Abstract
PPARα agonism impairs mitochondrial function, but the effect of PPARδ agonism on mitochondrial function is equivocal. Furthermore, PPARα and δ agonism increases muscle fatty acid oxidation, potentially via activation of FOXO1 signalling and PDK4 transcription. Since FOXO1 activation has also been suggested to increase transcription of MAFbx and MuRF-1, and thereby the activation of ubiquitin–proteasome mediated muscle proteolysis, this raises the possibility that muscle fuel selection and the induction of a muscle atrophy programme could be regulated by a single common signalling pathway. We therefore investigated the effect of PPARδ (delta) agonist, GW610742, administration on muscle mitochondrial function, fuel regulation, and atrophy and growth related signalling pathways in vivo. Twenty-four male Wistar rats received vehicle or GW610742 (5 and 100 mg per kg body mass (bm)) orally for 6 days. Soleus muscle was used to determine maximal rates of ATP production (MRATP) in isolated mitochondria, gene and protein expression, and enzyme activities. MRATP were unchanged by GW610742. Muscle PDK2 and PDK4 mRNA expression increased with GW610742 (100 mg (kg bm)−1) compared to vehicle (P < 0.05), and was paralleled by a twofold increase in PDK4 protein expression (P < 0.05). The activity of β-hydroxyacyl-CoA dehydrogenase increased with GW610742 (P < 0.05). Muscle MuRF1 and MAFbx mRNA expression was increased by GW610742 (100 mg (kg bm)−1) compared to vehicle (P < 0.05), and was matched by increased protein expression (P < 0.001), whilst Akt1 protein declined (P < 0.05). There was no effect of GW610742 on 20S proteasome activity and mRNA expression, or the muscle DNA: protein ratio. GW610742 switched muscle fuel metabolism towards decreased carbohydrate use and enhanced lipid utilization, but did not induce mitochondrial dysfunction. Furthermore, GW610742 initiated a muscle atrophy programme, possibly via changes in the Akt1/FOXO/MAFbx and MuRF1 signalling pathway.
Peroxisome proliferator-activated receptors (PPARs) are members of the nuclear hormone receptor superfamily of ligand-activated transcription factors that interact with a number of endogenous lipids and drugs normally used to treat metabolic diseases in humans (Berger & Wagner, 2002). There are three distinct PPAR subtypes that are commonly designated PPARα, PPARδ (also known as PPARβ) and PPARγ (Lee et al. 2003). PPARα (fibrates) and PPARγ agonists are now used widely in the treatment of insulin resistance and dyslipidaemia. Furthermore, PPARδ agonists have been identified as also being a viable treatment strategy since they can reduce body fat content and insulin resistance in obese mice by apparently increasing fatty acid oxidation in skeletal muscle (Tanaka et al. 2003).
PPARα agonist administration has been reported to impair mitochondrial function, at least in vitro, and several different mechanisms have been proposed to explain this phenomenon, including membrane depolarization, uncoupling of respiration, induction of mitochondrial permeability transition, and direct inhibition of mitochondrial respiration (Keller et al. 1992; Zhou & Wallace, 1999; Qu et al. 2001). Evidence that PPARδ agonist administration impairs mitochondrial function is less clear. It has recently been shown that administration of the PPARδ agonist, GW501516, uncouples oxidative phosphorylation in isolated mitochondria, albeit when administered at a relatively high concentration (Brunmair et al. 2006). Increased mitochondrial uncoupling is usually accompanied by increased reactive oxygen species (ROS) generation, leading to deleterious effects on mitochondrial function (Fosslien, 2001). Conversely, GW501516 has also been reported to enhance mitochondrial biogenesis in skeletal muscle (Hondares et al. 2007), and to induce a switch in fast twitch muscle towards a slow phenotype (Wang et al. 2004). It would seem therefore that the influence of PPARδ agonism on mitochondrial function in skeletal muscle is currently unresolved.
An increase in fatty acid oxidation induced by PPARα and δ agonism (Motojima & Seto, 2003; Abbot et al. 2005) and starvation (Wu et al. 2001) has been shown to result in the transcriptional activation of muscle pyruvate dehydrogenase kinase 4 (PDK4), the enzyme that phosphorylates, and therefore inactivates, the pyruvate dehydrogenase complex (PDC), the rate limiting step in muscle carbohydrate oxidation. The mechanism of PDK4-induced PDC inhibition in starvation has been attributed to either increased fatty acid-mediated activation of the forkhead transcription factor FOXO1, followed by direct binding to the promoter region of the PDK4 gene (Furuyama et al. 2003), or through free fatty acid-mediated activation of the PPARα receptor (Wu et al. 2001). Irrespective of the mechanism, PDC inhibition could clearly alter fuel selection in skeletal muscle towards fat oxidation, when free fatty acid availability is increased, and thereby explain some of the reported effects of PPAR agonists on muscle fuel utilization.
The principal regulator of muscle proteolysis is the ATP-dependent ubiquitin–proteasome system. This system includes obligatory substrate ‘tagging’ by ubiquitination, after which proteins are recognized and degraded by the 26S proteasome complex. Ubiquination involves the interaction of three classes of proteins: ubiquitin activating (E1), ubiquitin conjugating (E2) and ubiquitin ligating enzymes (E3) or ligases. Two muscle specific ubiquitin ligases, muscle ring finger 1 (MuRF-1) and muscle atrophy F-box (MAFbx), also called Atrogin-1 (Gomes et al. 2001), have been identified and, similar to PDK4, both appear to be regulated by the Akt1–FOXO1 signalling pathway (Leger et al. 2006). Both MAFbx and MURF-1 have consistently been found to be up-regulated prior to the onset of atrophy in multiple models of muscle wasting, including disuse (Bodine et al. 2001a) and cachexia (Gomes et al. 2001). The relevance of this to the current study is that, given the reported increase in fatty acid oxidation following PPARδ agonist administration (Abbot et al. 2005), and the proposed related activation of FOXO1 signalling and PDK4 transcription (Furuyama et al. 2003), it is not unreasonable to suggest this might also result in increased transcription of MAFbx and MURF-1, and thereby the activation of ubiquitin–proteasome mediated muscle proteolysis, which to our knowledge has not been investigated.
The novel aims of the present study therefore were to concurrently determine in vivo the effects of GW610742, a high affinity PPARδ agonist (Sznaidman et al. 2003; Abbot et al. 2005), on mitochondrial function (ATP production capacity), the regulation of muscle fuel selection, and the expression of genes and proteins known to regulate ubiquitin–proteasome mediated muscle proteolysis. This would allow us to elucidate for the first time whether PPARδ agonism results in the impairment of mitochondrial function, and potentially regulates muscle fuel selection and the induction of a muscle atrophy programme via a single common signalling pathway.
Methods
Animals and tissue collection
Twenty-four male Wistar (Charles River, Margate, UK) rats (average mass 310 g) were randomly gavage fed with the PPARδ agonist GW610742 on a daily basis for 6 days, at a dose of 5 (n = 8) or 100 mg per kg body mass (bm) (n = 8), or an equivalent volume of a 0.1% vehicle solution (hydroxylpropylmethyl cellulose; n = 8). Food and water were available ad libitum, and animals were kept at a constant ambient temperature with light–dark cycles of 12 h. Following the seventh day, animals were terminally anaesthetized with sodium thiobutabarbital (Inactin, 120 mg (kg bm)−1; i.p.) and the soleus muscle of each limb was exposed and removed. One muscle was immediately snap frozen in liquid nitrogen, whilst the other was immersed in an ice-cold saline solution, after which each animal was humanely killed (according to the UK Home Office Guidelines). All experimental use of animals was approved by the UK Home Office and conducted in accordance with UK legislation governing the use of animals in research (Animals (Scientific Procedures) Act 1986).
Mitochondrial function
Approximately 50 mg of tissue was excised from the middle part of each non-frozen soleus muscle, weighed and used immediately for the bio-luminometric determination of maximal rates of ATP production (MAPR) in isolated mitochondria, using a variety of substrates (Wibom et al. 1990). The same mitochondrial sample was used at a later date for the measurement of glutamate dehydrogenase (GluDH) activity and mitochondrial protein content (see below). The yield of mitochondria for each mitochondrial preparation was determined from the relationship between GluDH activity in the whole muscle sample and in the mitochondrial suspension.
Muscle metabolites and DNA
Snap frozen soleus muscle was subsequently divided into two parts whilst under liquid nitrogen. One part was freeze-dried, dissected free from visible connective tissue and blood, and powdered. Five to ten milligrams of this muscle powder was then extracted with 0.5 mol l−1 perchloric acid containing 1 mmol l−1 EDTA, and, after centrifugation, the supernatant was neutralized with 2.2 mol l−1 KHCO3. Free carnitine and acetylcarnitine were measured in the neutralized extract by enzymatic assays that made use of a radioisotopic substrate, as previously described (Cederblad et al. 1990). Muscle ATP, PCr, creatine and lactate concentrations were determined fluorometrically using a modification of the method of Harris et al. (1974). Muscle DNA and total alkaline soluble protein concentrations were determined as previously described (Forsberg et al. 1991).
Muscle enzyme measurements
Snap frozen muscle (5–10 mg) was homogenized on ice in a glass homogeniser containing a solution with 50 mmol l−1 KH2PO4, 1 mmol l−1 EDTA and 0.05% Triton X-100. The muscle homogenates were then analysed for enzyme activity as described by Opie & Newsholme (1967) and Zammit & Newsholme (1976). The chymotrypsin peptide-hydrolysing activity of the 20S proteasome was measured using the fluorogenic substrate N-Suc-Leu-Leu-Val-Tyr-7-amido-4-methyl-coumarin (Sigma) (Dawson et al. 1995).
Protein assay
Protein concentration was determined in the mitochondrial and enzyme suspensions using the Bradford method (Bio-Rad, USA).
Real-time PCR
Total RNA was isolated from snap frozen soleus muscle using RNA plus (Qbiogene) according to the manufacturer's protocol. First strand cDNA was then synthesized from 1 μg RNA sample using random primers (Promega) and PowerScript Reverse Transcriptase (BD Biosciences).
Taqman PCR was carried out using an ABI prism 7000 sequence detector (Applied Biosystems, USA), with 2 μl of cDNA, 18 μmol l−1 of each primer, 5 μmol l−1 probe, and Universal Taqman 2× PCR Mastermix (Eurogentec) in a 25 μl final volume. Each sample was run in triplicate, in duplex reactions. Cyclophilin A labelled with the fluorescent dye VIC was used as internal control, while all genes of interest were labelled with the fluorescent reporter FAM. The thermal cycling conditions used were: 2 min at 50°C, 10 min at 95°C, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Ct values of the target gene were normalized to Ct values of the internal control cyclophilin A, and final results were calculated according to the 2−ΔΔCt method. The group of vehicle treated rats was used as calibrators with a value of 1. The choice of using cyclophilin A as housekeeping gene was based upon data showing stability of this gene in atrophy models such as ageing and muscle unloading (Lowe et al. 2000; Alway et al. 2001).
Immunoblotting
Each muscle sample was homogenized in Tris buffer (50 mmol l−1 Tris– 1 mmol l−1 EDTA, pH 7.5) supplemented with inhibitors of proteases and phosphatases (Sigma P2850). After homogenation each muscle extract was centrifuged for 15 min at 10 000 g. The supernatant was collected and stored at −80°C. Protein concentration in the supernatant was measured using the Bradford assay. Protein samples were run on a 4–12% Bis–Tris acrylamide gel (Invitrogen, UK) for 2 h at constant 200 V and transferred on a polyvinylidene difluoride membrane (PVDF) overnight at constant 100 mA, in ice-cold buffers (4°C).
The protein transfer was checked with Ponceau S red staining, before blocking the membrane in BSA–TBS–Tween for 1 h at RT. Membranes were probed with the primary antibody overnight at 4°C. The antibodies used in this study were purchased from Cell Signalling Technology (USA), with the exception of MuRF1, MaFbx and PDK4, which were produced in-house by Pfizer Inc. (USA) and Astra-Zeneca (UK) and provided as a gift. Membranes were washed in TBS-T and incubated with the appropriate secondary antibody, either horseradish peroxidase (HRP)-linked anti-mouse IgG (Dako, UK), or anti-rabbit IgG (Amersham). The membranes were washed again in the same buffer, incubated with ECL chemiluminescence detection (Pierce, UK) and exposed to X-ray film.
Densitometry
Blots were scanned using a Duo Scan T1200 Agfa scanner. Bands were identified using the Quantity One programme from Bio-Rad, and the optical density volume was adjusted by subtracting the local background. All values were normalized to an actin protein control.
Statistics
All data are expressed as means ±s.e.m. To investigate the treatment effect, one-way analysis of variance (ANOVA) was applied. When a significant F-ratio was obtained, a LSD post hoc test was applied to locate specific differences. Significance was set at the P < 0.05 level of confidence.
Results
The increase in body mass over 6 days of treatment with either vehicle control (0.1% hydroxy-propylmethylcellulose) or GW610742 was no different between treatment groups (data not shown).
Muscle metabolites
Table 1 shows muscle ATP, PCr, creatine, total creatine, lactate, acetylcarnitine, DNA and total protein concentrations in rat soleus muscle after vehicle or GW610742 treatment. No differences were observed between treatment groups.
Table 1.
Muscle metabolite and DNA and alkaline soluble protein concentrations in rat soleus skeletal muscle after six days of gavage feeding of GW610742 at 5 (n = 8), and 100 mg (kg bm)−1 (n = 8) or with a 0.1% vehicle solution (hydroxypropylmethylcellulose; n = 8)
| Vehicle | 5 mg kg−1 | 100 mg kg−1 | |
|---|---|---|---|
| ATP (mmol (kg DM)−1) | 18.08 ± 1.03 | 19.62 ± 1.14 | 19.59 ± 0.44 |
| PCr (mmol (kg DM)−1) | 55.31 ± 2.57 | 57.22 ± 3.58 | 63.60 ± 1.31 |
| Creatine (mmol (kg DM)−1) | 45.86 ± 2.66 | 43.00 ± 3.39 | 41.11 ± 1.88 |
| Total creatine (mmol (kg DM)−1) | 101.2 ± 3.9 | 100.2 ± 3.7 | 104.7 ± 2.2 |
| Lactate (mmol (kg DM)−1) | 0.70 ± 0.05 | 0.76 ± 0.09 | 0.87 ± 0.11 |
| Acetylcarnitine (mmol (kg DM)−1) | 0.93 ± 0.05 | 0.97 ± 0.03 | 0.92 ± 0.09 |
| DNA (g (kg DM)−1) | 4.17 ± 0.48 | 4.73 ± 0.43 | 4.16 ± 0.26 |
| Protein (g (kg DM)−1) | 449.1 ± 32.5 | 497.4 ± 18.4 | 481.1 ± 13.2 |
| DNA/protein | 115.5 ± 18.1 | 108.2 ± 11.6 | 116.7 ± 4.3 |
Values expressed as means ±s.e.m. (n = 8). DM, dry muscle.
Muscle energy metabolism
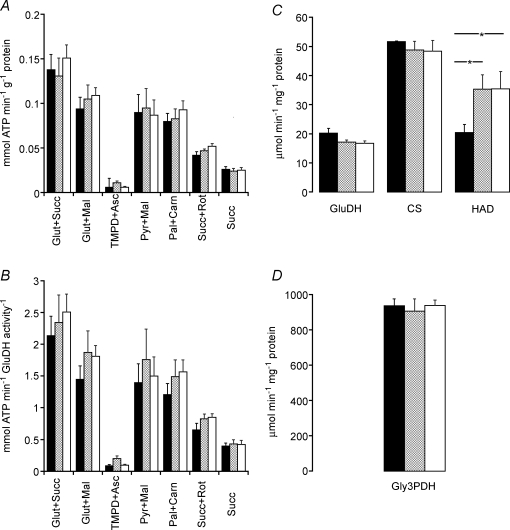
The maximal rate of mitochondrial ATP production, using a variety of substrates, was not different across treatment groups, irrespective of the reference base used to normalize mitochondrial data (i.e. mitochondrial protein, Fig. 1A, or mitochondrial density, as determined by glutamate dehydrogenase activity, Fig. 1B).
Figure 1.
Maximal rates of ATP production in skeletal muscle A and B, maximal rate of mitochondrial ATP production at 25°C, using a variety of substrates, normalized to either mitochondrial protein content (A) or to mitochondrial density (activity of glutamate dehydrogenase; B) measured in soleus muscle of rats fed with either vehicle (hydroxypropylmethylcellulose) (filled columns) or GW610742 at 5 mg (kg bm)−1 (cross-hatched columns) and 100 mg (kg bm)−1 (open columns). C and D, enzymatic activities of glutamate dehydrogenase (GluDH), citrate synthase (CS), β-hydroxyl acyl coenzyme A dehydrogenase (HAD) (C) and glyceraldehyde-3-phosphate dehydrogenase (Gly3PDH, D) normalized to the protein content measured in soleus muscle of rats fed with either vehicle (hydroxypropylmethylcellulose) (filled columns) or GW610742 at 5 mg (kg bm)−1 (cross-hatched columns) and 100 mg (kg bm)−1 (open columns). *Significantly different from the corresponding control (vehicle) group (P < 0.05).
The activities of muscle glutamate dehydrogenase and citrate synthase (both markers of mitochondrial density; Fig. 1C), and glyceraldehyde-3-dehydrogenase (a marker of glycolysis; Fig. 1D) were unchanged following 6 days of GW610742 treatment. However, the activity of muscle β-hydroxyacyl-CoA dehydrogenase (a marker for mitochondrial β-oxidation) was significantly increased in both treatment groups compared with vehicle control, and by a similar magnitude (75%, P < 0.05; Fig. 1C).
Muscle mRNA and protein expression
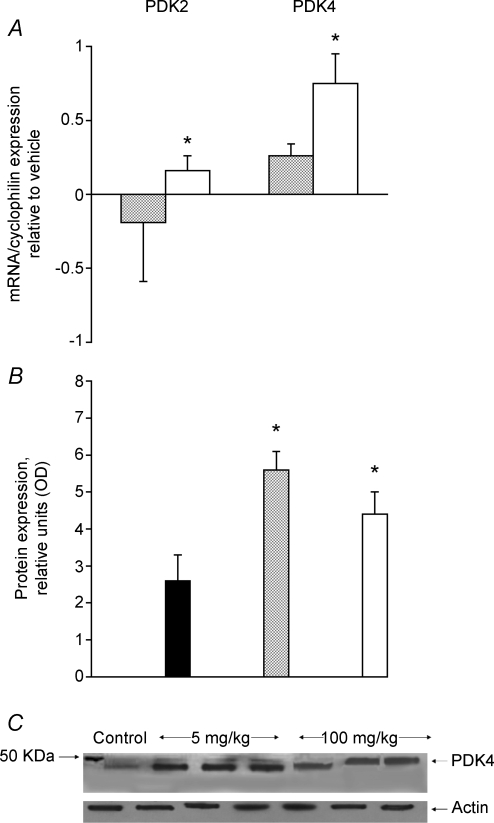
Figure 2A illustrates the change in mRNA expression from control of the two muscle specific pyruvate dehydrogenase kinase isoforms, PDK2 and PDK4, after 6 days of treatment with GW610742. The mRNA levels of PDK2 and PDK4 increased significantly in the 100 mg (kg bm)−1 treated group compared with control (1.3 ± 0.1, P < 0.05, and 1.8 ± 0.2, P < 0.05, respectively). This was accompanied by almost twofold greater PDK4 protein expression (100 mg (kg bm)−1) or more (5 mg (kg bm)−1) in the GW610742 treated groups when compared with control (both P < 0.05, Fig. 2B). A typical Western blot of PDK4 protein detected at 46 kDa is presented in Fig. 2C. The mRNA expression of two PDC phosphatase isoforms, PDP1 and PDP2, whose effect opposes that induced by PDK2 and PDK4, remained unchanged across the groups (data not shown).
Figure 2.
PDK2 and 4 mRNA and protein expression in skelatal muscle A, the relative expression of pyruvate dehydrogenase kinase (PDK) 2 and 4 mRNAs measured in soleus muscle of rats fed with either vehicle (hydroxypropylmethylcellulose) or GW610742 at 5 mg (kg bm)−1 (cross-hatched columns) and 100 mg (kg bm)−1 (open columns). The values are expressed as 2−ΔΔCt normalized to endogenous cyclophilin. The vehicle group was used as calibrator with a value of 1. *Significantly different from the corresponding control (vehicle) group (P < 0.05). B, the average density of pyruvate dehydrogenase kinase PDK4 protein bands, measured by Western blot in soleus muscle of rats fed with either vehicle (hydroxypropylmethylcellulose) (filled columns) or GW610742 at 5 mg (kg bm)−1 (cross-hatched columns) and 100 mg (kg bm)−1 (open columns). *Significantly different from the corresponding control (vehicle) group (P < 0.05). C, a representative Western blot of PDK4 protein detected at 46 KDa.
The administration of the GW610742 did not cause any change in PPARα, PPARγ or PPARδ mRNA expression (data not shown).
Muscle atrophy molecular pathway
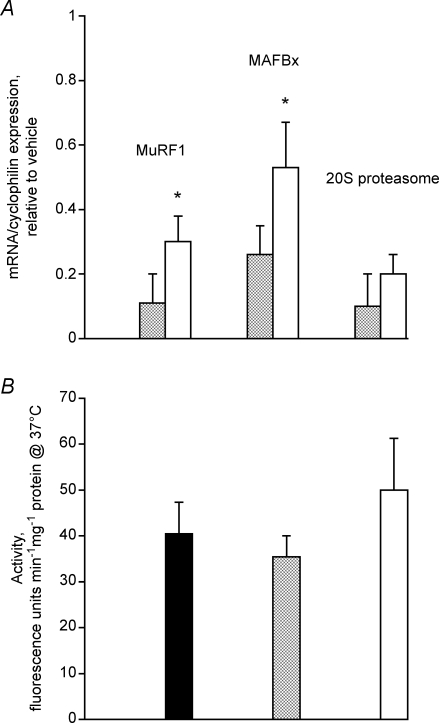
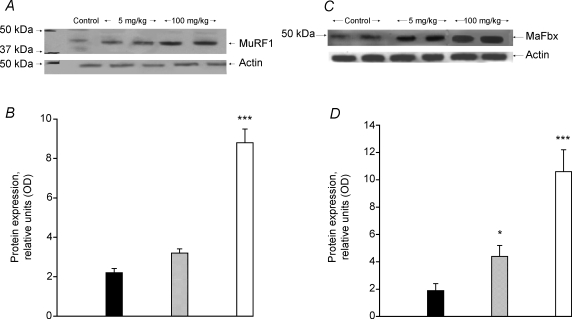
The transcriptional levels of ubiquitin ligases, MuRF1 and MAFbx were significantly increased in the 100 mg (kg bm)−1 group compared with control (> 30% increase in the MuRF1 mRNA, P < 0.05; and > 50% increase in MAFbx mRNA, P < 0.05; Fig. 3A). Measurements of the transcriptional (Fig. 3A) and enzymatic (expressed as 20S chymotrypsin-like activity, Fig. 3B) activity of the α subunit of the 20S proteasome were unchanged from control following GW610742 administration. Figure 4A illustrates a typical Western blot for MuRF1 (band at 42 kDa) showing elevated levels of protein at the highest dose of agonist administered. The overall level of MuRF1 protein at the 100 mg (kg bm)−1 dose of GW610742 increased fourfold compared with the control group (Fig. 4B; P < 0.001). MAFbx protein was identified as a band at 50 kDa (Fig. 4C). Similar to MuRF1, there was an increase in the level of MAFbx protein expression with the highest dose of the GW610742 administered (Fig. 4D), with the average band intensity in the 100 mg (kg bm)−1 group being sixfold greater than in the control group (P < 0.001).
Figure 3.
Skelatal muscle proteasome system A, the relative expressions of MuRF1, MAFbx and 20S proteasome mRNAs measured in soleus muscle of rats fed with either vehicle (hydroxypropylmethylcellulose) or GW610742 at 5 mg (kg bm)−1 (cross-hatched columns) and 100 mg (kg bm)−1 (open columns). The values are expressed as 2−ΔΔCt normalized to endogenous cyclophilin. The group of vehicle treated rats was used as calibrator with a value of 1. *Significantly different from the corresponding control (vehicle) group (P < 0.05). B, the 20S proteasome chymotrypsin-like enzymatic activity measured in soleus muscle of rats fed with either vehicle (hydroxypropylmethylcellulose) (filled columns) or GW610742 at 5 mg (kg bm)−1 (cross-hatched columns) and 100 mg (kg bm)−1 (open columns).
Figure 4.
MuRFI and MaFbx protein expression in skeletal muscle A, a representative Western blot of MurF1 protein detected at 42 kDa. B, the average density of MuRF1 protein bands, measured by Western blot in soleus muscle lysates obtained from rats fed with either vehicle (hydroxypropylmethylcellulose) (filled columns) or GW610742 at 5 mg (kg bm)−1 (cross-hatched columns) and 100 mg (kg bm)−1 (open columns). ***Significantly different from the corresponding control (vehicle) group (P < 0.001). C, a representative Western blot of MAFbx protein detected at 50 kDa. D, the average density of MAFbx protein bands, measured by Western blot in soleus muscle lysates obtained from rats fed with either vehicle (hydroxypropylmethylcellulose) (filled columns) or GW610742 at 5 mg (kg bm)−1 (cross-hatched columns) and 100 mg (kg bm)−1 (open columns). *Significantly different from the corresponding control (vehicle) group (P < 0.05). ***Significantly different from the corresponding control (vehicle) group (P < 0.001).
Muscle hypertrophy molecular pathway
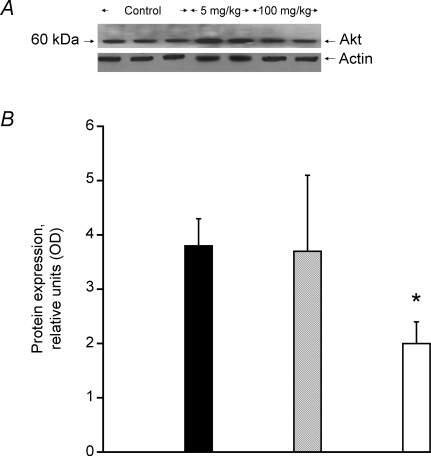
The level of total Akt1 protein detected at 60 kDa (Fig. 5A), a key component of the mTOR anabolic signalling pathway, was found to be 50% less than control with the highest dose of GW610742 administered (P < 0.05, Fig. 5B). However, the corresponding percentage of phosphorylated Akt was similar across the groups.
Figure 5.
Akt1 protein expression in skeletal muscle A, a representative Western blot of Akt1 protein detected at 60 kDa. B, the average density of Akt1 protein bands, measured by Western blot in soleus muscle lysates obtained from rats fed with either vehicle (hydroxypropylmethylcellulose) (filled columns) or GW610742 at 5 mg (kg bm)−1 (cross-hatched columns) and 100 mg (kg bm)−1 (open columns). *Significantly different from the corresponding control (vehicle) group (P < 0.05).
Discussion
The results of the present study clearly point to acute (6 days) PPARδ agonist administration inducing a switch in muscle fuel metabolism towards decreased carbohydrate use and enhanced lipid oxidation. This is illustrated, firstly, by the increase in PDK4 mRNA and protein expression, which is indicative of the inhibition of carbohydrate flux via the PDC and, secondly, by the simultaneous increase in β hydroxy acyl-CoA dehydrogenase (HAD) activity, indicating increased capacity for β-oxidation. There was, however, no evidence that PPARδ agonism impaired mitochondrial function under the current experimental setting.
A second novel finding was that GW610742 administration resulted in the transcriptional and translational initiation of the muscle specific E3 ligases, MAFbx and MuRF1, suggesting that a muscle atrophy programme had been initiated. The up-regulation of MAFbx and MuRF1, which precedes ubiquitin–proteasome dependent muscle proteolysis, has been consistently linked to muscle catabolism (Lecker et al. 2004). It is possible that changes in the Akt1–FOXO signalling axis may explain the simultaneous increase in PDK4, MAFbx and MuRF1 mRNA and protein expression seen in the present study following GW610742 administration (see below).
PPARδ agonism and the regulation of muscle fuel selection
The activity of PDC, which catalyses the oxidative decarboxylation of carbohydrate-derived pyruvate, is regulated by a covalent mechanism involving competition between a kinase (PDK1-4) and a phosphatase (PDP1-2). Skeletal muscle contains two PDK isoforms, PDK2 and PDK4, that inhibit the activity of PDC (Holness et al. 2000). In the present study, both of these isoforms were significantly increased at the transcriptional level by agonist administration, particularly in the group that received the highest dose of GW610742 (Fig. 2A). In keeping with this, a previous study using cultured human muscle cells has also shown a specific up-regulation of PDK4 mRNA following GW610742 administration (Abbot et al. 2005). Therefore, it can be concluded that activation of PPARδ receptor induces an up regulation of PDK4 mRNA. Furthermore, PDK4 was increased at the protein level in both treatment groups (Fig. 2B). Since the transcription of PDP1 and PDP2, which dephosphorylate and activate PDC, were unchanged (results not shown), it is pertinent to suggest that down-regulation of carbohydrate oxidation will have occurred following the GW610742 administration, particularly at the highest dose of agonist.
Interestingly, the apparent down-regulation of carbohydrate oxidation in the GW610742 treated groups of the present study was matched by an increase in the activity of HAD (Fig. 1C), the rate limiting step in β-oxidation, and by a tendency of fat derived mitochondrial ATP production (Pal + Carn, Fig. 1A and B) to be higher in the treated groups suggesting greater reliance on fat oxidation. We propose therefore that the decline in energy provision following GW610742 mediated inactivation of PDC was matched by an increase in fat derived energy provision. The similarity between treatment groups in in vitro mitochondrial ATP generation (Fig. 1) does show, however, that mitochondrial function was not impaired by GW610742 administration, which is also supported by the similarity in muscle ATP and PCr concentrations across treatment groups. Both of these observations strongly detract from any premise that myopathic changes in mitochondrial function occurred as a result of PPARδ agonism.
PPARδ agonism and the induction of a muscle atrophy programme
Loss of muscle mass (atrophy) is a common feature of illness, such as diabetes, HIV, sepsis, trauma and cancer (Price, 2003) and of more ‘normal’ physiological insults, such as starvation, immobilization and ageing. In ‘normal’ healthy (non-atrophying) muscle, protein mass is maintained by a balance of protein synthesis and proteolysis. However, unique proteolytic mechanisms are activated during skeletal muscle atrophy that involves an increase in muscle proteolysis, and usually in conjunction with a decline in the rate of muscle protein synthesis (Glass, 2005). As outlined above, one major proteolytic pathway that has been studied in relation to wasting is the ATP-dependent ubiquitin-proteasome system. In the present study, we were able to show for the first time that mRNA and protein expression of the two muscle specific E3 ubiquitin ligases, MuRF1 and MAFbx, were significantly up regulated, particularly with the highest dose of the agonist (Figs 3A and 4). MAFbx and MuRF1 select and target other muscle proteins for degradation by the 26S proteasome, as part of the ‘degradation programme’. Up-regulation of MAFbx and MuRF1 expression has been shown to be a fundamental feature of muscle atrophy in a range of animal models of wasting, including sepsis (Dehoux et al. 2003), fasting (Lecker et al. 2004), immobilization (Jones et al. 2004) and denervation (Bodine et al. 2001b).
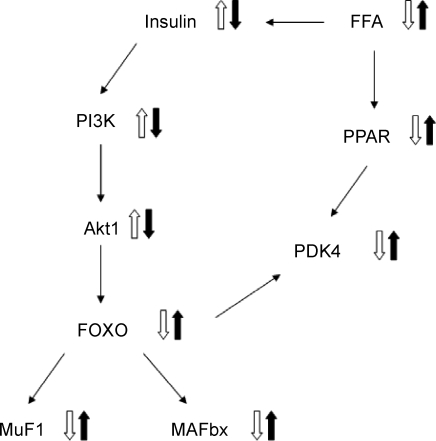
Recent cell and in vivo based evidence points towards a functional coupling of the up-regulation of MAFbx and MuRF1 and the suppression of muscle protein synthesis in catabolic and insulin resistant states via the Akt1–mTOR–FOXO signalling axis (Hoffman & Nader, 2004; Wang et al. 2006). In particular, if Akt1 activity is suppressed, protein synthesis is inhibited. Furthermore, FOXO becomes dephosphorylated, thereby transcriptionally activating MAFbx and MURF-1, and inducing protein degradation (see Fig. 6).
Figure 6.
A schematic diagram of the proposed mechanism of PPARδagonist action Open arrow, up-regulation; filled arrow, down-regulation.
Based on the present data therefore, it is appropriate to conclude that acute (six days) administration of GW610742 initiated the activation of a ubiquitin–proteasome proteolytic-dependant atrophy programme in skeletal muscle. However, although it is widely accepted that up-regulation of MAFbx and MuRF1 is a common feature of muscle proteolysis, we were unable to demonstrate an increase in muscle 20S proteasome transcription (Fig. 3A) or its chymotrypsin-like activity (Fig. 3B), or a change in the muscle protein: DNA ratio (Table 1), implying muscle proteolysis had not occurred. It could be argued that an increase in cellular protein ubiquitination might not necessarily be followed by an increase in the activity of the 26S proteasome. Furthermore, although it has been suggested that transcription of several subunits of the 19S and 20S proteasome should increase in a coordinated manner during atrophy, some do not change in several catabolic states (Lecker et al. 2004).
Regardless of these observations, it would be sensible to extend the current study beyond a period of 6 days to determine if proteasome activity is increased and accompanied by proteolysis during GW610742 administration, or whether these changes in muscle E3 ligase expression merely occur secondary to changes in PI3 kinase signalling via Akt1 and FOXO. Furthermore, from a clinical perspective inclusion of a relevant animal model, such as insulin resistance, combined with additional measures of muscle proteolysis might provide further insight into whether the chronic effects of GW610742 on molecular signalling translate into physiological muscle atrophy.
Does a single signalling pathway simultaneously regulate muscle fuel selection and protein turnover?
Akt1 is a constitutive protein found in muscle cells, and when over-expressed causes hypertrophy (Bodine et al. 2001b). Furthermore, activation of Akt1 in cell based studies has been shown to block the atrophy associated increases in MAFbx and MuRF1 transcription through the phosphorylation of members of the family of FOXO transcription factors (Sandri et al. 2004). It has been shown in myotubes that FOXO transcription factors are phosphorylated by Akt1, and as a result become extruded from the nucleus to the cytoplasm and incapable of activating MuRF1 and MAFbx transcription (Hoffman & Nader, 2004). In the present study, we saw a significant decrease in Akt1 protein at the highest dose of GW610742 (Fig. 5), suggesting the activation of the muscle atrophy programme related to the ubiquitin-dependent muscle pathway was triggered by a down-regulation of the Akt1 pathway. Furthermore, the increase in PDK4 mRNA and protein expression we observed following GW610742 administration, in conjunction with the up-regulation in MAFbx and MuRF1 mRNA and protein expression, implicates a single signalling pathway (via Akt1 and FOXO) in the simultaneous regulation of muscle protein turnover and muscle fuel use (Fig. 6). In line with this view, food deprivation in mice has been shown to increase FOXO1 mRNA expression, thereby increasing PDK4 transcription (Furuyama et al. 2003), which would be expected to decrease carbohydrate use and increase fat oxidation (and proteolysis, which the authors did not address). We measured both the transcriptional and translational levels of FOXO1. However, FOXO1 mRNA could hardly be detected in our model (results not shown), whilst both total FOXO1 protein, and its phosphorylated form measured in total muscle lysates, remained largely unchanged (results not shown). A reasonable explanation for these findings is that another member of the FOXO family of transcription factors could have been responsible for activating PDK4, MuRF1 and MAFbx transcription. Indeed, Sandri et al. (2004) have shown that constitutively active FOXO3 can also act on the MuRF1 promoter to cause MuRF1 transcription and atrophy of myotubes and muscle fibres. It has also been shown that the level of FOXO 3 mRNA increases in muscle atrophy caused by diabetes or fasting (Furuyama et al. 2003; Lecker et al. 2004).
In conclusion, it appears that the acute (6 days) administration of PPARδ agonist, GW610742, produced changes consistent with a switch in muscle fuel metabolism towards decreased carbohydrate use and enhanced lipid utilization. This we believe was achieved by a PPARδ agonist mediated increase in PDK4 protein expression, and thereby inhibition of the PDC, and the simultaneous up-regulation of β-oxidation. However, the energy status of the cell and the capacity of mitochondrial ATP production were unchanged suggesting the absence of any mitochondrial dysfunction. A second important finding of the present study was that GW610742 administration initiated the activation of a muscle atrophy programme, possibly via changes in the Akt1/FOXO/MAFbx and MuRF1 signalling pathway.
References
- Abbot EL, McCormack JG, Reynet C, Hassall DG, Buchan KW, Yeaman SJ. Diverging regulation of pyruvate dehydrogenase kinase isoform gene expression in cultured human muscle cells. FEBS J. 2005;272:3004–3014. doi: 10.1111/j.1742-4658.2005.04713.x. [DOI] [PubMed] [Google Scholar]
- Alway SE, Lowe DA, Chen KD. The effects of age and hindlimb supension on the levels of expression of the myogenic regulatory factors MyoD and myogenin in rat fast and slow skeletal muscles. Exp Physiol. 2001;86:509–517. doi: 10.1113/eph8602235. [DOI] [PubMed] [Google Scholar]
- Berger J, Wagner JA. Physiological and therapeutic roles of peroxisome proliferator-activated receptors. Diabetes Technol Ther. 2002;4:163–174. doi: 10.1089/15209150260007381. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001a;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001b;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Brunmair B, Staniek K, Dorig J, Szocs Z, Stadlbauer K, Marian V, Gras F, Anderwald C, Nohl H, Waldhausl W, Furnsinn C. Activation of PPAR-δ in isolated rat skeletal muscle switches fuel preference from glucose to fatty acids. Diabetologia. 2006;49:2713–2722. doi: 10.1007/s00125-006-0357-6. [DOI] [PubMed] [Google Scholar]
- Cederblad G, Carlin JI, Constantin-Teodosiu D, Harper P, Hultman E. Radioisotopic assays of CoASH and carnitine and their acetylated forms in human skeletal muscle. Anal Biochem. 1990;185:274–278. doi: 10.1016/0003-2697(90)90292-h. [DOI] [PubMed] [Google Scholar]
- Dawson SP, Arnold JE, Mayer NJ, Reynolds SE, Billett MA, Gordon C, Colleaux L, Kloetzel PM, Tanaka K, Mayer RJ. Developmental changes of the 26 S proteasome in abdominal intersegmental muscles of Manduca sexta during programmed cell death. J Biol Chem. 1995;270:1850–1858. doi: 10.1074/jbc.270.4.1850. [DOI] [PubMed] [Google Scholar]
- Dehoux MJ, van Beneden RP, Fernandez-Celemin L, Lause PL, Thissen JP. Induction of MafBx and Murf ubiquitin ligase mRNAs in rat skeletal muscle after LPS injection. FEBS Lett. 2003;544:214–217. doi: 10.1016/s0014-5793(03)00505-2. [DOI] [PubMed] [Google Scholar]
- Forsberg AM, Nilsson E, Werneman J, Bergstrom J, Hultman E. Muscle composition in relation to age and sex. Clin Sci (Lond) 1991;81:249–256. doi: 10.1042/cs0810249. [DOI] [PubMed] [Google Scholar]
- Fosslien E. Mitochondrial medicine – molecular pathology of defective oxidative phosphorylation. Ann Clin Lab Sci. 2001;31:25–67. [PubMed] [Google Scholar]
- Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J. 2003;375:365–371. doi: 10.1042/BJ20030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DJ. A signaling role for dystrophin: inhibiting skeletal muscle atrophy pathways. Cancer Cell. 2005;8:351–352. doi: 10.1016/j.ccr.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci U S A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RC, Hultman E, Nordesjo LO. Glycogen, glycolytic intermediates and high-energy phosphates determined in biopsy samples of musculus quadriceps femoris of man at rest. Methods and variance of values. Scand J Clin Lab Invest. 1974;33:109–120. [PubMed] [Google Scholar]
- Hoffman EP, Nader GA. Balancing muscle hypertrophy and atrophy. Nat Med. 2004;10:584–585. doi: 10.1038/nm0604-584. [DOI] [PubMed] [Google Scholar]
- Holness MJ, Kraus A, Harris RA, Sugden MC. Targeted upregulation of pyruvate dehydrogenase kinase (PDK)-4 in slow-twitch skeletal muscle underlies the stable modification of the regulatory characteristics of PDK induced by high-fat feeding. Diabetes. 2000;49:775–781. doi: 10.2337/diabetes.49.5.775. [DOI] [PubMed] [Google Scholar]
- Hondares E, Pineda-Torra I, Iglesias R, Staels B, Villarroya F, Giralt M. PPARδ, but not PPARα, activates PGC-1α gene transcription in muscle. Biochem Biophys Res Commun. 2007;354:1021–1027. doi: 10.1016/j.bbrc.2007.01.092. [DOI] [PubMed] [Google Scholar]
- Jones SW, Hill RJ, Krasney PA, O'Conner B, Peirce N, Greenhaff PL. Disuse atrophy and exercise rehabilitation in humans profoundly affects the expression of genes associated with the regulation of skeletal muscle mass. FASEB J. 2004;18:1025–1027. doi: 10.1096/fj.03-1228fje. [DOI] [PubMed] [Google Scholar]
- Keller BJ, Yamanaka H, Thurman RG. Inhibition of mitochondrial respiration and oxygen-dependent hepatotoxicity by six structurally dissimilar peroxisomal proliferating agents. Toxicology. 1992;71:49–61. doi: 10.1016/0300-483x(92)90053-h. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- Lee CH, Olson P, Evans RM. Minireview: Lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144:2201–2207. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- Leger B, Cartoni R, Praz M, Lamon S, Dériaz O, Crettenand A, Gobelet C, Rohmer P, Konzelmann M, Luthi F, Russell AP. Akt signalling through GSK-3 beta mTOR and Fox01 is involved in human skeletal muscle hypertrophy and atrophy. J Physiol. 2006;576:923–933. doi: 10.1113/jphysiol.2006.116715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe DA, Degens H, Chen KD, Alway SE. Glyceraldehyde-3-phosphate dehydrogenase varies with age in glycolytic muscles of rats. J Gerontol A Biol Sci Med Sci. 2000;55:B160–B164. doi: 10.1093/gerona/55.3.b160. [DOI] [PubMed] [Google Scholar]
- Motojima K, Seto K. Fibrates and statins rapidly and synergistically induce pyruvate dehydrogenase kinase 4 mRNA in the liver and muscles of mice. Biol Pharm Bull. 2003;26:954–958. doi: 10.1248/bpb.26.954. [DOI] [PubMed] [Google Scholar]
- Opie LH, Newsholme EA. The activities of fructose 1,6-diphosphatase, phosphofructokinase and phosphoenolpyruvate carboxykinase in white muscle and red muscle. Biochem J. 1967;103:391–399. doi: 10.1042/bj1030391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SR. Increased transcription of ubiquitin-proteasome system components: molecular responses associated with muscle atrophy. Int J Biochem Cell Biol. 2003;35:617–628. doi: 10.1016/s1357-2725(02)00385-0. [DOI] [PubMed] [Google Scholar]
- Qu B, Li QT, Wong KP, Tan TM, Halliwell B. Mechanism of clofibrate hepatotoxicity: mitochondrial damage and oxidative stress in hepatocytes. Free Radic Biol Med. 2001;31:659–669. doi: 10.1016/s0891-5849(01)00632-3. [DOI] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sznaidman ML, Haffner CD, Maloney PR, Fivush A, Chao E, Goreham D, Sierra ML, LeGrumelec C, Xu HE, Montana VG, Lambert MH, Willson TM, Oliver WR, Jr, Sternbach DD. Novel selective small molecule agonists for peroxisome proliferator-activated receptor δ (PPARδ) – synthesis and biological activity. Bioorg Med Chem Lett. 2003;13:1517–1521. doi: 10.1016/s0960-894x(03)00207-5. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Yamamoto J, Iwasaki S, Asaba H, Hamura H, Ikeda Y, Watanabe M, Magoori K, Ioka RX, Tachibana K, Watanabe Y, Uchiyama Y, Sumi K, Iguchi H, Ito S, Doi T, Hamakubo T, Naito M, Auwerx J, Yanagisawa M, Kodama T, Sakai J. Activation of peroxisome proliferator-activated receptor δ induces fatty acid β-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci U S A. 2003;100:15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu Z, Du Hu J, J & Mitch WE. Insulin resistance accelerates muscle protein degradation: Activation of the ubiquitin-proteasome pathway by defects in muscle cell signaling. Endocrinology. 2006;147:4160–4168. doi: 10.1210/en.2006-0251. [DOI] [PubMed] [Google Scholar]
- Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARδ. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibom R, Lundin A, Hultman E. A sensitive method for measuring ATP-formation in rat muscle mitochondria. Scand J Clin Lab Invest. 1990;50:143–152. doi: 10.1080/00365519009089146. [DOI] [PubMed] [Google Scholar]
- Wu P, Peters JM, Harris RA. Adaptive increase in pyruvate dehydrogenase kinase 4 during starvation is mediated by peroxisome proliferator-activated receptor α. Biochem Biophys Res Commun. 2001;287:391–396. doi: 10.1006/bbrc.2001.5608. [DOI] [PubMed] [Google Scholar]
- Zammit VA, Newsholme EA. The maximum activities of hexokinase, phosphorylase, phosphofructokinase, glycerol phosphate dehydrogenases, lactate dehydrogenase, octopine dehydrogenase, phosphoenolpyruvate carboxykinase, nucleoside diphosphatekinase, glutamate-oxaloacetate transaminase and arginine kinase in relation to carbohydrate utilization in muscles from marine invertebrates. Biochem J. 1976;160:447–462. doi: 10.1042/bj1600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Wallace KB. The effect of peroxisome proliferators on mitochondrial bioenergetics. Toxicol Sci. 1999;48:82–89. doi: 10.1093/toxsci/48.1.82. [DOI] [PubMed] [Google Scholar]