Abstract
Candida dubliniensis and Candida albicans, the most common human fungal pathogen, have most of the same genes and high sequence similarity, but C. dubliniensis is less virulent. C. albicans causes both mucosal and hematogenously disseminated disease, C. dubliniensis mostly mucosal infections. Pulse-field electrophoresis, genomic restriction enzyme digests, Southern blotting, and the emerging sequence from the Wellcome Trust Sanger Institute were used to determine the karyotype of C. dubliniensis type strain CD36. Three chromosomes have two intact homologues. A translocation in the rDNA repeat on chromosome R exchanges telomere-proximal regions of R and chromosome 5. Translocations involving the remaining chromosomes occur at the Major Repeat Sequence. CD36 lacks an MRS on chromosome R but has one on 3. Of six other C. dubliniensis strains, no two had the same electrophoretic karyotype. Despite extensive chromosome rearrangements, karyotypic differences between C. dubliniensis and C. albicans are unlikely to affect gene expression. Karyotypic instability may account for the diminished pathogenicity of C. dubliniensis.
1. Introduction
The history of infectious disease is largely devoted to bacterial and viral infections, such as plague, tuberculosis, and smallpox, and it is only in the last 50 years or so that fungal infections have emerged as a serious medical problem. This emergence is due directly to the efficacy of medical care, which now has the resources to maintain highly debilitated patients with deficient immune systems. A significant number of these patients get infections with one or another candida species, and although Candida albicans is still the most commonly isolated pathogenic fungus, other species, such as Candida dubliniensis and Candida glabrata, are increasing in frequency among isolates (Kullberg and Filler 2002; Maschmeyer 2006).
C. dubliniensis is the species most closely related to C. albicans, but the two differ significantly in virulence properties. C. dubliniensis is found mostly as an oral pathogen and only rarely in disseminated infections (Gilfillan, Sullivan et al. 1998). However, it has been estimated that more than 88% of the genes of these closely related species have nucleotide homology greater than 80% (Moran, Stokes et al. 2004). In addition, the two species share the ability to grow as yeast, pseudohyphae, and hyphae and to make chlamydospores. Similarities have also been noted in their genomes: both are diploid and have a large repeated DNA sequence on most chromosomes (the Major Repeat Sequence or MRS). The origin and function of the MRS are not known, but it is found only in C. dubliniensis and C. albicans. It is the site of most of the translocations which have been mapped in C. albicans, and it is responsible for much of the chromosome length polymorphism which exists in that organism (Chibana, Beckerman et al. 2000).
The published genome sequence assembly of C. albicans released in 2004 (Assembly 19) was a diploid one, including separate alleles for many of the loci (Braun and Kuo 2005). Assembly 19 had 133 pairs of allelic contigs and 136 unique contigs and has been annotated by a large set of investigators (Jones, Federspiel et al. 2004). The genomic sequence of C. dubliniensis is virtually complete and is available (unpublished) at the Wellcome Trust Sanger Institute (SangerInstitute http://www.sanger.ac.uk/) but has not yet been annotated. Thus comparison between the two genomes is possible. The karyotype of C. albicans, determined using pulse-field electrophoresis and Southern blotting, demonstrated that there were 8 chromosomes (Magee, Koltin et al. 1988; Lasker, Carle et al. 1989; Wickes, Staudinger et al. 1991). A major tool in the further characterization of chromosome substructure was the use of the 8-base-pair-specific restriction enzyme SfiI, which cleaves the various chromosomes into from 2 to 8 fragments, ranging in size from 90 to >2200 kb. This SfiI macro-restriction map allowed markers to be localized to specific parts of the chromosomes (Chu, Magee et al. 1993). The majority of the SfiI restriction sites are in the MRS, which ranges in size from 18 kb to as much as 80 kb (Iwaguchi, Homma et al. 1992; Chibana, Iwaguchi et al. 1994; Chindamporn, Nakagawa et al. 1998). Furthermore, all of the reciprocal translocations analyzed in several strains seemed to take place between MRS regions (Chu, Magee et al. 1993; Navarro-Garcia, Perez-Diaz et al. 1995). C. dubliniensis also has an MRS (Joly, Pujol et al. 1999), but its role in chromosome organization and translocation has not been investigated.
Assembly of the C. dubliniensis sequence at the Wellcome Trust Sanger Institute is based on the karyotype of SC5314, the sequenced C. albicans strain, and is similarly organized into 8 chromosomes, each nominally representing one of a pair of homologues. However unlike C. albicans, where approximately half the strains have a standard karyotype, in C. dubliniensis the karyotypes differ in every isolate we have examined (Magee and Chibana 2002). Here we present the detailed karyotype of CD36, the C. dubliniensis strain whose genomic sequence was determined, and the pulse-field chromosome separations of some other C dubliniensis isolates compared to C. albicans. Although large blocks of DNA, corresponding in several cases to the SfiI restriction fragments in C. albicans, are similarly organized in the two species, they are much less frequently arranged as pairs of chromosome homologues in C. dubliniensis. As in C. albicans, the MRS in C. dubliniensis seems to be a major site for chromosome reorganization (Chibana, Beckerman et al. 2000) (Magee and Chibana 2002).
2. Materials and Methods
2.1 Strains
The strains of C. albicans and C. dubliniensis in this paper are listed in Table 1. Media and growth conditions were as previously described (Chu, Magee et al. 1993).
Table 1.
List of strains used
| Organism | Strain | Reference |
|---|---|---|
| Candida albicans | ||
| 1006 | (Goshorn, Grindle et al. 1992) | |
| SC5314 | (Jones, Federspiel et al. 2004) | |
| Candida dubliniensis | ||
| CD36 | (Sullivan, Westerneng et al. 1995) | |
| R3b | (Timmins, Howell et al. 1998) | |
| R1b | (Timmins, Howell et al. 1998) | |
| 16F | (Sullivan, Westerneng et al. 1995) | |
| 3225 | (Muller, Kasai et al. 1999) | |
| 3233 | (Joly, Pujol et al. 1999) | |
| 514 | (Gee, Joly et al. 2002) |
2.2 Pulse-field electrophoresis and Southern blotting
CHEF samples were made according to standard procedures (Chibana et al. 1998). Gels were run using the CHEF DRIII System (BioRad) at 14 C. Two separation protocols were used.
All chromosomes: 0.6% agarose (Amresco PFGE Grade III), in 0.5X TBE buffer (50mM Tris, 50mM boric acid, 1mM EDTA, pH8.3) 60-300sec switch ramp, 24hr, 4.5V/cm 120°; 720-900sec ramp, 12hr, 2.0V/cm, 106° (Figs 1B, 1C, 3A, 5).
Figure 1.
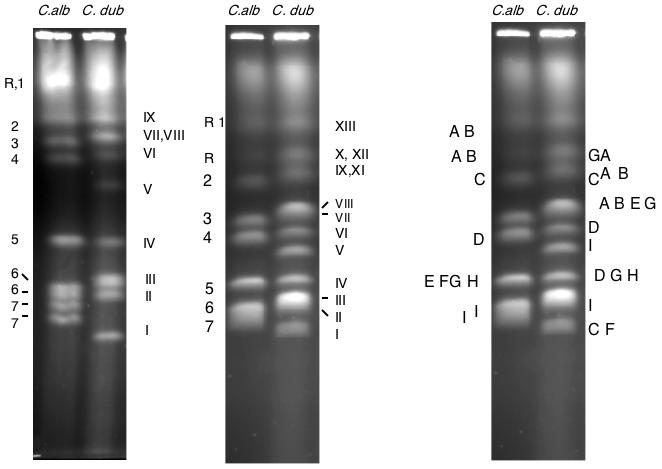
CHEF separation of chromosomes from C. albicans strain SC5314 and from C. dubliniensis strain CD36. Left: optimal separation conditions for the smaller chromosomes. Center: CHEF separation of the same strains under conditions optimal for the large chromosomes. Because the position of chromosome R on a pulse-field gel is variable, band XI (one homologue of chromosome R) sometimes runs ahead of band X (chromosome 2, as it does in this gel. The numbers and letter on the left identify the chromosomes of SC5314; the Roman numeral labels on the figure refer to the bands in the CD36 lane. Right: Location of several probes on the SC5314 and CD36 karyotypes. Probes: A, rDNA (SfiI fragments RS/RU); B, HCS1 (RB); C, p2A (2A); D, CPY52 (4F); E, MTLα (5I); F, MTLa (5I); G,HEX1 (5I); H, HIS1 (5I); I, p282 (7A).
Figure 3.
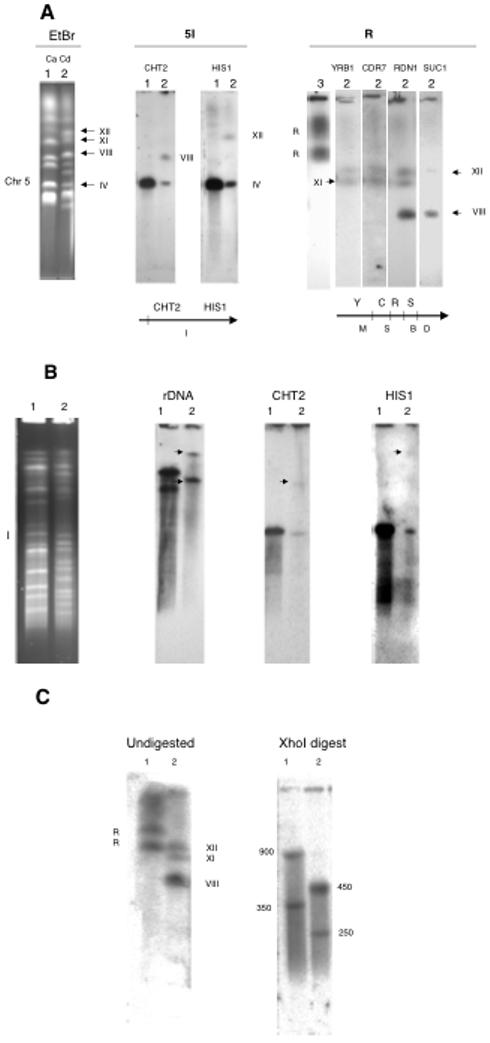
Assignment of chromosome R and chromosome 5 probes to the CD36 karyotype. Lane 1, SC5314; lane 2, CD36; lane 3, 1006. A. The karyotypes at the left were blotted and probed with the genes indicated. Beneath the blots are SfiI maps of portions of the chromosomes in C. albicans, with the SfiI fragment designations listed below and the genes above. B. SfiI digestion. The SfiI digests at the left were blotted and probed with the genes as indicated. Arrows point to the hybridizing bands. C. XhoI digests probed with RDN1. Numbers indicate the size in kb of the XhoI fragments.
Figure 5.
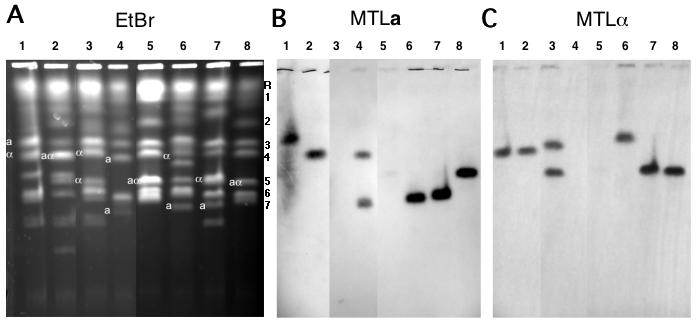
Candida dubliniensis pulse-field chromosome separations. A. CHEF separation of chromosomes from C. albicans strain 1006 and from 7 C. dubliniensis strains. Three lanes from the middle of the gel which contained irrelevant strains have been removed from the figure. The letters to the right of this figure indicate the positions of the C. albicans chromosomes used as molecular markers: 1, 3.2 Mb; 2, 2.2 Mb; 3, 1.8 Mb; 4, 1.6 Mb; 5, 1.2 Mb; 6, 1.03Mb; 7, 1.0 Mb. Chromosome R, as mentioned above, does not migrate strictly according to its molecular size in a CHEF gel. B. Southern blot probed with a sequence from the C. dubliniensis MTLa1 gene. C. Southern blot probed with a sequence from the C. dubliniensis MTLα1 gene. Lane 1, Strain R3b; 2, R1b; 3, 3225; 4, 3233; 5, 1006 (C. albicans); 6, CD36; 7, 16F; 8, 514.
Lower chromosomes: 0.9% BioRad Pulsed Field certified agarose, 60-120sec ramp 24hr, 6.0V/cm; 120-360sec ramp, 15hr, 4.5V/cm, 120° (Fig 1A).
SfiI and XhoI digests: 1.0% BioRad Pulsed Field certified agarose, 7-100 sec ramp, 20hr, 4.5V/cm; 80-400 sec ramp, 20hr, 3.5V/cm, 120° (Figs 2, 3B, 3C).
Figure 2.
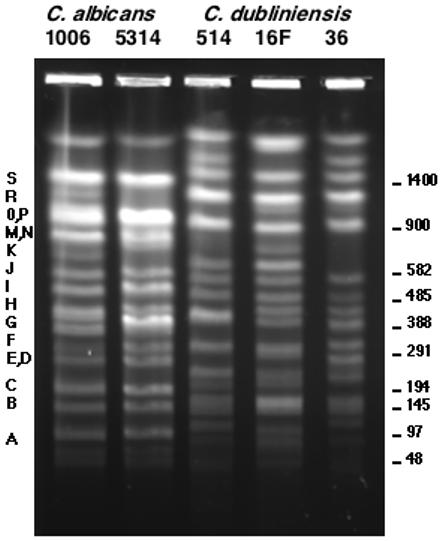
CHEF separation of the SfiI digests of C. albicans strains 1006 and SC5314 and three C. dubliniensis strains: 514, 16F, and CD36. The letters refer to the C. albicans bands described by Chu, et al (Chu, Magee et al. 1993). The size in kilobases is given on the right.
For Southern hybridization, the DNA on the gels was nicked (BioRad Gene Linker) and vacuum blotted (BioRad 785) to nylon membranes (MSI or Hybond) according to manufacturer’s specifications. DNA probes were labeled using 32P-dCTP with the Rediprime II kit (Amersham Pharmacia). Hybridizations were performed at 65 C in modified Church/Gilbert Buffer (7% SDS, 0.5M NaPhosphate (pH7.2), 1mM EDTA). Blots were washed to medium stringency according to the membrane manufacturer (final washes at 37 C).
Most C. albicans probes gave a poor signal with C.dubliniensis blots under standard conditions. We used DNA primers designed for C.albicans genes to amplify C. dubliniensis genomic DNA by PCR, using standard conditions. Most primers gave product. We also found that many C. albicans amplicons could be used directly as probes if medium stringency washes were employed. The MTL loci, which differ significantly between the two species, were amplified by PCR using C. dubliniensis primers to get probes. The primers used were MTLa1-F: 5′-ATG CGA AAA CAT TCT TAC AAT TAG AC-3′; MTLa1-R: 5′-AAG TTA TTT GTT CCA ATG TAA ATT-3′ and MTLalpha2-F: 5′-ATT AGC ATG TTA GTA GAG TTA CAA CG-3′; MTLalpha2-R: 5′-CGA TAT CAA AAA AAT GGT TTC TAT TAT-3′
Probes were assigned to EtBr-stained bands on C. dubliniensis CHEF gels by hybridization as described (Chibana, Beckerman et al. 2000), except that C. albicans probes hybridized to C. dubliniensis blots were washed at 37 C rather than 65 C. Bands on the C. dubliniensis CHEF electrophoretic separations sometimes contain more than one chromosome as described in the text.
2.3 SfiI analysis
Digestion with the restriction enzyme SfiI and subsequent analysis was according to the methods of Chibana and coworkers (Chibana, Beckerman et al. 2000). XhoI digestion and fragment analysis were carried out as described by Magee, et al (Magee, D’Souza et al. 1987).
3. Results
Because the chromosomal organization of the emerging sequence may contain clues to the differences in virulence between C. dubliniensis and C. albicans, we examined the karyotypic structure of the type strain, CD36, in detail. Early results suggested that not only the sequence of individual genes but the synteny of genes over distances of several hundred kilobases was similar in C. dubliniensis and C. albicans. We decided to use the well-characterized karyotype of the latter organism as a template to construct the chromosome organization of C. dublinensis. We will therefore describe the results in terms of their similarity to or difference from C. albicans.
Our experimental approach was to separate both intact chromosomes and restriction fragments from genomic digestion by the enzyme SfiI, use probes whose location is known in C. albicans, and assign them by blotting to chromosomes and SfiI fragments of CD36. This analysis was guided by our preliminary results which suggested that the organization of the genomes of the two closely related strains was somewhat similar and that most C. albicans probes will hybridize to C. dubliniensis chromosomes under moderately stringent conditions.
3.1 CHEF pulse-field electrophoresis of CD36
Although many strains of C. albicans have eight pairs of approximately equal-sized homologous chromosomes, giving 8 bands on pulse-field gel, CD36 has an electrophoretic karyotype with 12 ethidium bromide-staining bands. Figure 1 shows a comparison of the pulse-field electrophoresis chromosome separations of C. albicans strain SC5314 and the C. dubliniensis strain CD36. The chromosomes of SC5314 are labeled at the left. In these separations, the bands in CD36 are numbered from I to XIII in order of increasing size. The different numbering system is designed to minimize confusion in the discussion; C. albicans chromosomes have Arabic numbers; chromosomes with Roman numerals are from C. dubliniensis. Gels were run under two sets of conditions: one set was designed to separate the smaller chromosomes (left-hand panel) and the other the larger ones (center panel). The right-hand panel is a summary of several probes (some of which are genes) assigned to the chromosomes of SC5314 and CD36 by Southern blotting. Each of the probes hybridizes to one of the CHEF bands in SC5314, while in CD36 each hybridizes to at least two bands with the exception of MTLa and MTLα, which are present only once each in the diploid genome. The probes which hybridize to more than one band in C. dubliniensis come from 5 of the 8 chromosomes. We will show below that each of these chromosomes is involved in a translocation. Strain CD36 has three bands (XII, XI, and VII) which hybridize to the ribosomal DNA genes. The MTLa and MTLα clusters from chromosome 5 are separated in C. dubliniensis, as are the alleles of CPY52 (chromosome 4) and HEX1 (chromosome 5). The p2A probe (chromosome 2) lights up a band in C.dubiniensis that runs below the C. albicans chromosome 7 and a band that migrates at the same place as C. albicans chromosome 2, while p282 (chromosome 7) lights up a band in C. dubliniensis near chromosome 4 and a band in the chromosome 7 region.
3.2 Assignment of probes to SfiI fragments
Figure 2 shows an electrophoretic separation of the SfiI genomic digests of three C. dubliniensis strains, 514, 16F, and CD36. SfiI digests of two C. albicans strains, 1006 and SC5314, are shown for comparison. There are some minor differences between SC5314 and 1006, but for the most part the patterns are identical. The three C. dubliniensis isolates are more similar to each other than to C. albicans, but in general the SfiI patterns are more variable from strain to strain in C. dubliniensis compared to C. albicans. In particular, strain 514, which has a karyotype very similar to C. albicans (see below), has an SfiI pattern very similar to that for CD36..
We hybridized probes from each of the C. albicans chromosomes to gel separations of C. dubliniensis SfiI digests. Some of the SfiI bands identified by markers from C. albicans (e.g., 4H and 5I) are quite similar in size in the two species, while others differ significantly. Table 2 provides a summary of the hybridization of various unique probes to the karyotype and SfiI digest of CD36. The locations of the probes in C. albicans are given for comparison. It can be seen that probes that are syntenic on a particular SfiI fragment in C. albicans are often but not always syntenic in C. dubliniensis. This seems in all but one case to be due to the differences in the location of SfiI sites found in unique DNA in SC5314 and CD36. The predicted size of the C. dubliniensis SfiI fragments based on the sequence assembly is also given in Table 2, and the agreement is quite good.
Table 2.
Assignment of probes to the C. dubliniensis electrophoretic bands and SfiI fragments
| Probe | C. dubliniensis electrophoretic band | C. albicans assignment | SfiI band size (kb) (from hybridization data) | Predicted SfiI band (kb) (from assembly) | |
|---|---|---|---|---|---|
| C. albicans | C. dubliniensis | ||||
| rDNA | VIII, XI, XII | rS, rU | 1200, 1400 | 1300, >2300 | <22381 |
| CDR7 | XI,XII | rS, rU | 1200, 1400 | <22381 | |
| HCS1 | VIII, XII | rB | 140 | S, U | <22381 |
| YRB1 | XI,XII | rM, rS | 750, 1200 | <22381 | |
| CDC23 | XI,XII | rS, rU | 1200, 1400 | <22381 | |
| HGT1 | 1S | 1400 | 1200 | 1218 | |
| SER3 | XIII | 1L | 700 | 1487 | |
| ENO1 | XIII | 1J | 520 | R | 1487 |
| CPH1 | XIII | 1B | 145 | 145 | 127 |
| GLY1 | XIII | 1E | 290 | 1487 | |
| PCHR2A | I, IX | 2A | 90 | 90 | 93 |
| SAS2 | 2U | 2200 | 2200 | 2200 | |
| NPS1 | VII | 3O | 900 | >400 | 257 |
| PKC1 | 3P | 920 | 1030 | 1161 | |
| LYS1 | IV, VI | 4F | 340 | 340 | 369 |
| YJR61 | IV, VI | 4H | 430 | 430 | 402 |
| CDR3 | V, VI | 4O | 840 | 840 | 842 |
| HIS1 | IV, XI | 5I | 480 | 480, >2300 | 469 |
| CHT2 | IV, VIII | 5I | 480 | 480, 1300 | 469 |
| GCD11 | I, VIII | 5M | 750 | 520 | 541 |
| MTLa | I | 5M | 750 | 520 | 541 |
| MTLα | VIII | 5M | 750 | 520 | 541 |
| 6Cfc | III | 6C | 190 | 190 | 202 |
| ARD1 | III | 6O | 900 | 900 | 874 |
| LEU2 | II, V | 7C | 190 | 180 | 149 |
| YPL12 | II, V | 7A | 100 | 127 | |
| ARG4 | II, X | 7G | 390 | 280 | 265 |
The figure of 2238 kb does not include the rDNA repeat.
3.3 Evidence for a translocation involving rDNA
Figure 3A shows a CHEF separation of the karyotypes of SC5314 and CD36 probed with sequences located on chromosomes R and 5 in C. albicans. The rDNA probe hybridizes to two closely migrating bands in SC5314, while there are three hybridizing bands in CD36, one of which, VIII, is much smaller than the other two (XII, XI). This is due to a reciprocal translocation of part of the rDNA cluster, the adjacent unique DNA, and the telomere of chromosome R with about 39 kb of the right end of chromosome 5; the translocation products are bands XI and VIII. Probing the gel with other C. albicans chromosome R probes shows that YRB1 and CDR7 hybridize with bands XII and XI and not with VIII. Probe SUC1, which is on chromosome R, telomere-proximal to the rDNA, hybridizes with XII and VIII but not with XI. Two probes from chromosome 5 in C. albicans also differ in their hybridization behavior; HIS1, located 36 kb from the right telomere of chromosome 5 in C. albicans, hybridizes to bands IV and XI, while CHT2 lights up IV and VIII. These results indicate that chrR and chr5 have exchanged their right-hand ends, yielding bands XI and VIII. Figure 3B shows an SfiI digest probed with rDNA, CHT2, and HIS1. In CD36, the rDNA probe hybridizes strongly to a band at the top of gel; this band contains XII and XI, which do not change their migration rates significantly after SfiI digestion (Table 2). rDNA also lights up a fragment of 1300 kb. CHT2 hybridizes to a band of 480 kb and the one of about 1300 kb. HIS1, on the other hand, hybridizes to the 480 kb band and the one at the top of the gel. The 1300 kb fragment is a digestion product of band VIII, containing most of 5I and the right hand end of chr R, and the band at the top is the digestion product of chrR translocated with the right-hand tip of chr 5.
In order to determine the sizes of the three blocks of rDNA repeats in CD36, we used the enzyme XhoI, which does not cut in the rDNA sequence of C. albicans (Magee, D’Souza et al. 1987) nor of C. dubliniensis (data not shown), to isolate the rDNA clusters. Figure 3C shows the XhoI genomic digests of SC5314 and CD36, blotted and probed with the C. albicans rDNA probe. In the C. albicans strain, the two chromosome homologues carry repeats of 900 and 350 kb, respectively, whereas in CD36 the repeats are 450 and 250 kb. The fact that there are only two XhoI bands in CD36 suggests that two of the chromosomes carry the same size rDNA cluster. This is supported by the fact that the 450 kb band is faintly visible on the stained gel (not shown). It seems most likely that band VIII carries one of the 450 kb blocks of rDNA, while XII carries the other, and the 250 kb block is located on band XI. This is based on the sizes of the fragment and band VIII. SfiI fragment 5I is 480 kb in size. HIS1, which is on the piece translocated to R, is about 40 kb from the telomere. Thus, the 5I sequence remaining on VIII must be about 440 kb, while the portion of chromosome R telomere-proximal to the rDNA in C. albicans is about 400 kb. Summing the rough estimates of 5I (440 kb), the rDNA block (450), and the unique telomere-adjacent part of chromosome R (400) gives a size of 1290 kb for the predicted SfiI fragment, very near the measured 1300 kb size. This conclusion is further supported by the size of band VIII, which is about 2 Mb. Chromosome 5 in C. albicans is about 1.2 Mb in size. The C. dubliniensis assembly suggests that if chromosome 5 were intact it would be 1.23 Mb. Thus, the translocation must add about 800 kb to chromosome 5 to give band VIII; this is close to 850 kb, the calculated size of the translocated piece of R.
3.4 Inferred chromosome arrangements in C. dubliniensis strain CD36
The karyotype of CD36 is highly complex compared to that of SC5314, with only 3 pairs of unrearranged homologues. However, the subchromosomal fragments of the two species have a great deal of synteny. Because of the similarity of the genome sequence to that of C. albicans, and because the karyotype of SC5314 is well established (Chu, Magee et al. 1993; Jones, Federspiel et al. 2004), we propose to name the chromosomes based on the karyotype of C. albicans. In fact, we argue in the Discussion that the two species’ chromosomes can be considered orthologous in the genetic sense, having evolved from a common precursor. Figure 4 shows a cartoon of the inferred karyotype.
Figure 4.
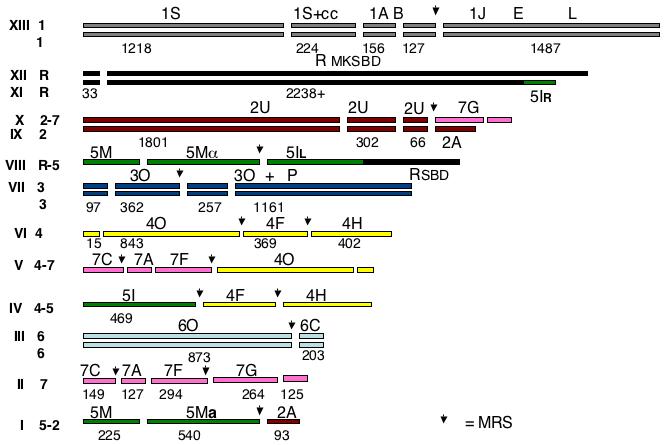
Cartoon of the karyotype of the C. dubliniensis karyotype. The chromosomes are given Arabic numerals and color-coded according to the orthologous chromosome in C. albicans. Chromosome R and its translocated piece are coded black, chr 1 is grey, 2 is violet, 3 is dark blue, 4 is yellow, 5 is green, 6 is light blue, and 7 is red. The breaks in the bars indicate the SfiI sites identified in the sequence assembly and the numbers under the bars give the size of the SfiI fragment calculated from the sequence assembly. When C. dubliniensis has SfiI sites which do not exist in C. albicans, the C. albicans letter designation is retained for the group of fragments. The arrows mark the sites of MRSs. The Roman numerals indicate the bands in the CHEF gels in Figure 1.
Chromosome 1
(C. albicans SfiI fragments: 1S, 1CC, 1A, 1B, 1J, 1E, 1L) Probes for four chromosome 1 genes hybridize only to band XIII. Band XIII is about 3500 kb, comparable to the size of chromosome 1 in C. albicans. We conclude that this band contains both homologues of chromosome 1.
Chromosome R
(C. albicans SfiI fragments: rM, rK, rS, rB, rD) Bands XII and XI hybridize with probes YRB1 and CDR7, both telomere distal to the rDNA in C. albicans. Probe SUC1 lights up XII and VIII. Band XII is about 2200 kb, XI is slightly smaller, and VIII is about 2000 kb.
SfiI digestion does not change the gel positions of bands XI and XII, but it does affect the rDNA-hybridizing band in VIII. Thus, in contrast to C. albicans, the larger bands appear to lack SfiI sites. As shown by the sequence determination (See supplemental material) there is only one Sfi site in chromosome R of C. dubliniensis; the site is located 33 kb from the left telomere. Digestion with SfiI therefore does not change the migration rate of chromosome R under the electrophoresis conditions we use. Bands XII, XI, and VIII all hybridize with the rDNA probe. Bands XI and XII hybridize with CDR7 and YRB1, while VIII does not. Band XI does not hybridize with HCS1 or SUC1 (rB), probes which are telomere-proximal to the rDNA in C. albicans, but XII and VIII do. We conclude that band XII contains a chromosome which is quite similar to that of chromosome R in C. albicans, but it lacks a Major Repeat Sequence and has only one SfiI site. Comparing the sequence around the point where the MRS is found in C. albicans with the C. dubliniensis shows that the arrangement of ORFs excluding the MRS is identical in the two species. The arrangement of the other homologue, involved in a translocation with chromosome 5, is discussed above.
Chromosome 2
(C. albicans SfiI fragments: 2U, 2A) Probe 2B6, specific for 2U in C. albicans, hybridizes to bands IX (2260 kb) and X (2560 kb). Probe 2A, specific for the right end of chromosome 2, hy bridizes to band IX and to band I (860 kb). ARG4, a probe specific for C. albicans band 7G, hybridizes to band X and to band II (960 kb). Band X contains a 2U-7G translocation and band IX an intact chromosome 2 homologue. MTLa and 2117, probes from fragment 5M in C. albicans, hybridize to band I. Band I contains a 5M-2U translocation.
Chromosome 3
(C. albicans SfiI fragments: 3P, 3O) Probes URA3 and NPS1 hybridize to band VII (1900 kb). In C. albicans, these probes hybridize to only one SfiI fragment, 3O, but in C. dubliniensis NPS1 lights up a band smaller than 400 kb. Chromosome 3 in C. dubliniensis differs from C. albicans in that it has an SfiI site 98 kb away from the right telomere as well as a Major Repeat Sequence (at around base 455, 000), while in C. albicans, chromosome 3 is the only chromosome that lacks an MRS. The MRS splits fragment 3O into two pieces of 460 and 257 kb, and the 460 piece is split again into a 360 and a 98 kb piece by the sequence-generated SfiI site. The piece corresponding to 3P is 1,161 kb long. PKC1 hybridizes to this band. Band VII contains both homologues of chromosome 3.
Chromosome 4
(C. albicans SfiI fragments: 4O, 4F, 4H) Probes CDR3 and S1C4, specific for 4O, hybridize to bands VI (1600 kb) and V (1400 kb) and probes CYP52 and LYS1, specific for 4F, hybridize to VI and IV (1240 kb). Probe YJR61 (4H) hybridizes to IV and VI. CHT2, a 5I probe, also hybridizes to IV. YPL12, a 7A probe, lights up bands II and V as do LEU2 (7C) and pCHR7 (7F). Band VI thus contains a complete chromosome 4, while band V contains a translocation of 4O with 7A, 7C, and 7F. Band IV contains a translocation involving 4F, 4H, and 5I.
Chromosome 5
(C. albicans SfiI fragments: 5M, 5I) CHT2 and HIS1, probes for 5I, both hybridize with band IV; CHT2 lights up band VIII and HIS1 lights up XI. The MTLa probe, specific for 5M, hybridizes with band I and MTLα lights up band VIII. GCD11, a 5M probe, lights up bands VIII and I. Bands VIII and XI are described above (chromosome R). Band IV is a translocation product of fragment 5I and 4FH, and band I contains a translocation of 5M with 2A.
Chromosome 6
(C. albicans SfiI fragments: 6O, 6C) Probes specific for 6O (2002) and a mixed set of probes for 6C both hybridize to band III (1080 kb) only. Two complete homologues of chromosome 6 are found in this band.
Chromosome 7
(C. albicans SfiI fragments: 7C, 7A, 7F, 7G) All chromosome 7 probes hybridize to band II. This band comprises the complete chromosome 7. LEU2 and YPL12, found on C. albicans fragments 7C and 7A respectively, hybridize with bands II and V. Fragment 7G is translocated to 2U in band X as described above. A translocation product of fragments 7CAF and 4O constitutes band V. In C. albicans, chromosome 7 has two MRSs, one located between Sfi fragments F and G and one between A and F. CD36 also has two MRSs, but one is located between SfiI fragments C and A, rather than between A and F. The second MRS is between F and G as in SC5314.
3.5 SfiI sites derived from the unique DNA in the genomic sequence
The C. dubliniensis sequence assembly allows us to locate the MRS equivalents as well as to identify other SfiI sites in the chromosomes in C. dubliniensis chromosomes. Not surprisingly, these latter sites frequently differ between the two species. For example, although chromosome 3 in C. albicans has only one SfiI site, in the C. dubliniensis assembly there are three, dividing the chromosome into fragments of 97, 362, 257, and 1161 kb. The SfiI sites in the assembly for each chromosome are given in the Supplementary Data. Table 2 summarizes some hybridization results using blots of SfiI digests. For the most part, the measured fragment size and the predicted fragment size were in agreement. However, there were some discrepancies. Probe PKC1 hybridizes to a band of 1030 kb (920 in C. albicans), indistinguishable from the predicted 1161 kb. In C. albicans, chromosome 5 has one SfiI site, dividing it into fragments of 750 (M) and 480 kb (I). The C. dubliniensis genome assembly predicts that the 750 5M fragment should have an internal SfiI site, giving products of 224 and 542 kb. Probe PUT1 hybridizes to a 520 kb band from C. dubliniensis, and GCD11 to a band of the same size. Although we have not verified all the SfiI sites in the assembly by digestion, we have included them in the karyotype map in Fig. 4.
3.6 The karyotype of CD36
Figure 4 is a graphic depiction of the CD36 karyotype. The chromosomes are colored to indicate the relationship of the DNA organization to that of the C. albicans karyotype. The MRS sites derived from the sequence assembly are indicated by downward arrows, and the breaks in the bars indicate SfiI sites. The karyotype has pairs of homologues for chromosomes 1, 3, and 6, and one intact homologue for each of chromosomes R, 2, 4, and 7. Four translocations have occurred at MRSs (as is the commonest case in C. albicans), but they are not reciprocal, although they do not affect the ploidy of the strain (A. Selmecki, personal communication). There is a fifth chromosomal rearrangement, which seems to have occurred by translocation between the right end of chromosome R and the right end of chromosome 5, leading to an exchange of telomere-proximal sequences.
Except for chromosomes 4 and 7, the SfiI order was determined by examination of the sequence. For 4 and 7, where multiple MRSs make determination of order based on the sequence impossible, the following logic was used. For chromosome 4, the order O-F was assigned based on the 7-4 translocation which leads to bands V and IV. The F-H order was based on C. albicans. For chromosome 7, the order F-G was based on the translocations in bands V and X. The order C-A was based on the MRS which in C. dubliniensis occurs between C and A, and the sequence, which shows that A is adjacent to F.
3.7 Electrophoretic karyotypes of other strains
The electrophoretic karyotypes of several isolates of C. dubliniensis were determined using the standard conditions for analysis of the C. albicans chromosomes. Figure 5 shows that in comparison to C. albicans strain 1006 (lane 5; this karyotype is the same as that of SC5314, the source of the DNA for the C. albicans genome sequence), the karyotypes of the C. dubliniensis strains are highly varied. In comparison to the 8 bands of C. albicans, strain R3b (lane 1) has 7, with two which look like they include multiple chromosomes and one smaller than 900 kb. R1b (lane 2) has 11 bands, including two smaller than C. albicans chr 7. Strain 3225 (lane 3) has 11 or possibly 12 bands, 2 of which appear to include multiple chromosomes. Strain 3233 in lane 4 has 8 bands. Strain 16F (lane 7) has 9 bands, two or three of them possibly multiple. Strain 514 (lane 8) has a karyotype similar to 1006, but its SfiI restriction digest pattern is clearly that of C. dubliniensis (Fig. 2, lane 3). Strain 514 belongs to genotype group 3 of C. dubliniensis, while CD36 (lane 6) has a group 1 genotype (Gee, Joly et al. 2002).
The divergent karyotypes are not the result of simple chromosome length polymorphism of the sort studied by Chibana, et al (Chibana, Beckerman et al. 2000), as shown by the location of the MTL locus, which is found in two heteroallelic states, a and α, on chromosome 5 in C. albicans. The results of probing a Southern blot of the gel with the MTLa and α probes are indicated on the figure. Only in 1006, 514, and R1b do the the two homologues of chr 5 migrate together. In 3225 and 3233 the MTL locus is homozygous, but the two identical alleles are on separable electrophoretic bands. Thus a karyotype with few paired homologues and several translocations, like that of CD36, is quite common in C. dubliniensis.
4. Discussion
C. dubliniensis is very closely related to C. albicans. The two species differ by 13 nucleotides in the D1/D2 region of the large ribosomal subunit RNA (Kurtzman and Robnett 1998) and can share more than 98% identity between orthologous genes. In addition, interspecific hybrids can be formed by mating (Pujol, Daniels et al. 2004). Thus, it is not surprising that the arrangement of DNA in the genomes of these sister species is similar. However, their biological properties are distinct (Sullivan and Coleman 1998). C. dubliniensis is more sensitive to elevated temperatures (Sullivan and Coleman 1998), it makes chlamydospores profusely under conditions where C. albicans does not make them (Staib and Morschhauser 1999), and, most importantly from the point of view of infectious disease, it is less virulent than C. albicans (Gilfillan, Sullivan et al. 1998). A pioneering microarray study suggested that 247 C. albicans genes are missing or are less than 60% homologous in C. dubliniensis (Moran, Stokes et al. 2004). With the achievement of the sequence of C. dubliniensis it is important to examine the chromosome structure to determine whether karyotypic differences play a role in the divergent properties.
We have shown that there are strong similarities between the chromosome structure found in C. albicans (Chu, Magee et al. 1993) and that of C. dubliniensis. Most of the contigs assembled by Jones and coworkers (Jones, Federspiel et al. 2004) are highly homologous to the C. dubliniensis genome sequence. However, the large scale organization of the chromosomes differs not only from the most common karyotype found in C. albicans but also among all the C. dubliniensis isolates we have examined. Although C. albicans does show considerable karyotypic variation, the standard karyotype is found in 50% of isolates (Magee and Chibana 2002). We have examined 10 C. dubliniensis strains and found no karyotypes in common. Although one strain, 514, has a karyotype which resembles the standard C. albicans one, its SfiI restriction fragment pattern clearly identifies it as C. dubliniensis and its MTL loci hybridize with the C. dubliniensis probes. We have chosen to determine the molecular details of the karyotype of CD36, the type strain and the source of the genome sequence. In contrast to the evidence for variation in vivo (and rarely in vitro) among C. dubliniensis strains (Gee, Joly et al. 2002), the karyotype of CD36 has remained constant as determined by pulse-field electrophoresis for more than 10 years in our laboratory. During this time, three separate isolates have been maintained, including freezing, defrosting, growing, and refreezing, without any sign of change.
Since the numbering system of C. albicans is most useful for this discussion, we will distinguish between the C. albicans chromosomes (CAchr) and the C. dubliniensis orthologues (CDchr). Three of the chromosomes of CD36, CDchr 1 (Band XIII), 3 (VII), and 6 (III), have homologues which, at the level of resolution used, appear not to be rearranged. CDchrs 1 and 3 have patterns of SfiI sites which differ from their orthologues in C. albicans, while the SfiI arrangement in chr6 (III) is the same in both species, with the only sites located in the MRS. The location of the chr 6 MRS, about 90 kb away from the telomere, is also similar in the two species. In CAchr 1, the MRS is about 1692 kb from the left telomere, while in CDchr 1 (XIII) it is at 1723 kb. CDchr 3 (VII) has a pair of unrearranged homologues, like CAchr 3, but it differs in having an MRS. CAchr 3 is the only chromosome in C. albicans lacking a complete MRS.
CDchr R (XII) differs from CAchr R in one important way: it has no MRS. In fact, it has only one SfiI site, 33kb from the left end of the chromosome. One chr R homologue in CD36 has undergone a reciprocal translocation with the right terminus of chr 5 (Bands XI and VIII). The break point for this translocation is within the cluster of rDNA repeats; this explains the fact that the translocation was not detected by sequencing. Since there are approximately 90 rDNA repeats in the diploid genome only one in 90 rDNA traces would have shown it. The translocation results in three dissimilar chromosomes carrying rDNA (Table 2, Fig 4): the unchanged homologue of R and the two 5-R translocation products. The total amount of rDNA, about 1150 kb, is not significantly different from that of C. albicans (van Het Hoog, Rast et al. 2007).
The other translocations in CD36 apparently occur at the MRS, as is common in C. albicans. The best-studied C. albicans strain with multiple translocations is WO-1. Each of the translocations in WO-1 seems to be the result of a single event: all are reciprocal and occur at the MRS, so they do not result in any significant alteration in ploidy (Magee and Chibana 2002). In CD36, several of the translocations seem to involve more than two chromosomes. For example, a piece of chromosome 7 has translocated to chromosome 2, while the other piece of 7 is part of a 4,7 translocation. The other half of chromosome 4 has translocated with chromosome 5, and the remaining part of chromosome 5 has translocated with the end of chromosome 2. A minimum of three sequential events and the R-5 reciprocal translocation would be required for the present karyotype to arise.
The karyotypes of C. dubliniensis and C. albicans are thus highly similar in their basic construction: blocks of largely unique DNA broken up by the large intermediate repeat, the MRS, which appears in one or more copies on all but one chromosome. But they differ in the chromosome lacking the MRS (chr3 in C. albicans; chrR in C. dubliniensis). Since the two karyotypes must have evolved from a similar ancestor, it seems most likely that the ancestral karyotype resembled the standard C. albicans one, with pairs of homologues. Both species tend to undergo translocations, but the frequency of occurrence is much greater in the population represented by the C. dubliniensis isolates than in C. albicans.
The presence of a larger number of RPSs (a subunit of the MRS) in C. dubliniensis compared to C. albicans has been noted, and data presented that C. dubliniensis strains have a relatively high frequency of rearrangements of chromosome 7 (Joly, Pujol et al. 2002). This is in accord with our data showing that chromosome rearrangement in CD36 is extensive and that only one of eight randomly chosen strains has a complete set of unaltered homologous chromosome pairs. Can the increased frequency of chromosome rearrangement be attributed to the increase in RPS number? Since no one has been able to study the process of chromosome translocation, no answer to the question is available at this time. Two pieces of evidence suggest that the situation may be more complicated than just the amount of near-homologous DNA on non-homologous chromosomes. The first is the translocation in the rDNA, a chromosomal aberration which has not been reported in C. albicans but which has been found in another C. dubliniensis strain (B. Magee, unpublished). This suggests that the instability of the C. dubliniensis genome is due to more than a larger number of RPS subunits. It would be interesting to examine the genes involved in DNA repair, such as the homologous end-joining pathway, which has not yet been studied in C. dubliniensis, to see whether this pathway is deficient.
The second report with bears on the question of the effect of increased numbers of RPS subunits is a study of mitotic recombination on chromosome 5 in C. albicans. In this paper, the rate of recombination in the MRS was slightly lower than in unique DNA (Lephart and Magee 2006). This would suggest that increasing the number of RPS subunits in any MRS might not greatly increase the frequency of translocation.
The biological role of mating in C. albicans (and by analogy in C. dubliniensis) is still not understood, and various suggestions for its role in the biology of these organisms have been made, ranging from facilitation of commensality (Magee and Magee 2004) to importance in pathogenesis (Soll 2004). Whatever that role, the highly varied karyotypes we have found in C. dubliniensis would rule out meiosis except as a very infrequent event. Translocations lead to mispairing during meiosis, and mispairing in turn leads to aneuploidy. Although aneuploid strains of C. albicans have been reported (Magee and Magee 1997; Selmecki, Bergmann et al. 2005; Selmecki, Forche et al. 2006), they have relatively small deviations from perfect diploidy, all but one being trisomics, while meiosis between two strains with extensive translocations would be expected to lead to a more radical level of aneuploidy, including some regions of haploidy. Hence, it seems that C. dubliniensis, at least that part of the species represented by this isolate, does not undergo meiosis as a frequent part of its life cycle.
The present comparison does not shed light on the origin and function of the MRS. This sequence appears in these two species only; the relatively closely related Candida tropicalis lacks an MRS. The MRS from C. dubliniensis is from 81-95% identical to that from C. albicans. The maps of CDchr3 and CAchr3 are identical around the site of the MRS insertion in C. dubliniensis. Shotgun sequencing is unreliable in repetitive regions, but in this case the maps match closely on both sides of the MRS, which is inserted between ORF19.1653 and ORF19.1652 in Assembly 21. The lack of any sequences related to the MRS in this region in C. albicans supports the idea that the common ancestor of the two species lacked this MRS, since one would not expect an excision event to proceed leaving neither sequence homology nor a deletion. The situation is harder to determine on chromosome R, but once again the map outside the MRS region on CAchrR is identical to the map on CDchrR. Chromosome 7 shows another deviation from the MRS organization of C. albicans. In CD36, the MRS constitutes the boundary between Sfi fragments 7C and 7A, while in SC5314, it is about 100 kb telomere-distal and constitutes the boundary of Sfi fragments 7A and 7F. On the other chromosomes, the positions of the MRSs correspond in the two species. Thus the most reasonable hypothesis is that the common ancestor had MRSs at the sites common to the two species, but after they diverged, C. albicans had an insertion of the MRS on chromosome R, C. dubliniensis had an insertion on chromosome 3, and both had an insertion on chromosome 7, but in slightly different locations. The fact that the MRS has persisted in these two species suggests that it confers some selectable advantage.
The significance of the very high level of variation in karyotype among C. dubliniensis strains can only be a subject of speculation at this time. If karyotypic variation leads to phenotypic diversity, then one might expect C. dubliniensis to be better at colonizing diverse niches than C. albicans, but there is no evidence of this. In fact, the converse seems to be true; C. dubliniensis seems to be most common in the mouth and only rarely to cause disseminated disease (Sullivan, Moran et al. 2004). This raises the question of whether a genome rearranged by translocations at the MRS actually provides variation. Alteration of the context of coding DNA in C. albicans, for example, moving URA3 to new chromosome positions, does affect its expression (Lay, Henry et al. 1998; Sharkey, McNemar et al. 1999; Staab and Sundstrom 2003), but for genes on a chromosome arm involved in a translocation at the MRS, the context, as distinct from the chromosome location, is unaltered. Rearrangements have not been shown to alter transcription in the best-studied C. albicans strain, WO-1. This isolate has three chromosome translocations, and its transcriptome has been analyzed (Lan, Newport et al. 2002) but no changes in gene expression related to its chromosomal configuration were observed. WO-1 is virulent despite its rearrangements, but not hypervirulent (Kvaal, Srikantha et al. 1997). Only a comparison of the relative levels of transcription of genes adjacent to the MRS in WO-1 and a non-rearranged strain like SC5314 under identical conditions can determine whether MRS-based translocations affect gene regulation.
An alternative possibility is that very high levels of karyotypic instability in the host may affect the fitness of the yeast and diminish its ability to colonize niches other than the gastrointestinal tract. If this is true, the high level of rearrangement in C. dubliniensis would be the signature of this instability and the process could be one of the reasons behind the reduced virulence.
5. Conclusions
The karyotype of CD36, the strain of C. dubliniensis whose genome has been sequenced (unpublished) at the Wellcome Trust Sanger Institute, is composed of three pairs of homologues and 10 chromosome translocation products derived from homologues very similar to those of the related pathogenic yeast, C. albicans. The overall similarity in the organization of the chromosomes of these two yeasts is consistent with their close phylogenetic relationship. However, they differ in the details of genome organization and apparently in the frequency of translocation. While the differences in organization seem unlikely to be related to virulence, the frequency with which reorganization seems to occur may signal greater genomic instability in C. dubliniensis. Such a difference may affect the ability to adapt to more unfamiliar and demanding niches like the blood sream, thus helping to account for the contrasts in virulence.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by USPHS grant AI16567 from the National Institute of Allergy and Infectious Disease and by the University of Minnesota Life Sciences Summer Undergraduate Research Program. The sequence data for C. dubliniensis were produced by the Pathogen Sequencing Unit at the Wellcome Trust Sanger Institute (WTSI) and are available from the website http://www.sanger.ac.uk/sequencing/Candida/dubliniensis/ . DS, DEH, and MB are supported by the Wellcome Trust through their funding of WTSI. We thank Professor Judith Berman for helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Braun BR, van het Hoog M, d’Enfert C, Martchenko M, Dungan J, Kuo A, Inglis DO, Uhl MA, Hogues H, Berriman M, Lorenz M, Levitin A, Oberholzer U, BAchewich C, Harcus D, Marcil A, Dignard D, Iouk T, Zito R, Frangeul L, Tekaia F, Rutherford K, Wang E, Munro C, Bates S, Gow NA, Hoyer L, Kohler G, Morschhauser J, Newport G, Znaidi S, Raymond M, Turcotte B, Sherlock G, Constanzo M, Ihmels J, Berman J, Sanglard D, Agabian N, Mitchell A, Johnson AD, Whiteway M, Nantel A. A human-curated annotation of the Candida albicans genome. PLOS Genetics. 2005;1(1):36–57. doi: 10.1371/journal.pgen.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibana H, Beckerman JL, et al. Fine-resolution physical mapping of genomic diversity in candida albicans. Genome Res. 2000;10(12):1865–77. doi: 10.1101/gr.148600. [DOI] [PubMed] [Google Scholar]
- Chibana H, Iwaguchi S, et al. Diversity of tandemly repetitive sequences due to short periodic repetitions in the chromosomes of Candida albicans. Journal of Bacteriology. 1994;176(13):3851–8. doi: 10.1128/jb.176.13.3851-3858.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chindamporn A, Nakagawa Y, et al. Repetitive sequences (RPSs) in the chromosomes of Candida albicans are sandwiched between two novel stretches, HOK and RB2, common to each chromosome. Microbiology. 1998;144(4):849–857. doi: 10.1099/00221287-144-4-849. [DOI] [PubMed] [Google Scholar]
- Chu WS, Magee BB, et al. Construction of an SfiI macrorestriction map of the Candida albicans genome. Journal of Bacteriology. 1993;175(20):6637–51. doi: 10.1128/jb.175.20.6637-6651.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee SF, Joly S, et al. Identification of four distinct genotypes of Candida dubliniensis and detection of microevolution in vitro and in vivo. J Clin Microbiol. 2002;40(2):556–74. doi: 10.1128/JCM.40.2.556-574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan GD, Sullivan DJ, et al. Candida dubliniensis: phylogeny and putative virulence factors. Microbiology. 1998;144(Pt 4):829–38. doi: 10.1099/00221287-144-4-829. [DOI] [PubMed] [Google Scholar]
- Goshorn AK, Grindle SM, et al. Gene isolation by complementation in Candida albicans and applications to physical and genetic mapping. Infection & Immunity. 1992;60(3):876–84. doi: 10.1128/iai.60.3.876-884.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaguchi S, Homma M, et al. Isolation and characterization of a repeated sequence (RPS1) of Candida albicans. Journal of General Microbiology. 1992;138(Pt 9):1893–900. doi: 10.1099/00221287-138-9-1893. [DOI] [PubMed] [Google Scholar]
- Joly S, Pujol C, et al. Development and characterization of complex DNA fingerprinting probes for the infectious yeast Candida dubliniensis. J Clin Microbiol. 1999;37(4):1035–44. doi: 10.1128/jcm.37.4.1035-1044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly S, Pujol C, et al. Microevolutionary changes and chromosomal translocations are more frequent at RPS loci in Candida dubliniensis than in Candida albicans. Infect Genet Evol. 2002;2(1):19–37. doi: 10.1016/s1567-1348(02)00058-8. [DOI] [PubMed] [Google Scholar]
- Jones T, Federspiel NA, et al. The diploid genome sequence of Candida albicans. Proc Natl Acad Sci U S A. 2004;101(19):7329–34. doi: 10.1073/pnas.0401648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg BJ, Filler SG. Candidemia. In: Calderone R, editor. Candida and Candidiasis. ASM Press; Washington, DC: 2002. pp. 327–340. [Google Scholar]
- Kurtzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek. 1998;73(4):331–71. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- Kvaal CA, Srikantha T, et al. Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infection & Immunity. 1997;65(11):4468–75. doi: 10.1128/iai.65.11.4468-4475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan CY, Newport G, et al. Metabolic specialization associated with phenotypic switching in Candidaalbicans. Proc Natl Acad Sci U S A. 2002;99(23):14907–12. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasker BA, Carle GF, et al. Comparison of the separation of Candida albicans chromosome-sized DNA by pulsed-field gel electrophoresis techniques. Nucleic Acids Research. 1989;17(10):3783–93. doi: 10.1093/nar/17.10.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lay J, Henry LK, et al. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect Immun. 1998;66(11):5301–6. doi: 10.1128/iai.66.11.5301-5306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lephart PR, Magee PT. Effect of the major repeat sequence on mitotic recombination in Candida albicans. Genetics. 2006;174(4):1737–44. doi: 10.1534/genetics.106.063271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee BB, D’Souza TM, et al. Strain and species identification by restriction fragment length polymorphisms in the ribosomal DNA repeat of Candida species. Journal of Bacteriology. 1987;169(4):1639–43. doi: 10.1128/jb.169.4.1639-1643.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee BB, Koltin Y, et al. Assignment of cloned genes to the seven electrophoretically separated Candida albicans chromosomes. Molecular & Cellular Biology. 1988;8(11):4721–6. doi: 10.1128/mcb.8.11.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee BB, Magee PT. WO-2, a stable aneuploid derivative of Candida albicans strain WO-1, can switch from white to opaque and form hyphae. Microbiology. 1997;143(Pt 2):289–95. doi: 10.1099/00221287-143-2-289. [DOI] [PubMed] [Google Scholar]
- Magee PT, Chibana H. The Genomes of Candida albicans and other Candida species. In: Calderone R, editor. Candida and Candidiasis. ASM Press; Washington, DC: 2002. pp. 293–304. [Google Scholar]
- Magee PT, Magee BB. Through a glass opaquely: the biological significance of mating in Candida albicans. Curr Opin Microbiol. 2004;7(6):661–5. doi: 10.1016/j.mib.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Maschmeyer G. The changing epidemiology of invasive fungal infections: new threats. Int J Antimicrob Agents. 2006;27S1:3–6. doi: 10.1016/j.ijantimicag.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Moran G, Stokes C, et al. Comparative genomics using Candida albicans DNA microarrays reveals absence and divergence of virulence-associated genes in Candida dubliniensis. Microbiology. 2004;150(Pt 10):3363–82. doi: 10.1099/mic.0.27221-0. [DOI] [PubMed] [Google Scholar]
- Muller FM, Kasai M, et al. Transmission of an azole-resistant isogenic strain of Candida albicans among human immunodeficiency virus-infected family members with oropharyngeal candidiasis. J Clin Microbiol. 1999;37(10):3405–8. doi: 10.1128/jcm.37.10.3405-3408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Garcia F, Perez-Diaz RM, et al. Chromosome reorganization in Candida albicans 1001 strain. Journal of Medical & Veterinary Mycology. 1995;33(6):361–6. [PubMed] [Google Scholar]
- Pujol C, Daniels KJ, et al. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot Cell. 2004;3(4):1015–27. doi: 10.1128/EC.3.4.1015-1027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SangerInstitute ( http://www.sanger.ac.uk/)
- Selmecki A, Bergmann S, et al. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol Microbiol. 2005;55(5):1553–65. doi: 10.1111/j.1365-2958.2005.04492.x. [DOI] [PubMed] [Google Scholar]
- Selmecki A, Forche A, et al. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313(5785):367–70. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey LL, McNemar MD, et al. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J Bacteriol. 1999;181(17):5273–9. doi: 10.1128/jb.181.17.5273-5279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll DR. Mating-type locus homozygosis, phenotypic switching and mating: a unique sequence of dependencies in Candida albicans. Bioessays. 2004;26(1):10–20. doi: 10.1002/bies.10379. [DOI] [PubMed] [Google Scholar]
- Staab JF, Sundstrom P. URA3 as a selectable marker for disruption and virulence assessment of Candida albicans genes. Trends Microbiol. 2003;11(2):69–73. doi: 10.1016/s0966-842x(02)00029-x. [DOI] [PubMed] [Google Scholar]
- Staib P, Morschhauser J. Chlamydospore formation on Staib agar as a species-specific characteristic of Candida dubliniensis. Mycoses. 1999;42(910):521–4. doi: 10.1046/j.1439-0507.1999.00516.x. [DOI] [PubMed] [Google Scholar]
- Sullivan D, Coleman D. Candida dubliniensis: characteristics and identification. J Clin Microbiol. 1998;36(2):329–34. doi: 10.1128/jcm.36.2.329-334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DJ, Moran GP, et al. Comparison of the epidemiology, drug resistance mechanisms, and virulence of Candida dubliniensis and Candida albicans. FEMS Yeast Res. 2004;4(45):369–76. doi: 10.1016/S1567-1356(03)00240-X. [DOI] [PubMed] [Google Scholar]
- Sullivan DJ, Westerneng TJ, et al. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141(Pt 7):1507–21. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- Timmins EM, Howell SA, et al. Rapid differentiation of closely related Candida species and strains by pyrolysis-mass spectrometry and Fourier transform-infrared spectroscopy. J Clin Microbiol. 1998;36(2):367–74. doi: 10.1128/jcm.36.2.367-374.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Het Hoog M, Rast TJ, et al. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 2007;8(4):R52. doi: 10.1186/gb-2007-8-4-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes B, Staudinger J, et al. Physical and genetic mapping of Candida albicans: several genes previously assigned to chromosome 1 map to chromosome R, the rDNA-containing linkage group. Infection & Immunity. 1991;59(7):2480–4. doi: 10.1128/iai.59.7.2480-2484.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


