Abstract
Elucidating the molecular pathways linking electrical activity to gene expression is necessary for understanding the effects of exercise on muscle. Fast muscles express higher levels of MyoD and lower levels of myogenin than slow muscles, and we have previously linked myogenin to expression of oxidative enzymes. We here report that in slow muscles, compared with fast, 6 times as much of the MyoD is in an inactive form phosphorylated at T115. In fast muscles, 10 h of slow electrical stimulation had no effect on the total MyoD protein level, but the fraction of phosphorylated MyoD was increased 4-fold. Longer stimulation also decreased the total level of MyoD mRNA and protein, while the level of myogenin protein was increased. Fast patterned stimulation did not have any of these effects. Overexpression of wild type MyoD had variable effects in active slow muscles, but increased expression of fast myosin heavy chain in denervated muscles. In normally active soleus muscles, MyoD mutated at T115 (but not at S200) increased the number of fibres containing fast myosin from 50% to 85% in mice and from 13% to 62% in rats. These data establish de-phosphorylated active MyoD as a link between the pattern of electrical activity and fast fibre type in adult muscles.
Muscle fibres can be classified into distinct types based on which of the myosin heavy chain (MyHC) isoenzymes that are expressed. MyHC determines shortening velocity, but MyHC type is also correlated with properties determined by other enzyme systems such as twitch duration and metabolic properties. Typically, phenotypic characteristics range from type I fibres that are slow contracting, with a high capacity for oxidative metabolism and good endurance to type IIb fibres that are fast contracting, fatigable and relying mostly on glycolytic metabolism. In rodents IIa and IIx fibres are intermediate forms, so that the four types in most muscles constitute a functional slow-to-fast spectrum: I ↔ IIa ↔ IIx ↔ IIb.
The fibre type composition of an adult muscle is partly dependent on cell lineage, but fully differentiated post-mitotic muscle fibres can undergo dramatic phenotypic change without degeneration/regeneration when subjected to an altered pattern of electrical stimulation. Changes can occur in both directions, slow-to-fast and fast-to-slow sequentially along the I ↔ IIa ↔ IIx ↔ IIb spectrum (for example see Eken & Gundersen, 1988; Gorza et al. 1988; Windisch et al. 1998; Pette & Staron, 2001). Phenotypic changes are mainly caused by altered gene expression, in particular switching between different MyHC and other fast/slow isoenzymes related to contraction and metabolism.
Most of the research has focused on the fast-to-slow transformations. It has been suggested that the long trains of impulses evoked in slow motor units lead to sustained moderate levels of free intracellular calcium that binds to the calcium sensor calmodulin, which in turn activates calmodulin-dependent protein kinases (CaMKs) and the calmodulin-dependent protein phosphatase calcineurin. CaMKs may activate the Ras–Raf–MEK–ERK pathway (Agell et al. 2002), which seems to be involved in activity-dependent fast-to-slow transformations (Murgia et al. 2000). CaMKs can also activate peroxisome proliferator-activated receptor gamma coactivator 1α (PGC-1α) and its partner peroxisome proliferator activated receptor δ (PPARδ) that can induce slow muscle properties, both during development (Lin et al. 2002; Luquet et al. 2003; Wang et al. 2004), and in adult muscles (Lunde et al. 2007). Activation of calcineurin can switch on transcription factors such as NFAT and MEF-2, both of which have been reported to activate slow muscle genes (Bassel-Duby & Olson, 2006). Calcineurin might also activate the myogenin promoter (Friday et al. 2003), and myogenin has in somatic gene transfer experiments been shown to induce oxidative enzymes in adult fast muscles similar to what is obtained after endurance training (Ekmark et al. 2003).
We here present evidence that MyoD is a factor linking expression of fast muscle genes and electrical activity. MyoD, together with myogenin, MRF4 and myf-5, forms a family of muscle-specific basic helix–loop–helix (bHLH) transcription factors (myogenic regulatory factors (MRFs)) that govern differentiation of muscle cells during development. In adults, MyoD and myogenin show reciprocal expression patterns in vertebrates as diverse as mammals and fishes. MyoD is high in fast muscles and myogenin in slow muscles, and MyoD cis regulatory regions seem to restrict expression to IIx and IIb fibres (Hughes et al. 1993, 1997; Voytik et al. 1993; Rescan et al. 1995; Delalande & Rescan, 1999). Moreover, thyroid hormone treatment results in activation of both MyoD and fast MyHC gene expression (Hughes et al. 1993). MyoD null mutants fail to display any major global shift in MyHC expression, but show subtle shifts in fibre type of fast muscles toward a slower character, while slow muscle tends to shift to a faster phenotype (Hughes et al. 1997; Seward et al. 2001).
Myogenic factors have a conserved threonin residue in the DNA binding region which when phosphorylated seems to prevent DNA binding and transactivation (Li et al. 1992). In cultured fibroblasts grown under mitotic conditions, MyoD is phosphorylated, does not bind to DNA and does not induce MyHC. When switched to differentiation conditions MyoD is de-phosphorylated, bound to DNA and fast MyHC expression is induced. When a T115A mutation was introduced under mitotic conditions, the recombinant protein did not display phospho-threonin and it induced fast MyHC in cells cultured under growth conditions where the endogenous MyoD was inactive (Liu et al. 1998).
We here report that slow-patterned electrical activity causes rapid phosphorylation and inactivation of MyoD, and that a slow-to-fast transformation of MyHC fibre type is observed upon forced expression of a T115A mutated form of MyoD after somatic gene transfer in adult mice and rats.
Methods
Animals
The animal experiments were approved by the Norwegian Animal Research Authority and were conducted in accordance with the Norwegian Animal Welfare Act of December 20, 1975. The Norwegian Animal Research Authority provided governance to ensure that facilities and experiments were in accordance with Animal Welfare Act, National Regulations of January 15, 1996, and the European Convention for the Protection Vertebrate Animals Use for Experimental and Other Scientific Purposes of March 18, 1986.
The experiments were performed on female NMRI mice weighing 20–30 g and male Wistar rats weighing 200–300 g. All surgical procedures were performed under aseptic conditions and under deep anaesthesia. Before surgery an intraperitoneal injection of 5 μl (g body weight)−1 Equithesin (Ullevål Apotek, Norway) was administered. Equithesin contains 42.5 g l−1 chloralhydrat and 9.7 g l−1 pentobarbital. The depth of anaesthesia was monitored by checking for an extinguished withdrawal reflex by pinching the metatarsus region, and additional dosages of 1 μl g−1 were administered if necessary. Denervation was performed by excising several millimetres of the sciatic nerve in the thigh. The animals were physiologically and visually checked for re-innervation at the time of the terminal experiment, and no signs of re-innervation were observed.
Electrical stimulation
Chronic nerve stimulations of musculus extensor digitorum longus (EDL) muscles, with an electrical pattern that mimic slow motor units, were performed essentially as previously described (Windisch et al. 1998). In brief, un-insulated ends of two Teflon-coated steel electrodes were placed on each side of the sciatic nerve in anaesthetized animals. The other ends were placed under the skin and exiting through the head via a silicon tube fixed to the scalp. The electrodes were further connected to an electrical stimulator. A 20 Hz pulse train consisting of 200 bipolar pulses repeated every 30 s was used to mimic the electrical activity of slow type I motor units.
The chronic nerve stimulation of soleus muscles was delivered below a complete nerve block with tetrodotoxin (TTX) essentially as previously described (Reid et al. 2003; Martinov & Nja, 2005). In brief, the TTX was delivered with a capillary implanted beneath the epineurium of the sciatic nerve. Below the block the soleus nerve was stimulated with a 150 Hz pulse train consisting of 25 bipolar pulses repeated every 15 min in order to mimic the electrical activity in fast type IIb motor units. Muscles were stored at −80°C for further analyses. The contralateral leg was used as control in all experiments.
For the 10 h stimulation period the animals were kept under anaesthesia during the whole period, and they were killed by neck dislocation while still anaesthetized. Animals stimulated for 14 days were allowed to recover for 24 h before commencement of stimulation. The strength was then increased gradually over several hours for the animal to adapt to the involuntary movement. The stimulus strength was kept below the threshold for pain fibres at all times. The animal's health condition was evaluated at least twice daily by an experienced experimenter in addition to the routine monitoring by the animal facility staff. Procedures for killing if animals showed signs of pain or illness were in place, but the animals tolerated the stimulation well. The experiments were terminated by re-anaesthetizing the animals and excising the muscles before killing by neck dislocation.
RNA quantification
RNA was quantified by real-time PCR. Total RNA was isolated from whole rat muscles with 15 μl (mg tissue)−1 TRIzol reagent (Invitrogen). The quality of RNA was analysed using the 2100 Bioanalyser with RNA 6000 Nano Laboratory Chip Kit (Agilent), and quantity was determined using a NanoDrop spectrophotometer (Nanodrop Technologies). Reverse transcription was performed with 2 μg of Dnase-treated RNA, poly dT primers (see below) and Superscript III RT (Invitrogen). A lightcycler and a LightCycler FastStart DNA masterplus SYBR Green I kit (Roche) were used for the real-time PCR assay. All reactions were run in duplicates and mean values were used in further analysis. Data from the lightcycler PCRs were visualized in the LightCycler3 data Analysis Program (Roche) and results were expressed relative to the housekeeping gene β2-microglobulin (Mahoney et al. 2004). All procedures followed protocols specified by the manufactures.
The oligonucleotide primer sequences for the RT-PCR were as follows: myogenin (forward), AGAA-GTCACCCCAAGA; myogenin (reverse), GAACGAT-AGGGTTCCAAAG; MyoD (forward), GATCTCGGGT-GTAACAG; MyoD (reverse), CCCGCTTGAGGAA-TAAA; β2-microglobulin (forward), CTCCCCAAATT-CAAGTGT; b2-microglobulin (reverse), GTGAGCCAG-GATGTAG.
Northern blots were performed to confirm some of the RT-PCR results. RNAs were resolved on 1% agarose gel with denaturing conditions, transferred to Hybond membranes, which later were probed with 32P-labelled full-length MyoD or myogenin, and exposed to X-ray film.
Antibodies
A new antibody, anti-pMyoDT115, specific for MyoD phosphorylated at T115, was generated for this study. The phosphopeptide CDRRKAApTMRERRR, corresponding to residues 109–121 of rat MyoD with N-terminal cysteine added, was synthesized, coupled via the cysteine thiol to keyhole limpet haemocyanin and used to immunize a sheep. The immunization schedule and affinity purification of the antibody on a phosphopeptide column was as previously described (Sugden et al. 1999).
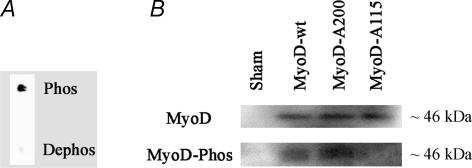
Samples of the phospho- and de-phosphopeptides used to generate the antibody (0.25 μg) were spotted onto nitrocellulose. The nitrocellulose was left to dry, washed briefly with Tris-buffered saline containing Tween (0.1%) then probed with affinity-purified antibody at 0.12 μg ml−1. Spots of phosphopeptide, but not dephosphopeptide, were still clearly detected down to < 10 ng (Fig. 1A). When used on protein extracts from MyoD-transfected cells anti-pMyoDT115 labelled a single band of the same size (∼46 kDa) as the band labelled with the antibody against total MyoD from Santa Cruz Biotechnology. The anti-pMyoDT115 did not label T115A or S200A mutated MyoD (Fig. 1B). The relevant peptide sequences for the other myogenic factors are very similar, but cross reactions were not observed in extracts from cells transfected with expression vectors for myogenin, MRF4 or myf-5. In addition, distinct identification of MyoD on Western blots is facilitated by MyoD being significantly larger (45–47 kDa) than the other myogenic factors (30–39 kDa).
Figure 1.
Anti-pMyoDT115 antibody is specific for MyoD phosphorylated at T115 A, spots of phosphopeptide, but not dephosphopeptide, were clearly detected down to < 10 ng. B, Western blot of nuclear extracts from Hek-293 cells after transfection with pCMS-EGFP expressing EGFP only (sham), pCMS-EGFP-MyoDwt expressing wild type MyoD, pSP3-MyoD expressing MyoD with a S200A mutation, or pCMS-EGFP-MyoDA115 expressing MyoD with a T115A mutation. The blots were stained with anti-MyoD or anti-pMyoDT115. Loading was adjusted for differences in transfection efficiency by adjusting the load to yield equal signals with anti-MyoD.
Antibodies against total myogenin and MyoD were from Santa Cruz Biotechnology.
Several antibodies specific for different types of myosin heavy chain were used. MY-32 (anti-all II) is from Sigma, while the following antibodies were gifts from Professor Stefano Schiaffino, Padova: BA-D5 (anti-I), SC-71 (anti-IIA), BF-F3 (anti-IIB) and BF-G6 (anti Embryonic).
Protein extraction and Western blotting
Extraction of nuclear protein from frozen muscles was performed as described elsewhere (Blough et al. 1999). Protein concentrations were determined with a Titertech multiscan spectrophotometer. Samples containing equal total protein (25 μg) were loaded onto 10% SDS-PAGE gels and run on a BIORAD electrophoresis set-up. Gels were electro-blotted onto a Hybond membrane (Amersham). Membranes were blocked with 5% low fat milk powder (Nestle) at 4°C overnight. MyoD and myogenin antibodies were diluted 1 : 500 and the anti-pMyoDT115 antibody was diluted 8 μg ml−1 in blocking solution, and incubated for 1 h at room temperature. Primary antibodies were detected by incubating with HRP-conjugated secondary antibodies to mouse, rabbit or sheep IgG at a 1 : 2000 dilution for 1 h at room temperature. Signal was developed with ECL kit (Amersham). All procedures were done according to the protocols provided by the manufactures. Developed films were scanned into a computer and quantified in ImageJ (NIH).
Plasmids
pCMS-EGFP (Clontech) encodes the reporter gene EGFP under control of a SV40 enhancer/promoter. pCMS-EGFP transfected alone was used in sham experiments. pCMS-EGFP-MyoDwt was constructed by insertion of mouse wild type MyoD in the EcoRI site of pCMS-EGFP. pCMS-EGFP-MyoD115A encodes mouse MyoD with an A to G mutation at position 343 of the coding sequence, resulting in a T115A mutation. Site-directed mutagenesis was performed using the QuickChange II Site-Directed Mutagenesis kit (Stratagene) with oligonucleotide primer 1 5′-CAAGGCCGCCGCCATGCGCGA 3′ and primer 2 5′-TCGCGCATGGCGGCGGCCTTG 3′. The mutated MyoD gene was ligated into the EcoRI site of pCMS-EGFP. DNA sequencing verified the mutation. MyoD in both pCMS-EGFP-MyoDwt and pCMS-EGFP-MyoDA115 are under the transcriptional control of a cytomegalovirus immediate-early promoter, and a SV40 polyadenylation signal.
pMyoD-200A (kindly provided by M. A. Harrington, Indiana University School of Medicine, IN, USA) contains MyoD mutated in serine200 (Song et al. 1998). pMyoD-200A does not express a reporter gene and was co-transfected with pCMS-EGFP. We have previously shown that such co-transfection results in almost 100% co-expression (Rana et al. 2004).
pCMS-EGFP-MyoD, pCMS-EGFP-MyoDA115 and pMyoD-200A were all tested in human embryonic kidney cells (Hek 293) after transfection with Lipofectamine 2000 (Invitrogen). After 48 h, nuclear protein was extracted (Andrews & Faller, 1991) and subjected to Western analysis (Fig. 1B). No MyoD was detected in sham-transfected Hek-293 cells. In transfected cells all the three plasmids displayed a single band at the expected ∼46 kDa. The phospho-specific antibody labelled only wild type and S200A MyoD.
In vivo transfection
Transfection of DNA was performed by electroporation essentially as previously described (Mathiesen, 1999; Rana et al. 2004). In brief, after anaesthesia the soleus muscle was exposed surgically, and 100 μl (rat) or 10 μl (mouse) of a DNA solution (1.0 μg μl−1 in 0.9% NaCl) was injected into the belly of the muscle. Immediately after, five trains of 1000 bipolar pulses (200 μs in each direction) with a peak to peak of 100 V (rat) or 20 V (mouse) were run across the muscle by two 10 mm × 1 mm silver electrodes spaced about 3 mm (rat) or 1 mm (mouse) apart. The animals were killed by neck dislocation under anaesthesia 14 days after transfection and muscles were subjected to immunohistological analysis.
Immunohistochemistry
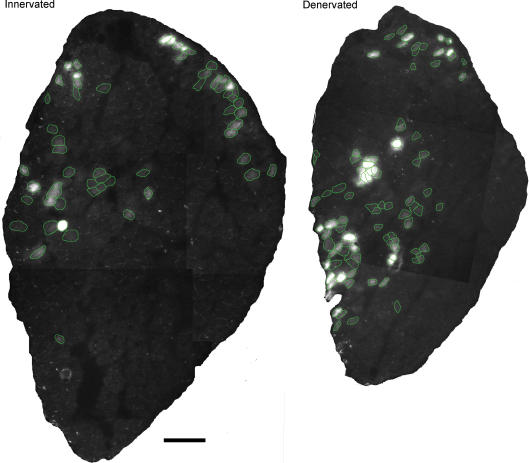
Muscles were frozen slightly stretched in isopentane at −160°C and stored at −80°C before being cryo-sectioned at 10 μm. One section was immediately dried with a hairdryer, which prevented lateral diffusion of EGFP. EGFP was observed in neighbouring sections stained for MyHC isoforms such that transfected and non-transfected fibres could be identified (Fig. 2). Fibre type was assigned according to Table 1. Sections were also stained with BF-G6 (anti-Embryonic myosin) to exclude regenerating fibres caused by damage related to DNA injection and electroporation. Unfixed sections were incubated with primary antibodies diluted 1 : 2000 in 1% BSA in PBS for 1 h at RT, washed in PBS, and incubated with a secondary antibody, anti-mouse IgG FITC (Sigma) or anti-mouse IgM Cy-3 (Jackson ImmunoResearch Laboratories), diluted 1 : 300 in 0.5% BSA in PBS for 30 min at 37°C.
Figure 2.
EGFP expression after electroporation Cross-sections of rat muscles electroporated with expression vectors containing EGFP. Fibres expressing EGFP are encircled in green for clarity. Both muscles were sectioned 14 days after electroporation. Scalebar, 50 μm.
Table 1.
MyHC antibodies and fibre type determination
| Antibody | Fibre type | |||||||
|---|---|---|---|---|---|---|---|---|
| MyHC | Name | Reference | I | I/IIa | IIa | IIx | I/IIx | IIb |
| Anti-I | BA-D5 | + | + | − | − | + | − | |
| Anti-IIa | SC-71 | Schiaffino et al. 1989 | − | + | + | − | − | − |
| Anti-IIb | BF-F3 | − | − | − | − | − | + | |
| Anti-all-II | MY-32 | Havenith et al. 1990 | − | + | + | + | + | + |
Statistics
Differences in mRNA and protein levels were compared pair-wise for experimental and control muscles from the same animal by a two-tailed Wilcoxon signed rank test, P≤ 0.05). A Fisher's exact test was used for comparison of number of fibres of a fibre type between normal and experimental fibres.
Supporting information
Supplementary Table S1 shows values of mRNA and protein levels in numbers (represented in Figs 3, 4 and 5 as bars), number of observations (n) and results from the statistical tests performed. Supplementary Table S2 shows the percentage fibre type distribution in numbers and results from the statistical tests performed.
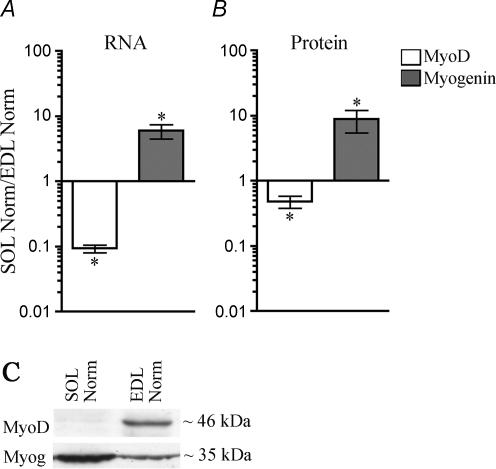
Figure 3.
MyoD and myogenin mRNA and protein levels are different in the slow soleus muscle and the fast EDL muscle A, The level of mRNA for MyoD and myogenin quantified by RT-PCR relative to expression of the housekeeping gene β2-microglobulin.B, the level of MyoD and myogenin proteins in nuclear extracts quantified on Western blots stained with anti-MyoD or anti-myogenin antibodies. C, examples of representative Western blots. Data are given as mean ±s.e.m. of the ratio between normal soleus muscles and normal EDL muscles on a logarithmic scale, n= 6–7. *P≤ 0.05. Numerical values are given in Table S1.
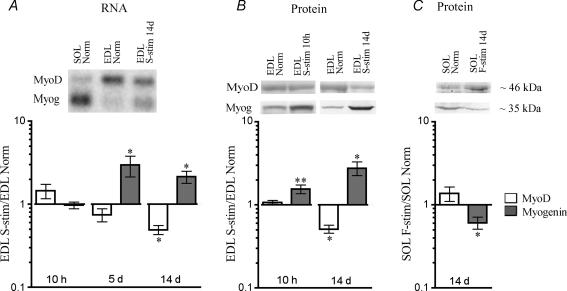
Figure 4.
MyoD and myogenin mRNA and protein levels change in response to electrical activity A, the level of mRNA for MyoD and myogenin quantified by RT-PCR relative to expression of the housekeeping gene β2-microglobulin (Myog). Some of the RT-PCR data were confirmed on Northern blots probed against MyoD and myogenin (upper panel). B and C, the level of MyoD and myogenin proteins in nuclear extracts quantified on Western blots stained with anti-MyoD or anti-myogenin antibodies. Examples of representative Western blots are shown above graphs. Data are given as the ratio between the levels in EDL muscles stimulated with a slow (S-stim) pattern, and contralateral normal EDLs (A and B), or as the ratio between fast-stimulated (F-stim) soleus muscles and contralateral normal solei (C). Data are given as mean ±s.e.m. of the stimulated versus normal muscles on a logarithmic scale, n= 6–10. *P≤ 0.05; **P≤ 0.01. Numerical values are given in Table S1.
Figure 5.
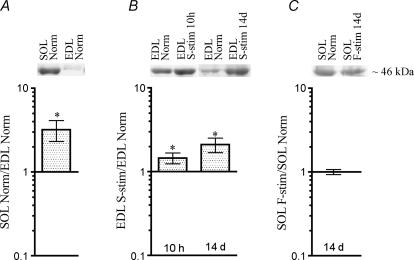
Levels of T115-phosphorylated MyoD differ between EDL and soleus, and are influenced by activity The level of MyoD T115-phosphorylated protein from nuclear extracts quantified on Western blots stained with the anti-pMyoDT115 antibody in normal solei relative to EDL (A), in normal EDL stimulated with a slow pattern (S-stim) 10 h and 14 days relative to normal EDL (B), and in soleus muscles stimulated with a fast pattern (F-stim) for 14 days relative to normal solei (C). Data are given as mean ±s.e.m. on a logarithmic scale, n= 6–11, and as representative Western blots (upper panels). *P≤ 0.05. Numerical values are given in Table S1.
Results
MyoD and myogenin are reciprocally expressed in fast and slow muscles
In order to confirm previous reports that MyoD and myogenin are differentially expressed in fast and slow muscles, we measured the level of mRNA and protein in the fast extensor digitorum longus (EDL) and the slow soleus muscle of rats. mRNA levels were measured by RT-PCR, and expressed relative to the housekeeping gene β2-microglobulin (see Methods). Measured in this way, MyoD mRNA was found to be 10-fold higher in EDL than in soleus, while the myogenin level was 6-fold higher in soleus than in EDL (Fig. 3A).
The level of MyoD protein measured by Western blotting was > 2-fold higher in EDL than in the soleus, while the myogenin level was 9-fold higher in soleus than EDL (Fig. 3B). Representative blots are shown in Fig. 3C.
MyoD and myogenin levels are reciprocally regulated by electrical activity
In order to investigate if MyoD and myogenin were regulated by electrical activity, we first subjected EDL to stimulation via the nerve with a pattern mimicking the activity in slow motor units (Hennig & Lømo, 1985; Eken & Gundersen, 1988).
The levels of mRNA for MyoD and myogenin showed no statistically significant change after 10 h (Fig. 4A). After 5 days, myogenin showed a 3-fold increase but although MyoD showed signs of reduction, this was not statistically significant. After 2 weeks, MyoD mRNA levels were reduced to half while myogenin was more than doubled compared with the normal contralateral leg (Fig. 4A).
Protein levels were investigated after 10 h and 2 weeks of stimulation (Fig. 4B). After 10 h the MyoD level was unaltered while myogenin was already increased by 50%. Since the mRNA for myogenin was unaltered at this stage, we speculate that the rapid effect of activity is on myogenin protein stability. After 2 weeks, MyoD protein levels were reduced to half; while myogenin levels were almost three times as high as in normal EDL muscles.
The converse experiment, to subject the slow soleus to a fast pattern was more complicated because of the high amounts of activity in slow motor units. Adding a fast pattern onto the background of slow activity was not an option, because high amounts of activity tend to induce slow muscle characteristics independent of frequency (Gundersen & Eken, 1992; DeNardi et al. 1993; Gundersen, 1998). We therefore combined nerve impulse block (see Methods) and stimulation. Protein levels of MyoD and myogenin were measured after 14 days. Although, on average, levels of MyoD were increased, the change was not statistically significant (Fig. 4C). The level of myogenin was significantly reduced by 39% (Fig. 4C).
To our knowledge the effects of specific patterns of electrical activity on MRF expression have not previously been investigated. Pette and collaborators found that hypothyroidism led to pronounced depression of MyoD, but only to small increases in myogenin mRNA in fast muscles, and adding electrical stimulation only slightly increased these effects (Kraus & Pette, 1997; Putman et al. 2000).
T115 phosphorylation of MyoD is differentially affected by different patterns of electrical stimulation
As described in the introduction, there is evidence that phosphorylation of T115 in MyoD can abolish its biological function by preventing it from binding to DNA (Liu et al. 1998). Thus, the total protein level might not reflect the level of active MyoD in muscles under different conditions. In order to investigate if the phosphorylation state of MyoD was related to muscle fibre type, or influenced by electrical activity, we made an antibody specific for MyoD phosphorylated at T115 (see Methods).
The level of phosphorylated, inactive MyoD was three times higher in soleus than in EDL (Fig. 5A), and since the total MyoD level was about half (Fig. 3A), the fraction of the total MyoD that is inactive must be six times higher in soleus than in EDL.
In the EDL, the amount of inactive phosphorylated MyoD (Fig. 5B) was significantly increased already after 10 h of slow stimulation, and after 2 weeks of stimulation it was more than doubled. At this time the level of total MyoD was reduced to half, so stimulation increased the fraction of inactive MyoD four times. The analysis does not allow a quantification of the fraction of MyoD that is active. However, the measured increase in phospho-MyoD and decrease in total MyoD protein by stimulation indicates that before stimulation the fraction of phosphorylated protein could not be higher than 1/4 of the total, and if it is this high, all the MyoD in the slow-stimulated EDL would be phosphorylated, and hence inactive.
When slow rat soleus was stimulated with a fast pattern for 2 weeks, the level of phosphorylated MyoD was virtually unchanged (Fig. 5C).
We conclude that there are significant pattern-specific alterations in the phosphorylation state at the functionally important amino acid T115 in muscles subjected to electrical stimulation. These findings suggest that slow stimulation patterns increase phosphorylation, and hence render a higher fraction of MyoD inactive.
Overexpression of wild type MyoD has variable effects in normally nerve-activated muscles
The battery of antibodies we used showed that normal soleus contained 50% type I, 49% type IIa and 1% type I/IIa in mice, and 87% type I, 12% type IIa 1% type I/IIa in rats. No type IIx or IIb fibres were found in either species. This is in agreement with previous observations (Windisch et al. 1998; Hughes et al. 1999). The results of the fibre typing of normal and experimental fibres are shown in Figs 6 and 7 and Table S2.
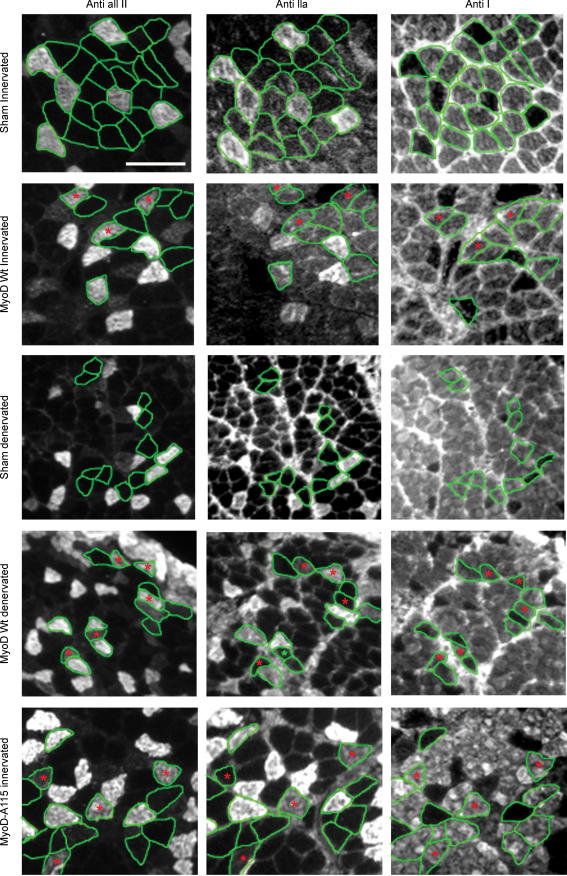
Figure 6.
MyHC immuno-histochemistry on neighbouring cross sections from rat soleus Transfected fibres are encircled in green and hybrid fibres are marked with red asterisks. Notice the increased amount of hybrid fibres in MyoD-positive denervated fibres and T115A-mutated MyoD fibres. All fibres were negative for IIb MyHC (not shown). Scalebar, 100 μm.
Figure 7.
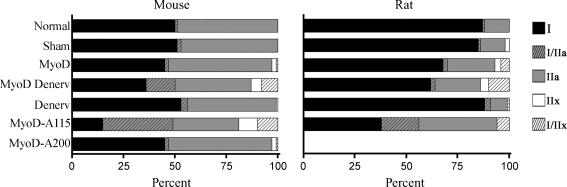
Active MyoD shifts fibre type distribution in the fast direction Serial cross sections from mice and rats were stained with antibodies against different MyHC isoforms and typed according to Table 1 (see Methods). Data are given for non-transfected fibres (Normal), non-transfected denervated fibres (Denerv); and 14 days after transfection with plasmids overexpressing EGFP only (Sham), or EGFP combined with wild type MyoD in innervated (MyoD) or denervated fibres (MyoD Denerv); T115A-mutated MyoD (MyoD-A115); or S200A-mutated MyoD (MyoD-A200). The mutated MyoDs were investigated in innervated fibres. Numerical values and statistical analysis are given in Table S2.
Transfection of reporter plasmid alone served as sham controls. Sham experiments had no effect on muscle fibre type in rats or mice, when observed 14 days after electroporation.
We then overexpressed the wild type form of MyoD in normally innervated mouse soleus. The fibre type distribution of expressing fibres was essentially the same as in the rest of the muscle, suggesting that higher levels of MyoD had no effect. In rats the overexpression fibres displayed a higher frequency of fast phenotype, most notably the number of type IIa fibres was doubled at the expense of type I fibres (Fig. 7).
Overexpression of wild type MyoD induces fast fibre types in denervated muscles
Given the minor effects of overexpression of wild type MyoD in normal mouse soleus, we hypothesized that the transgene MyoD might be rendered inactive by nerve activity-induced phosphorylation. In order to test this we abolished nerve activity by denervation. Denervation in itself had little effect on fibre type after 2 weeks (Fig. 7), but when denervation was combined with MyoD overexpression, MyoD had an effect on fibre type both in mice and rats. In mice, where overexpression of MyoD had no effect on normally innervated fibres, the number of fibres expressing type II myosin was increased from 50% to 64%. Type IIx MyHC was not detected in normal mouse soleus, but was now found in 13% of the fibres, either as pure type IIx, or as I/IIx hybrids. Similarly the number of hybrid I/IIa fibres was increased from 1% to 14%.
Similar results were obtained in the rat, but differed less from the effects obtained in innervated muscles (Figs 6 and 7). The largest difference was the higher number of type I/IIx fibres that increased from 4% in innervated MyoD fibres to 10% in denervated MyoD fibres.
We conclude that in the absence of nerve-evoked activity, forced expression of MyoD induces a fast phenotype in the slow soleus muscle in both mice and rats.
Overexpression of MyoD mutated at T115 induces fast fibre types in muscles receiving normal slow activity
We hypothesized that the lack of MyoD effects in normally active soleus muscles of the mouse was caused by the slow activity-inducing phosphorylation of T115. We therefore mutated T115 to an alanine (T115A), and hence abolished this phosphorylation site. The mutant form should be resistant to activity-mediated phosphorylation and inactivation, and thus efficient in transforming even normally active slow muscles.
We found that when mutated MyoD was transfected into normally active solei, it was highly efficient in transforming fibre type both in the mice and rats. The number of MyHC type II positive fibres increased from 50% in normal fibres, to 85% in MyoD-115A fibres of mice, and from 13% to 62% in rats. Thus, the effect of MyoD-115A was much stronger than wild type MyoD even after denervation. While normal mice had only 1% hybrid fibres, transfected fibres displayed 44% type I/II hybrids, and 9% of the fibres expressed type IIx alone. In the rat, 6% I/IIx hybrids appeared, while the number of IIa fibres increased from 12% to 38%. Due to lack of a IIx specific antibody, it cannot be excluded that IIa fibres also express IIx myosin.
All muscles were probed with anti-IIb antibody, but we never detected IIb expression in normal rat or mouse solei or any solei subjected to any of our experimental protocols. In control experiments using the BF-F3 antibody on mouse and rat EDL numerous IIb fibres were detected, thus validating our methodology.
Overexpression of MyoD mutated at S200 is inefficient in inducing fast fibre type in active mouse soleus
Cdk-1 and Cdk-2 phosphorylate MyoD at S200. The phosphorylated form is less stable, and a MyoD mutant that cannot be phosphorylated at this site displayed a 2- to 5-fold higher ability to transactivate muscle specific genes including MyHC in vitro (Song et al. 1998; Kitzmann et al. 1999). We hypothesized that the relative inefficiency of wild type MyoD in inducing effects could be related to an activity-induced phosphorylation and subsequent destabilization of the transgenic protein. To test this we used a MyoD that was mutated to alanine at position 200 (S200A) (Song et al. 1998). However, when this form was transfected into mouse muscles, only a minor increase in IIx fibres from 0% to 2% was observed, and the effect was virtually identical to the effects of wild type MyoD (Fig. 7). Thus, the relative inefficiency of MyoD in transforming normally active muscles is not related to a phosphorylation of S200, leading to a destabilization of the protein.
Discussion
Our data show that within hours, a slow pattern of electrical activity delivered to a fast muscle inactivates MyoD by phosphorylation at T115, and with more prolonged treatment reduced the mRNA and protein levels of MyoD. When active MyoD was overexpressed, a slow-to-fast fibre type transformation occurred, and we suggest that MyoD is a link between electrical activity and expression of fast muscle genes. Myogenin was regulated in the opposite direction and slow activity increased the protein level within hours. We have previously shown that forced expression of myogenin in adult muscles elevates oxidative enzymes in fast glycolytic fibres to levels typically found in slower fibre types, or in fast fibres subjected to slow activity (Ekmark et al. 2003). We hypothesize that a balance between myogenin and MyoD activity regulates adult fibre type. MyoD and myogenin are potent regulators of muscle-specific genes during early muscle differentiation, and might play a similar role in adults muscle to fine tune the muscle specific gene expression to changes in functional demands.
In the slow soleus muscle, the increased level of MyoD by fast stimulation was not significant, and the level of phosphorylated MyoD was unaltered. It is therefore possible that MyoD is more critical in maintaining normal fast properties in fast muscles than in inducing them in slow muscles. Fast-to-slow transformation upon slow stimulation of fast muscles, on the other hand, might be explained by reduced MyoD activity; first by phosphorylation, and subsequently by reduced expression of MyoD protein. The reduced MyoD activity might shut down expression of fast MyHC and genes for other fast isoenzymes.
Myogenin, on the other hand, was significantly decreased upon fast stimulation in the slow soleus and might explain the shift away form oxidative metabolism observed during slow-to-fast transformations (Gundersen et al. 1988). Myogenin has so far not been implicated in regulating of MyHC fibre type, and overexpression of wild type myogenin in transgenic animals, or in adults after somatic gene transfer failed to alter MyHC expression in normally active muscles (Hughes et al. 1999; Ekmark et al. 2003). Myogenin has, however, also an inhibitory phosphorylation site that is influenced by activity (Blagden et al. 2004), and the role of de-phosphorylated myogenin in regulating MyHC fibre type should be investigated in light of the present findings for MyoD. In fact, few of the large number of papers dealing with MRFs have considered their state of phosphorylation.
Reciprocal regulation of MyoD and myogenin in vivo
One of the most striking findings in the present study is the inverse response of MyoD and myogenin. In vitro experiments have suggested that the different MRFs regulate each other positively (Olson, 1990; Weintraub, 1993), but, as we have discussed previously (Gundersen et al. 1995), the situation in vivo might be different. Thus, in transgenic mice starting to overexpress myogenin in differentiated muscle cells, MyoD expression is decreased (Gundersen et al. 1995), and in mice with reduced MyoD expression, myf-5 expression is increased in a dose-dependent manner (Rudnicki et al. 1992). It is possible that, in addition to reacting differently to activity, MyoD and myogenin cross regulate each other negatively. Such a negative coupling would tend to stabilize fibres as ‘MyoD’ or ‘myogenin’ fibres, respectively, and hence as fast or oxidative fibres, respectively. Altered activity or forced expression of one of the factors would undermine the stability, and transform the phenotype such as we demonstrate here.
When muscles are subjected to denervation the expression of all the MRFs are increased ∼10-fold or more (Buonanno et al. 1992; Voytik et al. 1993). In spite of this we found little effect on MyHC fibre type after 2 weeks of denervation. It is possible that the ratio of MyoD/myogenin is critical, rather than the absolute levels of either protein. This would be the case, for example, if MyoD and myogenin were competing for a limiting co-factor. MRFs are dependent on E-proteins to form efficient transactivating heterodimers, and E47 might be limiting since expression is only increased 2-fold after denervation (Carlsen & Gundersen, 2000). In fast muscles the MyoD/myogenin ratio decreases after denervation (Walters et al. 2000) and with longer denervation times slower MyHCs can be detected (Patterson et al. 2006). The effect of denervation on MRF levels is less pronounced in slow muscles, and the MyoD/myogenin mRNA ratio is unaltered (Walters et al. 2000). It is possible, however, that more MyoD is in the active form in denervated muscles with no nerve-evoked activity, thus increasing the MyoD/myogenin activity ratio. This might explain why after 50 days of denervation the slow soleus display more type IIa and less type I MyHC (Patterson et al. 2006).
Upstream factors: how does activity influence MyoD?
Fast muscle fibres typically receive small amounts of action potentials from the CNS delivered in brief, high-frequency trains, while slow fibres are subjected to long trains of low-frequency activity (Hennig & Lømo, 1985). If denervated fibres are subjected to a native activity pattern they largely maintain their normal properties, while switching to an alternate pattern changes the muscle phenotype (Eken & Gundersen, 1988). Thus, the differential patterns of gene expression are somehow achieved by decoding the patterns of action potentials into an intracellular signal.
In the present study, 10 s long trains of low-frequency activity rendered MyoD less active within 10 h by phosphorylating T115, while 0.17 s brief high-frequency trains delivered every 15 min had no effect (Fig. 5). T115 is in a good context for protein kinase C (PKC) phosphorylation (Ma et al. 1994), and a constitutively active PKC form prevented MyoD from transactivating the muscle creatin kinase promoter in cell culture co-transfection studies (Li et al. 1992). Various PKC isoforms are differently driven by calcium-induced translocation and discylglycerol-mediated kinase activity that operates at the timescale of seconds (Oancea & Meyer, 1998; Mogami et al. 2003). Hence, PKC might act as a pattern-specific signal decoder of nerve-evoked activity. PKC has been demonstrated to be activated in rat muscle by large amounts of electrical stimulation (Cleland et al. 1989), and atypical PKC isoforms (aPKC) are activated within minutes following the onset of a single bout of endurance training in both mice (Chen et al. 2002) and men (Nielsen et al. 2003; Perrini et al. 2004; Rose et al. 2004).
Another good MyoD kinase candidate is calmodulin-dependent protein kinase-II. (CaMKII). CaMKII is also activated by contraction (Rose & Hargreaves, 2003) and can decode the frequency of calcium spikes (De Koninck & Schulman, 1998).
Also for myogenin, PKC and CaMKII have been implied in activity-dependent phosphorylation (Blagden et al. 2004). For electrical activity to regulate MyoD and myogenin activity differentially, however, the state of phosphorylation for these two factors should differ when the muscle is subjected to either fast or slow activity. MyoD and myogenin could either be phosphorylated by different kinases that are selectively activated by slow and fast patterns of activity, respectively, or alternatively activity-dependent phosphatases might have substrate specificity for MyoD versus myogenin.
Downstream factors: how does MyoD influence expression of fibre type-specific genes?
Our findings suggest that MyoD and myogenin work as different gene regulatory factors. Like other bHLH molecules, both MyoD and myogenin bind to the consensus sequence CANNTG, called an E-box, and the MRFs can to some extent substitute for each other in trans-activating muscle-specific genes. Nevertheless, differential trans-activation ability has been reported, and the position and number of E-boxes, their flanking sequences, and the specific proteins that interact with them, are all factors that can combine to give selectivity for various bHLH molecules (Munoz et al. 2002).
Our data suggest that type II MyHC promoters are activated in MyoD fibres. In mice the IIa and IIb gene each has two E-boxes, while the IIx gene has seven (Allen et al. 2001), but after overexpression of MyoD in C2C12 cells only the IIb, and not the IIa and IIx, promoter is activated (Allen et al. 2001). This demonstrates that MyoD can work selectively on different MyHC promoters in myotubes in culture, and might provide a mechanism to explain how MyoD could maintain IIb expression in normal EDL muscles. In the soleus fibres overexpressing MyoD only IIa and IIx were induced, and no IIb was detected. There are limitations preventing slow muscles from acquiring all the properties of a fast muscle even after prolonged electrical stimulation (Eken & Gundersen, 1988; Westgaard & Lømo, 1988), probably related to cell lineage (Kalhovde et al. 2005). Thus, no IIb MyHC was detected in soleus after more than 2 months of fast electrical stimulation (Ausoni et al. 1990), and even cross-innervation for more than a year failed to produce shortening velocities consistent with a high IIb-fibre content (Close, 1969; Eken & Gundersen, 1988). Interestingly, our data indicate that the ability of MyoD to induce IIb MyHC in the soleus is subjected to similar restrictions.
A differential effect of MyoD and myogenin has been demonstrated also for several genes relevant to fibre type other than MyHC. In the fast troponin I gene from quail the affinity of MyoD for various E-boxes was found to correlate with the ability to trans-activate, and substitution of E-boxes from other muscle-specific genes conferred less binding of MyoD, and less expression when introduced into the fast troponin I gene (Yutzey & Konieczny, 1992). The fast myosin light chain 1 has an upstream regulatory region where the distal part contains two E-boxes which are acting as an enhancer responsive to MyoD, while the proximal part contain a single E-box responsive to myogenin (Asakura et al. 1993). In this study the differences in transactivation were not attributed to binding ability, partly because MyoD and myogenin dimerizing with E12 were found to bind equally well to the two target sequences. For both the MLC1 and the troponin gene it was suggested, however, that flanking sequences of the E-boxes are involved in the target sequence specificity for different MRFs.
While several pathways have been suggested to link slow electrical activity to activation of slow muscle genes, we link de-phosphorylated MyoD to expression of fast genes. To our knowledge only one previous report has dealt with regulation of fast genes: overexpression of Six1 and its partner Eya1 induced a slow-to-fast transition of muscle phenotype (Grifone et al. 2004). It was, however, unclear how the Eya–Six system could be connected to electrical activity. MyoD can activate Six1 in proliferating myoblasts in vitro (Ishibashi et al. 2005), and if MyoD has the same ability in adult muscle fibres, our data suggest that MyoD could be linking Six1 expression to specific patterns of muscle activity.
We conclude that de-phosphorylated MyoD could be responsible for activating or maintaining a gene program for fast fibre type, either by interacting directly and differentially with promoters of fast muscle-specific genes, or by interacting with Six/Eya1 or other transcription factors. So far MyoD and Six/Eya1 are the only transcription factors implied in a fast fibre-type program for gene expression in adult muscle.
Acknowledgments
We thank Dr Maureen Harrington, Indianapolis, for donating the pMyoD-200A plasmid, Dr Stefano Schiaffino, Padova, for donating MyHC antibodies and Victoria Tudor Edwards, Tove Klungervik Larsen and Guro Sandvik for technical assistance. We are especially grateful to Dr Arild Njå, Oslo, for teaching us the TTX technique and for the generous donation of the TTX capillary containers. K.G. and M.E. were supported by grant 170473 from The Research Council of Norway. D.G.H. was supported by EXGENESIS, an Integrated Project (LSHM-CT-2004-005272) funded by the European Commission.
Supplementary material
Online supplemental material for this paper can be accessed at:
http://jp.physoc.org/cgi/content/full/jphysiol.2007.141457/DC1 and http://www.blackwell-synergy.com/doi/suppl/10.1113/jphysiol.2007.141457
References
- Agell N, Bachs O, Rocamora N, Villalonga P. Modulation of the Ras/Raf/MEK/ERK pathway by Ca2+, and calmodulin. Cell Signal. 2002;14:649–654. doi: 10.1016/s0898-6568(02)00007-4. [DOI] [PubMed] [Google Scholar]
- Allen DL, Sartorius CA, Sycuro LK, Leinwand LA. Different pathways regulate expression of the skeletal myosin heavy chain genes. J Biol Chem. 2001;276:43524–43533. doi: 10.1074/jbc.M108017200. [DOI] [PubMed] [Google Scholar]
- Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura A, Fujisawa SA, Komiya T, Nabeshima Y, Nabeshima YI. MyoD and myogenin act on the chicken myosin light-chain 1 gene as distinct transcriptional factors. Mol Cell Biol. 1993;13:7153–7162. doi: 10.1128/mcb.13.11.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausoni S, Gorza L, Schiaffino S, Gundersen K, Lomo T. Expression of myosin heavy chain isoforms in stimulated fast and slow rat muscles. J Neurosci. 1990;10:153–160. doi: 10.1523/JNEUROSCI.10-01-00153.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- Blagden CS, Fromm L, Burden SJ. Accelerated response of the myogenin gene to denervation in mutant mice lacking phosphorylation of myogenin at threonine 87. Mol Cell Biol. 2004;24:1983–1989. doi: 10.1128/MCB.24.5.1983-1989.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blough E, Dineen B, Esser K. Extraction of nuclear proteins from striated muscle tissue. Biotechniques. 1999;26:202–204. doi: 10.2144/99262bm05. 206. [DOI] [PubMed] [Google Scholar]
- Buonanno A, Apone L, Morasso MI, Beers R, Brenner HR, Eftimie R. The MyoD family of myogenic factors is regulated by electrical activity: isolation and characterization of a mouse Myf-5 cDNA. Nucleic Acids Res. 1992;20:539–544. doi: 10.1093/nar/20.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen H, Gundersen K. Helix-loop-helix transcription factors in electrically active and inactive skeletal muscles. Muscle Nerve. 2000;23:1374–1380. doi: 10.1002/1097-4598(200009)23:9<1374::aid-mus8>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Chen HC, Bandyopadhyay G, Sajan MP, Kanoh Y, Standaert M, Farese RV, Jr, Farese RV. Activation of the ERK pathway and atypical protein kinase C isoforms in exercise- and aminoimidazole-4-carboxamide-1-β-D-riboside (AICAR)-stimulated glucose transport. J Biol Chem. 2002;277:23554–23562. doi: 10.1074/jbc.M201152200. [DOI] [PubMed] [Google Scholar]
- Cleland PJ, Appleby GJ, Rattigan S, Clark MG. Exercise-induced translocation of protein kinase C and production of diacylglycerol and phosphatidic acid in rat skeletal muscle in vivo. Relationship to changes in glucose transport. J Biol Chem. 1989;264:17704–17711. [PubMed] [Google Scholar]
- Close RI. Dynamic properties of fast and slow skeletal muscles of the rat after nerve cross-union. J Physiol. 1969;204:331–346. doi: 10.1113/jphysiol.1969.sp008916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koninck P, Schulman H. Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science. 1998;279:227–230. doi: 10.1126/science.279.5348.227. [DOI] [PubMed] [Google Scholar]
- Delalande JM, Rescan PY. Differential expression of two nonallelic MyoD genes in developing and adult myotomal musculature of the trout (Oncorhynchus mykiss) Dev Genes Evol. 1999;209:432–437. doi: 10.1007/s004270050274. [DOI] [PubMed] [Google Scholar]
- DeNardi C, Ausoni S, Moretti P, Gorza L, Velleca M, Buckingham M, Schiaffino S. Type 2X-myosin heavy chain is coded by a muscle fiber type-specific and developmentally regulated gene. J Cell Biol. 1993;123:823–835. doi: 10.1083/jcb.123.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eken T, Gundersen K. Electrical stimulation resembling normal motor-unit activity: Effects on denervated fast and slow rat muscles. J Physiol. 1988;402:651–669. doi: 10.1113/jphysiol.1988.sp017227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekmark M, Gronevik E, Schjerling P, Gundersen K. Myogenin induces higher oxidative capacity in pre-existing mouse muscle fibres after somatic DNA transfer. J Physiol. 2003;548:259–269. doi: 10.1113/jphysiol.2002.036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friday BB, Mitchell PO, Kegley KM, Pavlath GK. Calcineurin initiates skeletal muscle differentiation by activating MEF2 and MyoD. Differentiation. 2003;71:217–227. doi: 10.1046/j.1432-0436.2003.710303.x. [DOI] [PubMed] [Google Scholar]
- Gorza L, Gundersen K, Lømo T, Schiaffino S, Westgaard RH. Slow-to-fast transformation of denervated soleus muscles by chronic high-frequency stimulation in the rat. J Physiol. 1988;402:627–649. doi: 10.1113/jphysiol.1988.sp017226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R, Laclef C, Spitz F, Lopez S, Demignon J, Guidotti JE, Kawakami K, Xu PX, Kelly R, Petrof BJ, Daegelen D, Concordet JP, Maire P. Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol Cell Biol. 2004;24:6253–6267. doi: 10.1128/MCB.24.14.6253-6267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen K. Determination of muscle contractile properties: the importance of the nerve. Acta Physiol Scand. 1998;162:333–341. doi: 10.1046/j.1365-201X.1998.0336e.x. [DOI] [PubMed] [Google Scholar]
- Gundersen K, Eken T. The importance of frequency and amount of electrical stimulation for contractile properties of denervated rat muscles. Acta Physiol Scand. 1992;145:49–57. doi: 10.1111/j.1748-1716.1992.tb09335.x. [DOI] [PubMed] [Google Scholar]
- Gundersen K, Leberer E, Lømo T, Pette D, Staron RS. Fibre type, calcium-sequestering proteins and metabolic enzymes in denervated and chronically stimulated muscles of the rat. J Physiol. 1988;398:177–189. doi: 10.1113/jphysiol.1988.sp017037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen K, Rabben I, Klocke BJ, Merlie J. Overexpression of myogenin in muscles of transgenic mice: interaction with Id-1, negative crossregulation of myogenic factors and induction of extrasynaptic acetylcholine receptor expression. Mol Cellular Biol. 1995;15:7127–7134. doi: 10.1128/mcb.15.12.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenith MG, Visser R, Schrijvers-van Schendel JMC, Bosman FT. Muscle fiber typing is routinely processed skeletal muscle with monoclonal antibodies. Histochemistry. 1990;93:497–499. doi: 10.1007/BF00266407. [DOI] [PubMed] [Google Scholar]
- Hennig R, Lømo T. Firing patterns of motor units in normal rats. Nature. 1985;314:164–166. doi: 10.1038/314164a0. [DOI] [PubMed] [Google Scholar]
- Hughes S, Chi MM, Lowry OH, Gundersen K. Myogenin induces a shift of enzyme activity from glycolytic to oxidative metabolism in muscles of transgenic mice. J Cell Biol. 1999;145:633–642. doi: 10.1083/jcb.145.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SM, Koishi K, Rudnicki M, Maggs AM. MyoD protein is differentially accumulated in fast and slow skeletal muscle fibres and required for normal fibre type balance in rhodents. Mech Dev. 1997;61:151–163. doi: 10.1016/s0925-4773(96)00631-4. [DOI] [PubMed] [Google Scholar]
- Hughes SM, Taylor JM, Tapscott SJ, Gurley CM, Carter WJ, Peterson CA. Selective accumulation of myoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Dev. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- Ishibashi J, Perry RL, Asakura A, Rudnicki MA. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions. J Cell Biol. 2005;171:471–482. doi: 10.1083/jcb.200502101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhovde JM, Jerkovic R, Sefland I, Cordonnier C, Calabria E, Schiaffino S, Lomo T. ‘Fast’ and ‘slow’ muscle fibres in hindlimb muscles of adult rats regenerate from intrinsically different satellite cells. J Physiol. 2005;562:847–857. doi: 10.1113/jphysiol.2004.073684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmann M, Vandromme M, Schaeffer V, Carnac G, Labbe JC, Lamb N, Fernandez A. cdk1- and cdk2-mediated phosphorylation of MyoD Ser200 in growing C2 myoblasts: role in modulating MyoD half-life and myogenic activity. Mol Cell Biol. 1999;19:3167–3176. doi: 10.1128/mcb.19.4.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus B, Pette D. Quantification of MyoD, myogenin, MRF4 and Id-1 by reverse-transcriptase polymerase chain reaction in rat muscles – effects of hypothyroidism and chronic low-frequency stimulation. Eur J Biochem. 1997;247:98–106. doi: 10.1111/j.1432-1033.1997.t01-1-00098.x. [DOI] [PubMed] [Google Scholar]
- Li L, Zhou J, James G, Heller-Harrison R, Czech MP, Olson EN. FGF inactivates myogenic helix-loop-helix proteins through phosphorylation of a conserved protein kinase C site in their DNA-binding domains. Cell. 1992;71:1181–1194. doi: 10.1016/s0092-8674(05)80066-2. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Liu LN, Dias P, Houghton PJ. Mutation of Thr115 in MyoD positively regulates function in murine fibroblasts and human rhabdomyosarcoma cells. Cell Growth Differ. 1998;9:699–711. [PubMed] [Google Scholar]
- Lunde IG, Ekmark M, Rana ZA, Buonanno A, Gundersen K. PPARδ expression is influenced by muscle activity and induces slow muscle properties in adult rat muscles after somatic gene transfer. J Physiol. 2007;582:1277–1287. doi: 10.1113/jphysiol.2007.133025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquet S, Lopez-Soriano J, Holst D, Fredenrich A, Melki J, Rassoulzadegan M, Grimaldi PA. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. Faseb J. 2003;17:2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- Ma PC, Rould MA, Weintraub H, Pabo CO. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- Mahoney DJ, Carey K, Fu MH, Snow R, Cameron-Smith D, Parise G, Tarnopolsky MA. Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol Genomics. 2004;18:226–231. doi: 10.1152/physiolgenomics.00067.2004. [DOI] [PubMed] [Google Scholar]
- Martinov VN, Nja A. A microcapsule technique for long-term conduction block of the sciatic nerve by tetrodotoxin. J Neurosci Metheds. 2005;141:199–205. doi: 10.1016/j.jneumeth.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Mathiesen I. Electropermeabilization of skeletal muscle enhances gene transfer in vivo. Gene Ther. 1999;6:508–514. doi: 10.1038/sj.gt.3300847. [DOI] [PubMed] [Google Scholar]
- Mogami H, Zhang H, Suzuki Y, Urano T, Saito N, Kojima I, Petersen OH. Decoding of short-lived Ca2+ influx signals into long term substrate phosphorylation through activation of two distinct classes of protein kinase C. J Biol Chem. 2003;278:9896–9904. doi: 10.1074/jbc.M210653200. [DOI] [PubMed] [Google Scholar]
- Munoz E, Brewer M, Baler R. Circadian transcription. Thinking outside the E-box. J Biol Chem. 2002;277:36009–36017. doi: 10.1074/jbc.M203909200. [DOI] [PubMed] [Google Scholar]
- Murgia M, Serrano L, Calabria E, Pallafacchina G, Lømo T, Schiaffino S. Ras is involved in nerve-activity-dependent regulation of muscle genes. Nature Cell Biol. 2000;2:142–147. doi: 10.1038/35004013. [DOI] [PubMed] [Google Scholar]
- Nielsen JN, Frosig C, Sajan MP, Miura A, Standaert ML, Graham DA, Wojtaszewski JF, Farese RV, Richter EA. Increased atypical PKC activity in endurance-trained human skeletal muscle. Biochem Biophys Res Commun. 2003;312:1147–1153. doi: 10.1016/j.bbrc.2003.11.041. [DOI] [PubMed] [Google Scholar]
- Oancea E, Meyer T. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell. 1998;95:307–318. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- Olson EN. MyoD family: a paradigm for development? Genes Dev. 1990;4:1454–1461. doi: 10.1101/gad.4.9.1454. [DOI] [PubMed] [Google Scholar]
- Patterson MF, Stephenson GM, Stephenson DG. Denervation produces different single fiber phenotypes in fast- and slow-twitch hindlimb muscles of the rat. Am J Physiol Cell Physiol. 2006;291:C518–C528. doi: 10.1152/ajpcell.00013.2006. [DOI] [PubMed] [Google Scholar]
- Perrini S, Henriksson J, Zierath JR, Widegren U. Exercise-induced protein kinase C isoform-specific activation in human skeletal muscle. Diabetes. 2004;53:21–24. doi: 10.2337/diabetes.53.1.21. [DOI] [PubMed] [Google Scholar]
- Pette D, Staron RS. Transitions of muscle fiber phenotypic profiles. Histochem Cell Biol. 2001;115:359–372. doi: 10.1007/s004180100268. [DOI] [PubMed] [Google Scholar]
- Putman CT, Dusterhoft S, Pette D. Satellite cell proliferation in low frequency-stimulated fast muscle of hypothyroid rat. Am J Physiol Cell Physiol. 2000;279:C682–C690. doi: 10.1152/ajpcell.2000.279.3.C682. [DOI] [PubMed] [Google Scholar]
- Rana ZA, Ekmark M, Gundersen K. Coexpression after electroporation of plasmid mixtures into muscle in vivo. Acta Physiol Scand. 2004;181:233–238. doi: 10.1111/j.1365-201X.2004.01282.x. [DOI] [PubMed] [Google Scholar]
- Reid B, Martinov VN, Nja A, Lomo T, Bewick GS. Activity-dependent plasticity of transmitter release from nerve terminals in rat fast and slow muscles. J Neurosci. 2003;23:9340–9348. doi: 10.1523/JNEUROSCI.23-28-09340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescan PY, Gauvry L, Paboeuf G. A gene with homology to myogenin is expressed in developing myotomal musculature of the rainbow trout and in vitro during the conversion of myosatellite cells to myotubes. FEBS Lett. 1995;362:89–92. doi: 10.1016/0014-5793(95)00215-u. [DOI] [PubMed] [Google Scholar]
- Rose AJ, Hargreaves M. Exercise increases Ca2+-calmodulin-dependent protein kinase II activity in human skeletal muscle. J Physiol. 2003;553:303–309. doi: 10.1113/jphysiol.2003.054171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AJ, Michell BJ, Kemp BE, Hargreaves M. Effect of exercise on protein kinase C activity and localization in human skeletal muscle. J Physiol. 2004;561:861–870. doi: 10.1113/jphysiol.2004.075549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of myoD in mice leads to up-regulation of myogenic HLH gene myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Gorza L, Sartore S, Saggin L, Ausoni S, Vianello M, Gundersen K, Lomo T. Three myosin heavy chain isoforms in type 2 skeletal muscle fibres. J Muscle Res Cell Motil. 1989;10:197–205. doi: 10.1007/BF01739810. [DOI] [PubMed] [Google Scholar]
- Seward DJ, Haney JC, Rudnicki MA, Swoap SJ. bHLH transcription factor MyoD affects myosin heavy chain expression pattern in a muscle-specific fashion. Am J Physiol Cell Physiol. 2001;280:C408–C413. doi: 10.1152/ajpcell.2001.280.2.C408. [DOI] [PubMed] [Google Scholar]
- Song A, Wang Q, Goebl MG, Harrington MA. Phosphorylation of nuclear MyoD is required for its rapid degradation. Mol Cell Biol. 1998;18:4994–4999. doi: 10.1128/mcb.18.9.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C, Crawford RM, Halford NG, Hardie DG. Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5′-AMP. Plant J. 1999;19:433–439. doi: 10.1046/j.1365-313x.1999.00532.x. [DOI] [PubMed] [Google Scholar]
- Voytik SL, Przyborsky M, Badylak SF, Koenieczny SF. Differential expression of muscle regulatory factor genes in normal and denervated adult rat hindlimb muscles. Dev Dynamics. 1993;198:214–224. doi: 10.1002/aja.1001980307. [DOI] [PubMed] [Google Scholar]
- Walters EH, Stickland NC, Loughna PT. The expression of the myogenic regulatory factors in denervated and normal muscles of different phenotypes. J Muscle Res Cell Motil. 2000;21:647–653. doi: 10.1023/a:1005683825960. [DOI] [PubMed] [Google Scholar]
- Wang Y-X, Zhang CL, Tou RT, Cho HK, Nelson MC, Bayagua-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPAR-δ. PLOS Biol. 2004;2:1–8. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. The myoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- Westgaard RH, Lømo T. Control of contractile properties within adaptive ranges by patterns of impulse activity in the rat. J Neurosci. 1988;8:4415–4426. doi: 10.1523/JNEUROSCI.08-12-04415.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windisch A, Gundersen K, Szabolcs MJ, Gruber H, Lomo T. Fast to slow transformation of denervated and electrically stimulated rat muscle. J Physiol. 1998;510:623–632. doi: 10.1111/j.1469-7793.1998.623bk.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutzey KE, Konieczny SF. Different E-box regulatory sequences are functionally distinct when placed within the context of the troponin I enhancer. Nucleic Acids Res. 1992;20:5105–5113. doi: 10.1093/nar/20.19.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.