding (1). We observed no phosphorylation in this mutant (Fig. 2B), demonstrating the necessity of the G1 box for histidine kinase activity of ETR1. The G1-box mutation also helps eliminate the possibility that phosphorylation of ETR1 arises from copurification of a yeast histidine kinase (2, 4) capable of phosphorylating the ETR1 fusion protein; the conserved histidine residue of ETR1 (His-353) is still present, but is not phosphorylated. Western blot analysis was performed to confirm equivalent protein loading.
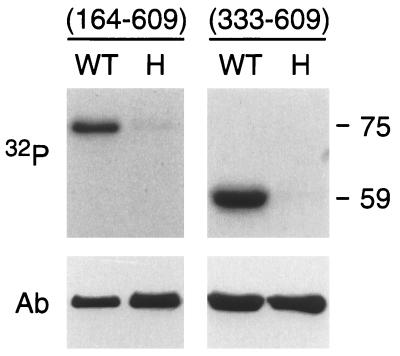
To further define the region of ETR1 containing histidine kinase activity, we made two truncations within the soluble portion of ETR1. ETR1(164–609) lacks the response regulator domain, whereas ETR1(333–609) lacks the region upstream of the predicted histidine kinase domain as well as the response regulator domain. Purified GST-ETR1(164–609) had an apparent molecular mass of 75 kDa when analyzed by SDS/PAGE, and purified GST-ETR1(333–609) an apparent molecular mass of 59 kDa. This is consistent with respective molecular masses of 76 kDa and 57 kDa predicted from the amino acid sequences. Both fusions were capable of autophosphorylation when assayed for histidine kinase activity (Fig. 3).
Figure 3.
In vitro phosphorylation of truncated versions of ETR1. GST fusions with either ETR1(164–609) or ETR1(333–609) were examined for the ability to autophosphorylate. Both wild-type (WT) and the His-353–Gln (H) site-directed mutation of ETR1 were tested in each case, and an autoradiograph of the alkali-resistant phosphorylation is shown (32P). Relative size of the phosphorylated proteins, as based on molecular mass standards, is indicated in kDa. After autoradiography, the presence of each fusion protein was confirmed by Western blot using a polyclonal antibody directed against GST (Ab).
Cation and pH Dependence of Histidine Kinase Activity.
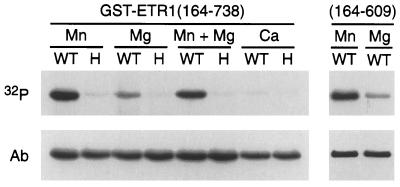
Bacterial histidine kinases require a divalent cation for enzymatic activity. In bacteria, Mg2+ is most commonly used, but other divalent cations such as Mn2+ are favored by some histidine kinases (26). The ETR1 histidine kinase also was found to be dependent on divalent cations for activity and demonstrated specificity for Mn2+ (Fig. 4). Minimal phosphorylation was observed in the presence of 5 mM Mg2+, and no phosphorylation was observed in the presence of 5 mM Ca2+. Lowering the Mg2+ concentration to 1 mM did not lead to greater effectiveness (results not shown), nor did the presence of Mg2+ and Mn2+ together yield any more phosphorylation than Mn2+ alone (Fig. 4), indicating that these cations do not act synergistically. It was conceivable that the low levels of phosphorylation observed with Mg2+ could be caused by transfer of the phosphate group from phosphorylated histidine to aspartic acid in the response regulator domain, phosphoaspartic acid being less stable than phosphohistidine. However, this is not the case as ETR1(164–609), which lacks the response regulator domain, still shows greater levels of phosphorylation with Mn2+ than with Mg2+ (Fig. 4).
Figure 4.
Cation dependence for autophosphorylation of ETR1. GST fusions with either ETR1(164–738) or ETR1(164–609) were examined for autophosphorylation in the presence of various cations (32P). Wild-type (WT) and the His-353–Gln (H) site-directed mutation of ETR1 were tested with 5 mM MnCl2, 5 mM MgCl2, 5 mM CaCl2, or a mixture of 5 mM MnCl2 and 5 mM MgCl2. After autoradiography, the presence of each fusion protein was confirmed by Western blot using a polyclonal antibody directed against GST (Ab).
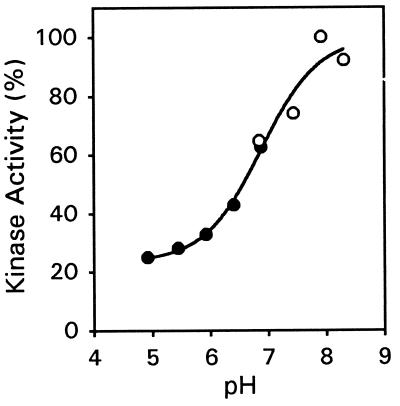
The pH dependence for histidine kinase activity also was examined. The level of autophosphorylation was examined in preparations of ETR1(333–609) buffered across a range of pH values from 4.9 to 8.3 (Fig. 5). The level of autophosphorylation sharply increased between pH 6 and 8, with the pH for 50% of this response being 6.9. The data can be fit to a sigmoidal curve and are most consistent with the titratable group being the imidazole group of histidine; in proteins, the pKa of histidine is dependent on the local environment and can vary across several pH units, with a characteristic pK value near 7 (27). The bacterial histidine-kinase CheA has a similar titratable group, and it has been suggested that this could be the histidine that is autophosphorylated (28). Based on the reaction mechanism for histidine phosphorylation, the deprotonated imidazole nitrogens of histidine should be better capable of nucleophilic attack on the γ-phosphoryl group of ATP (28). In addition, because the pH effect occurs over a physiologically relevant range, the kinase activity of ETR1 may vary within the cell depending on the local pH.
Figure 5.
pH dependence for autophosphorylation of ETR1. GST-ETR1(333–609) was incubated with 32P-ATP at the indicated pH, subjected to SDS/PAGE, and transferred to nylon membrane. Alkali-resistant phosphate was visualized by autoradiography and quantified densitometrically. For pH 4.9 to 6.9 (•), the kinase assay mixture was buffered with 50 mM Mes, 100 mM Tris. For pH 6.9 to 8.3 (○), the kinase assay mixture was buffered with 100 mM Tris. The data were fit to a sigmoidal curve using sigmaplot. Data points represent the mean of duplicate samples.
DISCUSSION
By use of two independent approaches, acid-base stability of the phosphoamino acid and site-directed mutagenesis, we have demonstrated that the ETR1 ethylene receptor of plants contains intrinsic histidine kinase activity. The biochemical characteristics of this plant kinase activity closely resemble those of the better-characterized bacterial histidine kinases (1, 26). Histidine kinase activity of ETR1 is contained within a discrete module of the polypeptide, which retains activity when independently expressed. Histidine autophosphorylation of ETR1 can be eliminated by mutations within either the putative ATP binding site (e.g., the G1 box) or at the presumptive site of autophosphorylation (His-353). Furthermore, kinase activity is dependent on the presence of a divalent cation, Mn2+ in the case of ETR1. This report thus extends the known organisms in which histidine kinases function from bacteria (1) and yeast (4) to plants.
Histidine kinase activity of ETR1 showed a preference for Mn2+ over Mg2+. This is in contrast to many of the bacterial histidine kinases (26) and the recently characterized SLN1 histidine kinase of yeast (4), where Mg2+ best fulfills the requirement for a divalent cation. Mn2+ is preferred by some bacterial histidine kinases such as FrzE (29). Although cation requirements for other plant histidine kinases are not yet known, Mn2+ is favored over Mg2+ by a variety of receptor-like serine/threonine kinases (30). Mn2+ dependence may represent the norm for membrane-associated kinases in plants.
ETR1 contains a modular structure similar to that of many bacterial histidine kinases, with a sensory domain located near the amino terminus and a histidine kinase domain located in the carboxyl-terminal half of the protein (11, 12). By analogy to the bacterial sensors (1), the binding of ethylene to ETR1 could modulate the level of histidine kinase activity. It is not known, nor can it be predicted at this point, whether binding of the ethylene ligand would serve to stimulate or repress the level of histidine kinase activity, as examples of both responses can be found in bacterial systems (1). One particularly relevant example may be the FixL protein of Rhizobium meliloti, which, like ETR1, is a histidine kinase that binds a gaseous ligand (31). FixL is an oxygen sensor, and binding of oxygen serves to repress the normally active histidine kinase domain (31).
ETR1 also contains a response regulator-like domain (11). In bacteria, response regulators autophosphorylate on a conserved aspartic acid residue, using a phosphorylated histidine kinase as the phosphodonor (1, 3). We did not find any evidence of phosphotransfer between the histidine kinase and response regulator domains of ETR1 (results not shown). The cause of this negative result may be our inability to determine the correct conditions for phosphotransfer or a requirement for other proteins in the complex. It is also possible that the response regulator domain of ETR1 performs a regulatory or interactive role that does not require phosphorylation.
The finding that the ETR1 protein has histidine kinase activity raises the question as to how this activity might function in ethylene signal transduction. Proteins similar to ETR1, representing additional putative ethylene receptors, exist in Arabidopsis (13, 32). Therefore, this question has implications for a family of proteins. The ERS1 protein, like ETR1, contains a histidine kinase domain with the conserved residues required for activity (13). On the other hand, the ERS2 protein (GenBank accession no. AC000104, bases 76079–78088) has a diverged histidine kinase domain and lacks many of the residues considered essential for activity. These residues include the histidine residue that is autophosphorylated and the G1 box that is involved in binding ATP, whose necessity for histidine kinase activity has been demonstrated by our analysis of ETR1. Genetic analysis of ERS2, like that of ETR1 and ERS1, indicates a role for ERS2 in ethylene signal transduction (Jian Hua and Elliot Meyerowitz, personal communication). Although ERS2 might represent a noncanonical histidine kinase, its existence raises the possibility that histidine kinase activity is not required for responses typically associated with ethylene signal transduction. In such a case, histidine kinase activity may play a more subtle role, for example in adaptation and desensitization to ethylene (33). Alternatively, histidine kinase activity might allow a subset of the ethylene receptors to participate in additional pathways, thereby allowing for discrete responses within certain tissue types. For example, the pathway for ethylene signal transduction is known to interact with pathways for other plant hormones (8, 34). Finally, it is possible that, analogous to phosphorylation of serine, threonine, and tyrosine residues in eukaryotic receptors, phosphorylation of histidine residues could serve to recruit specific proteins into the formation of signaling complexes (35). With ETR1, autophosphorylation could potentially regulate interactions with other proteins involved in ethylene signal transduction such as CTR1, a member of the Raf family of serine/threonine protein kinases (36).
The evidence presented here indicates that plant signaling pathways may use histidine kinases in a manner analogous to their bacterial counterparts. Evidence now exists that both the ethylene and cytokinin signaling pathways use proteins with the conserved features of histidine kinases. In ethylene signal transduction, Arabidopsis contains the ETR1 ethylene receptor (11) examined in this report, as well as the closely related ERS1 protein (13). Other plants, notably tomato, contain similar proteins involved in ethylene signaling (15, 16), indicating that they also may use histidine kinases in signal transduction. In cytokinin signal transduction, the CKI1 protein has been identified in Arabidopsis; CKI1 has all of the conserved residues required for histidine kinase activity (17). By analogy to the bacterial systems, one might expect these proteins to participate in a phosphorelay mechanism, whereby the phosphate from the phosphohistidine is passed on to downstream components such as phosphorelay proteins and response regulators (1, 37). The recent identification of response-regulator proteins in Arabidopsis (38) lends support to this concept. Further research should provide a clearer picture as to how this evolutionarily ancient signaling system has been adapted to the specific needs of a multicellular eukaryote.
Acknowledgments
We thank Estelle Hrabak, Andy Laudano, and Jennifer Findell for critical reading of the manuscript, and Rick Cote for assistance with sigmaplot. This work was supported by grants from the National Science Foundation (MCB-9603679) and the Norris Cotton Cancer Center. This is Scientific Contribution no. 1982 from the New Hampshire Agricultural Experiment Station.
ABBREVIATION
- GST
glutathione S-transferase
References
- 1.Parkinson J S. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 2.Swanson R V, Alex L A, Simon M I. Trends Biochem. 1994;19:485–490. doi: 10.1016/0968-0004(94)90135-x. [DOI] [PubMed] [Google Scholar]
- 3.Stock J B, Surette M G, Levit M, Park P. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 25–51. [Google Scholar]
- 4.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 5.Popov K M, Zhao Y, Shimomura Y, Kuntz M J, Harris R A. J Biol Chem. 1992;267:13127–13130. [PubMed] [Google Scholar]
- 6.Popov K M, Kedishvili N Y, Zhao Y, Shimomura Y, Crabb D W, Harris R A. J Biol Chem. 1993;268:26602–26606. [PubMed] [Google Scholar]
- 7.Bleecker A B, Schaller G E. Plant Physiol. 1996;111:653–660. doi: 10.1104/pp.111.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abeles F B, Morgan P W, Saltveit M E., Jr . Ethylene in Plant Biology. San Diego: Academic; 1992. [Google Scholar]
- 9.Ecker J R. Science. 1995;268:667–674. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- 10.Bleecker A B, Estelle M A, Somerville C, Kende H. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- 11.Chang C, Kwok S F, Bleecker A B, Meyerowitz E M. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 12.Schaller G E, Bleecker A B. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- 13.Hua J, Chang C, Sun Q, Meyerowitz E M. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- 14.Yen H-C, Lee S, Tanksley S D, Lanahan M B, Klee H J, Giovannoni J J. Plant Physiol. 1995;107:1343–1353. doi: 10.1104/pp.107.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkinson J Q, Lanahan M B, Yen H-C, Giovannoni J J, Klee H J. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- 16.Zhou D, Mattoo A K, Tucker M L. Plant Mol Biol. 1996;30:1331–1338. doi: 10.1007/BF00019564. [DOI] [PubMed] [Google Scholar]
- 17.Kakimoto T. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell D A, Marshall T K, Deschenes R J. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 19.Schaller G E, Ladd A N, Lanahan M B, Spanbauer J M, Bleecker A B. J Biol Chem. 1995;270:12526–12530. doi: 10.1074/jbc.270.21.12526. [DOI] [PubMed] [Google Scholar]
- 20.Schiestl R H, Gietz R D. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 21.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Strohl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 22.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Parkinson J S. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 9–24. [Google Scholar]
- 24.Duclos B, Marcandier S, Cozzone A J. Methods Enzymol. 1991;201:10–21. doi: 10.1016/0076-6879(91)01004-l. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Inouye M. J Mol Biol. 1993;231:335–342. doi: 10.1006/jmbi.1993.1286. [DOI] [PubMed] [Google Scholar]
- 26.Hess J F, Bourret R B, Simon M I. Methods Enzymol. 1991;200:188–204. doi: 10.1016/0076-6879(91)00139-n. [DOI] [PubMed] [Google Scholar]
- 27.Edsall J T, Wyman J. Biophysical Chemistry. New York: Academic; 1958. [Google Scholar]
- 28.Conley M P, Berg H C, Tawa P, Stewart R C, Ellefson D D, Wolfe A J. J Bacteriol. 1994;176:3870–3877. doi: 10.1128/jb.176.13.3870-3877.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCleary W R, Zusman D R. J Bacteriol. 1990;172:6661. doi: 10.1128/jb.172.12.6661-6668.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaller G E, Bleecker A B. FEBS Lett. 1993;333:306–310. doi: 10.1016/0014-5793(93)80676-l. [DOI] [PubMed] [Google Scholar]
- 31.Monson E K, Weinstein M, Ditta G S, Helsinki D R. Proc Natl Acad Sci USA. 1992;89:4280–4284. doi: 10.1073/pnas.89.10.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang C, Meyerowitz E M. Proc Natl Acad Sci USA. 1995;92:4129–4133. doi: 10.1073/pnas.92.10.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Q G, Bleecker A B. Plant Physiol. 1995;108:597–607. doi: 10.1104/pp.108.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann-Benning S, Kende H. Plant Physiol. 1992;99:1156–1161. doi: 10.1104/pp.99.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawson T, Scott J D. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 36.Kieber J J, Rothenberg M, Roman G, Feldman K A, Ecker J R. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 37.Appleby J L, Parkinson J S, Bourret R B. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 38.Imamura A, Hanaki N, Umeda H, Nakamura A, Suzuki T, Ueguchi C, Mizuno T. Proc Natl Acad Sci USA. 1998;95:2691–2696. doi: 10.1073/pnas.95.5.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]