Abstract
Ubiquitination is an essential process regulating turnover of proteins for basic cellular processes such as the cell cycle and cell death (apoptosis). Ubiquitination is initiated by ubiquitin-activating enzymes (E1), which activate and transfer ubiquitin to ubiquitin-conjugating enzymes (E2). Conjugation of target proteins with ubiquitin is then mediated by ubiquitin ligases (E3). Ubiquitination has been well characterized using mammalian cell lines and yeast genetics. However, the consequences of partial or complete loss of ubiquitin conjugation in a multi-cellular organism are not well understood. Here, we report the characterization of Uba1, the only E1 in Drosophila. We found that weak and strong Uba1 alleles behave genetically differently with sometimes opposing phenotypes. Whereas weak Uba1 alleles protect cells from cell death, clones of strong Uba1 alleles are highly apoptotic. Strong Uba1 alleles cause cell cycle arrest which correlates with failure to reduce cyclin levels. Surprisingly, clones of strong Uba1 mutants stimulate neighboring wild-type tissue to undergo cell division in a non-autonomous manner giving rise to overgrowth phenotypes of the mosaic fly. We demonstrate that the non-autonomous overgrowth is caused by failure to downregulate Notch signaling in Uba1 mutant clones. In summary, the phenotypic analysis of Uba1 demonstrates that impaired ubiquitin conjugation has significant consequences for the organism, and may implicate Uba1 as a tumor suppressor gene.
Keywords: Uba1, E1, Ubiquitin-activating enzyme, Apoptosis, Proliferation, Drosophila, Autonomous control, Non autonomous control
INTRODUCTION
Ubiquitination refers to the covalent attachment of the small protein ubiquitin to target proteins. This modification usually targets ubiquitinated proteins for proteolytic degradation by the proteasome (reviewed by Glickman and Ciechanover, 2002; Pickart, 2004; Welchman et al., 2005). In this capacity, ubiquitination is essential for turnover of proteins involved in many cellular processes including the cell cycle, cell death, signal transduction, etc. (Petroski and Deshaies, 2005). For example, ubiquitin-mediated degradation of cyclins is essential for progression through the cell cycle (reviewed by Pines, 2006). Inhibitor of apoptosis proteins (IAPs) need to be ubiquitinated and degraded in cells undergoing apoptosis (Herman-Bachinsky et al., 2007; Holley et al., 2002; Ryoo et al., 2002; Yang et al., 2000; Yoo et al., 2002) (reviewed by Cashio et al., 2005). However, ubiquitin-mediated degradation of caspases, the principal executioners of apoptosis, has been reported to protect cells from apoptosis (Cashio et al., 2005; Chai et al., 2003; Huang et al., 2000; Suzuki et al., 2001; Wilson et al., 2002). In addition, non-traditional functions of ubiquitination, which do not target proteins for proteolysis, have been reported (Chen, 2005; Mukhopadhyay and Riezman, 2007; Welchman et al., 2005). In this regard, it is noteworthy that activated cell surface signaling receptors are ubiquitinated, usually mono-ubiquitinated, for endocytosis and protein sorting at the early endosome (Gruenberg and Stenmark, 2004; Haglund and Dikic, 2005).
Alterations in the ubiquitination machinery are often associated with human diseases such as cancer, neurodegenerative disorders and inflammation (Devoy et al., 2005; Mukhopadhyay and Riezman, 2007; Reinstein and Ciechanover, 2006). On the other hand, targeting the ubiquitination system for therapeutic purposes holds promise in the clinic (Nalepa et al., 2006). Thus, a detailed understanding of the role of ubiquitination for proper homeostasis and physiology of multi-cellular organisms is critical.
E1 ubiquitin-activating enzymes catalyze the first step in the ubiquitination cycle, the ATP-dependent formation of a thioester bond between the C-terminal glycine residue of ubiquitin and the active site cysteine of the E1 (Haas and Siepmann, 1997; Pickart, 2001). This is followed by the transfer of ubiquitin from the E1 to a ubiquitin-conjugating enzyme (E2). The final step is the conjugation of ubiquitin to target proteins mediated by ubiquitin ligases (E3). The specificity of the ubiquitination process is conferred to by E3 ubiquitin ligases. The genomes of eukaryotic organisms contain hundreds of different E3-encoding genes required for the regulated protein turnover in many cellular processes (Hicke et al., 2005; Petroski and Deshaies, 2005). By contrast, there are considerably fewer E1 and E2 enzymes. For example, the Drosophila genome encodes only one E1 enzyme, termed Uba1 (Watts et al., 2003). This low complexity suggests that the primary function of the E1 enzyme is to provide activated ubiquitin for all ubiquitin-dependent reactions. This has indeed been observed in yeast. Genetic inactivation of the yeast gene Uba1 blocks most, if not all ubiquitin conjugation (Ghaboosi and Deshaies, 2007; McGrath et al., 1991; Swanson and Hochstrasser, 2000). There are mammalian cell lines containing temperature-sensitive alleles of E1. These cell lines have been of great importance for understanding the role of ubiquitin-mediated degradation of cyclins for progression through the cell cycle, and have further suggested an essential function of E1 enzymes to provide activated ubiquitin for conjugation of target proteins (Ciechanover et al., 1984; Ciechanover et al., 1985; Finley et al., 1984; Kulka et al., 1988; Salvat et al., 2000).
However, despite these valuable analyses of ubiquitin conjugation in single cell organisms and cell lines, a systematic analysis of partial or complete loss of ubiquitin conjugation in multi-cellular organisms has not been reported. This can be accomplished by reducing the activity of the only E1 enzyme in Drosophila, Uba1 (Watts et al., 2003). To date, a role of Drosophila Uba1 (from now on referred to as Uba1) has only been reported for axon pruning in the nervous system, and the precise mechanistic function of Uba1 in this process is unknown (Kuo et al., 2006; Watts et al., 2003). Here, we report the isolation and characterization of weak and strong alleles of Uba1 in Drosophila. We show that, depending on the strength of the Uba1 allele, different and sometimes opposing phenotypes are observed. For example, weak Uba1 alleles protect cells from apoptosis, whereas mutant clones of strong alleles are highly apoptotic. Strong Uba1 alleles which we show affect significantly ubiquitin conjugation, cause cell cycle arrest that correlates with increased cyclin levels. Unexpectedly, clones of strong Uba1 alleles induce cell proliferation in neighboring tissue, triggering non-autonomous overgrowth. These Uba1 clones fail to downregulate Notch activity which stimulates Jak/STAT signaling, and thus growth, in neighboring wild-type tissue. In summary, this analysis demonstrates that the lack of ubiquitin conjugation has significant consequences for the organism, and may implicate Uba1 as a tumor suppressor gene.
MATERIALS AND METHODS
Isolation of Uba1 alleles
The Uba1 alleles were obtained in the GMR-hid ey-FLP (GheF) suppressor screen for chromosome arm 2R, which is described elsewhere (Srivastava et al., 2007). Mapping was done relative to mapped P element insertions according to published procedures (Zhai et al., 2003). To identify the mutation in the Uba1 alleles, genomic DNA was amplified by PCR and prepared for DNA sequencing. Uba1s3484 and Uba103405 were obtained from the Bloomington Stock Center.
Fly stocks
Uba1D6, Uba1HH33 and Uba1H42 (this study); Uba1s3484 (Watts et al., 2003); Uba103405, Notch264-39 (obtained from Bloomington Stock Center); GMR-hid ey-FLP (GheF) (Xu et al., 2005); GMR-hid [w−] (Herz et al., 2006); UASp-p35 (Werz et al., 2005); ey-FLP; FRT42D P[ubi-GFP] and E(spl)m8 2.61-lacZ (obtained from G. Halder, MD Anderson Cancer Center, Houston, TX); stat9285c9 (obtained from E. Bach, New York University School of Medicine, NY).
Induction of Uba1 clones and immunohistochemistry
Uba1 clones were induced by the FLP/FRT-mitotic recombination system using either ey- or hs-FLP (Newsome et al., 2000; Xu and Rubin, 1993). If hs-FLP was used, heat-shock was induced in first instar larvae at 37°C for 1 hour in a water bath. Dissection and immunohistochemistry of larval and pupal discs was done using standard protocols (Fan et al., 2005). Antibodies against the following proteins were used: mono and poly-ubiquitylated conjugates (clone FK2; 1:100; Biomol); ubiquitin (Sigma, 1:10; or Chemicon, 1:500); cleaved caspase 3 (Cas3*; 1:100; Cell Signaling Technology); pSTAT (1:100; Cell Signaling Technology); Diap1 [1:500 (Ryoo et al., 2002)]; Dronc [1:500 (Wilson et al., 2002)]; BrdU (1:50; Becton Dickinson); β-gal (1:250; Promega); Discs large (Dlg) (1:250), cyclin A (1:20), Cyclin B (1:20), Elav (1:60), Notch (clone C17.9C6; 1:20) (all DHSB).
RESULTS
Uba1 alleles were identified as autonomous suppressors of GMR-hid
Expression of the pro-apoptotic gene hid [also known as Wrinkled (W) – FlyBase] under control of the eye-specific enhancer GMR (GMR-hid) causes a small eye phenotype due to excessive apoptosis (Fig. 1A) (Grether et al., 1995). Taking advantage of the GMR-hid small eye phenotype we have performed genetic mutagenesis screens aimed at identifying components of the cell death pathway in Drosophila (Srivastava et al., 2007; Xu et al., 2005; Xu et al., 2006). In these screens, we induced homozygous mutant clones of mutagenized chromosome arms using ey-FLP/FRT-induced mitotic recombination (Newsome et al., 2000; Xu and Rubin, 1993), and screened for suppressors of the small eye phenotype of GMR-hid (referred to as GheF screens for GMR-hid ey-FLP). In the GheF screen for chromosome arm 2R, we recovered a lethal complementation group consisting of three alleles (D6, H33, H42) as moderately strong recessive suppressors of GMR-hid (Fig. 1B,C).
Fig. 1. Uba1 mutants are autonomous suppressors of GMR-hid-induced apoptosis.
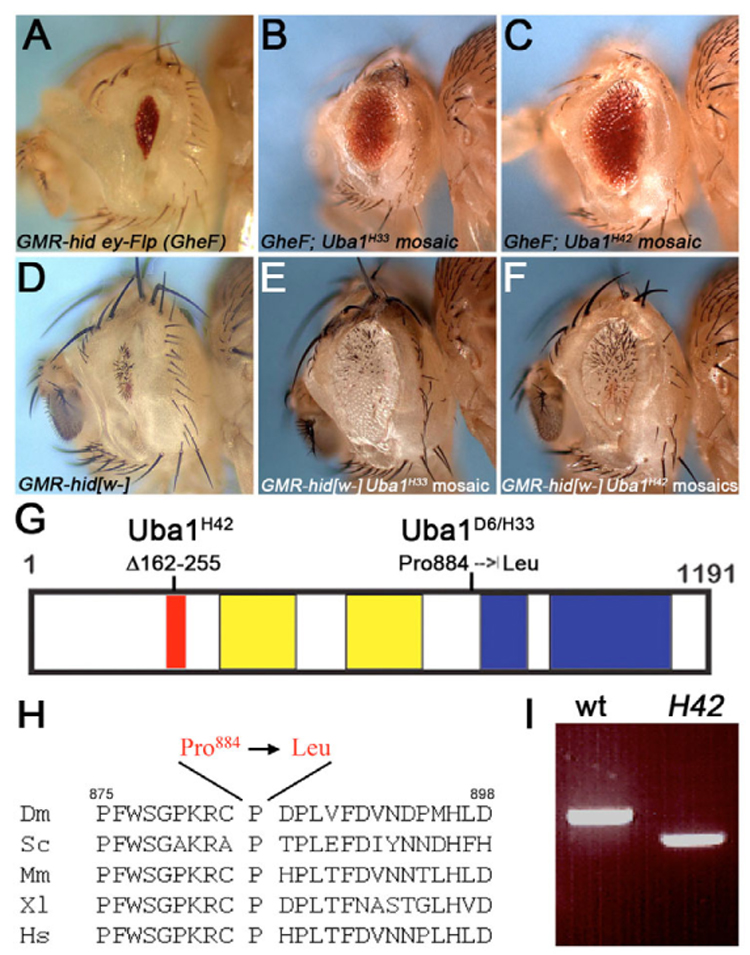
(A) The unmodified small eye phenotype caused by GMR-hid ey-FLP (GheF). (B,C) The GheF phenotype is moderately strongly suppressed by Uba1 alleles at 25°C. Genotype: GheF; FRT42D Uba1H33 and Uba1H42/FRT42 P[w+], respectively. (D) The unmodified small eye phenotype caused by a GMR-hid transgene that lacks the white+ marker gene (GMR-hid[w−]). Note, the GMR-hid transgene in (A) carries the white+ minigene. Genotype: ey-FLP; GMR-hid[w−]. (E,F) The suppression of GMR-hid[w−] is mediated by Uba1 mutant tissue which is phenotypically white–. Flies were incubated at 25°C. Genotype: ey-FLP; FRT42D Uba1H33 and Uba1H42/FRT42D P[w+]; GMR-hid[w−], respectively. (G) Outline of the Uba1 protein and locations of the mutations of the Uba1 alleles isolated in this study. The red box indicates the internal deletion of Uba1H42. The yellow boxes mark the Thif domains, and the blue boxes highlight the location of the catalytic Ube domains. (H) A highly conserved stretch of residues, incorporating Pro884 is present in range of species from yeast to humans. Dm, Drosophila melanogaster; Sc, Saccharomyces cerevisiae; Mm, Mus musculus; XI, Xenopus laevis; Hs, Homo sapiens. (I) RT-PCR of mRNA isolated from wild-type and Uba1H42 mutant larvae revealing an aberrantly spliced product. This results in an in frame deletion from amino acid residue 162 to 255 of Uba1H42 (see G). Primers were directed against the N-terminal third of Uba1.
To molecularly identify the gene, we meiotically mapped the mutant phenotype to 46A1 on the polytene chromosome. One of the genes mapping to this region is Uba1, encoding an E1 ubiquitin-activating enzyme. Two existing Uba1 alleles, Uba1s3484 and Uba103405, failed to complement the lethality of D6, H33 and H42. Uba1s3484 and Uba103405 are caused by P element insertions (www.flybase.org). Precise excision of Uba1s3484, which carries the P element insertion in the first intron (Watts et al., 2003), restores viability of Uba1s3484 itself, and viability in trans to D6, H33 and H42. This analysis demonstrates that the lethality of Uba1s3484 is indeed caused by the P element insertion in Uba1, and that the new mutants are allelic to Uba1. Sequencing of the Uba1 gene in these mutants revealed that Pro884 is changed to Leu in D6 and H33. Pro884 is a highly conserved residue in all E1 ubiquitin-activating enzymes ranging from yeast to humans (Fig. 1H). As determined by RT-PCR (Fig. 1I), the third allele, H42, contains a new splicing donor site, which results in inappropriate splicing causing an internal in-frame deletion of residues 162–255 (Fig. 1G). Finally, for clonal analysis, we recombined the P allele Uba1s3484 onto the FRT42D chromosome and obtained similar results to the analysis of the new alleles (see below; data not shown). Taken together, these results indicate that the suppressors of GMR-hid, D6, H33 and H42 encode new alleles of Uba1. We refer to these mutants as Uba1D6, Uba1H33 and Uba1H42.
We have recently shown that mutants obtained in our screens can either suppress GMR-hid in an autonomous or non-autonomous manner (Herz et al., 2006; Srivastava et al., 2007). Thus, we tested whether the new Uba1 alleles suppress GMR-hid in an autonomous or non-autonomous manner. A GMR-hid transgene that does not carry the white+ minigene (GMR-hid[w−]; Fig. 1D) allows determination of the genetic identity of the rescued eye tissue based on red/white pigment selection in a clonal analysis. This analysis showed that the rescued eye tissue in GMR-hid[w−]; Uba1 mosaics is genetically Uba1−, which is marked by the absence of red eye pigment, i.e. is phenotypically white− (Fig. 1E,F). Thus, the moderately suppressed GMR-hid eye phenotype is largely due to Uba1 mutant tissue, indicating that the suppression of GMR-hid-induced apoptosis occurs in an autonomous manner.
Strong Uba1 alleles cause non-autonomous overgrowth
To further characterize the Uba1 mutant phenotype, we performed a clonal analysis using ey-FLP without GMR-hid. When the flies were incubated at 25°C, ey-FLP-induced Uba1 mutant clones survived well and occasionally occupied an even larger area than the wild-type twin-spot (Fig. 2A–C). However, during the course of this work, we noticed that Uba1H33 and Uba1D6 are temperature-sensitive (ts) alleles, whereas the third allele, Uba1H42, is not. At 29°C, ey-FLP-induced clones of Uba1H33 (Fig. 2E) and Uba1D6 (not shown) could not be recovered in adult eyes whereas ey-FLP-induced clones of the non-ts allele Uba1H42 survived well (Fig. 2F). This suggests that Uba1D6 and Uba1H33 mutant clones are cell lethal at 29°C. As will be shown below, Uba1D6 and Uba1H33 clones undergo apoptosis at 29°C. Surprisingly, however, despite the absence of Uba1H33 and Uba1D6 clones, the eyes were often larger than normal and contained folds, indicative of overgrowth (Fig. 2D,E). The overgrowth phenotype also affected the size of the heads of Uba1D6 and Uba1H33 mosaics. Strikingly, we recovered several Uba1H33 and Uba1D6 mosaic animals (Fig. 2G) in which one half of the head did not contain Uba1 clones (as judged by the absence of twin-spots) and this half was normal in size, thus serving as an internal control (see right half of the head in Fig. 2G), whereas the other side of the head contained twin-spots and was overgrown (left half in Fig. 2G). Third instar larval eye-antennal imaginal discs of Uba1D6 and Uba1H33 mosaics reared at 29°C were overgrown compared to similarly staged wildtype controls (Fig. 2H,I). Interestingly, in imaginal discs clones with strong Uba1 alleles are present. However, although the entire disc was overgrown, Uba1H33 clones are relatively small compared with controls (Fig. 2H,I) and thus do not contribute to the overgrowth. To verify that the overgrowth phenotype is caused by mutations in Uba1 itself, and not by an unrelated ts mutation, we tested the independently obtained Uba1s3484 allele. In mosaic animals, this allele gives rise to a similar overgrowth phenotype at both 25°C and 29°C, i.e. in a temperature-independent manner (Fig. 2D). Uba1s3484 mosaic eye discs are also overgrown in a temperature-independent manner (data not shown). Precise excision of the P element in Uba1s3484 reverts the overgrowth phenotype (data not shown), suggesting that this phenotype is caused by genetic inactivation of the Uba1 locus.
Fig. 2. Strong Uba1 alleles cause non-autonomous overgrowth.
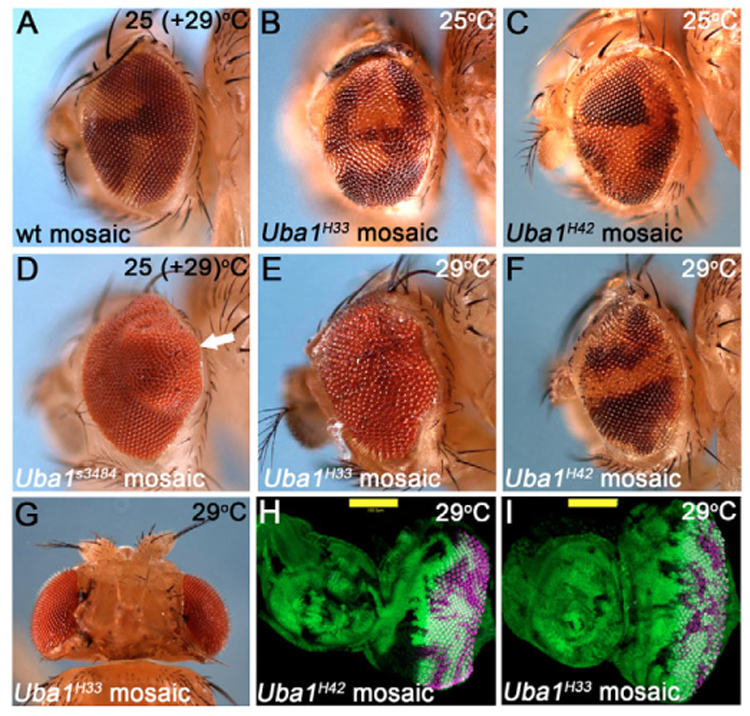
(A) Control mosaics: ey-FLP-induced clones in wild-type background. This phenotype is identical at 25°C and 29°C (shown is 25°C). Genotype: ey-FLP; FRT42D+/FRT42D P[w+]. (B,C) Clones of weak Uba1 mutants (phenotypically white−) develop normally. Genotype: ey-FLP; FRT42D Uba1H33 and Uba1H42/FRT42D P[w+], respectively. (D) Eye mosaics of the strong Uba1s3484 allele are overgrown with bulges (arrow). Genotype: ey-FLP;FRT42D Uba1s3484/FRT42D P[w+]. This phenotype was obtained at 25°C, and is similar at 29°C. (E) Eye mosaic of the ts allele Uba1H33 at 29°C. Uba1H33 clones, which would be phenotypically white−, are not detectable. The eyes appear rough, irregular and are often enlarged compared to wild-type eyes. Identical results were obtained for Uba1D6. Genotype as in B. (F) Eye mosaic of the weak Uba1H42 allele at 29°C. This allele does not behave as a ts allele. Phenotype and genotype are identical to those at 25°C (C). (G) Mosaic head of an Uba1D6 animal raised at 29°C. The left half of the Uba1H33 mosaic head is larger than the right half, suggesting that the overgrowth phenotype depends on the number and location of the Uba1 clones. Genotype as in B. (H,I) Eye-antennal imaginal discs of wild-type (H) and Uba1D6 (I) mosaic animals raised at 29°C. These discs were labeled with Elav antibody which labels photoreceptor neurons to identify discs of comparable age. Photoreceptor differentiation does occur in strong Uba1 clones, but is weakly delayed (see also Fig. S1 in the supplementary material). Genotype as in A and B, respectively. Size bar: 100 µm.
In summary, because the mutant clones of the ts alleles Uba1D6 and Uba1H33 do not survive to adulthood at 29°C, yet Uba1D6 and Uba1H33 mosaics display overgrowth phenotypes, we conclude that these Uba1 mutants cause tissue overgrowth in a non-autonomous manner. A similar conclusion can be drawn for Uba1s3484. Because Uba1s3484 and Uba1H42 are non-ts alleles, the temperature-sensitive nature of Uba1D6 and Uba1H33 is not an intrinsic feature of the process controlled by the Uba1 locus. Instead, it is likely that the ts nature of Uba1D6 and Uba1H33 reflects a transition from weak to strong loss-of-function alleles, since it is common for ts alleles to lose activity at the non-permissive temperature. This will also be confirmed in the next section. Therefore, at low temperature (<25°C) these alleles and the temperature-insensitive allele Uba1H42 behave as weak loss-of-function alleles, whereas Uba1s3484 and the ts alleles Uba1D6 and Uba1H33 at >29°C are strong loss-of-function alleles. It is interesting to note that the ts alleles Uba1D6 and Uba1H33 carry the same point mutation, causing a mutation of Pro884 to Leu (Fig. 1G,H), although both alleles were obtained in independent mutagenesis events. A Uba1 allele with exactly the same mutation and the same genetic behavior has recently been described by others (Pfleger et al., 2007). Because Uba1D6 and Uba1H33 behave genetically identically, we refer to them from now on as Uba1D6/H33.
Strong Uba1 alleles impair general ubiquitin conjugation
E1 ubiquitin-activating enzymes catalyze the first step in the ubiquitination cycle, the ATP-dependent formation of a thioester bond between the C-terminal glycine residue of ubiquitin and the active site cysteine of E1 (Haas and Siepmann, 1997; Pickart, 2001). Thus, E1 enzymes are required to provide most, if not all, activated ubiquitin for conjugation of target proteins. Uba1 encodes the only E1 ubiquitin-activating enzyme in Drosophila (Watts et al., 2003). Therefore, genetic inactivation of Uba1 should result in reduction of general ubiquitination.
To test this prediction, we analyzed the Uba1 mutants for loss of ubiquitin conjugation using the FK2 antibody which specifically recognizes mono- and poly-ubiquitinated proteins, but not unconjugated ubiquitin (Fujimuro et al., 1994; Fujimuro et al., 1997). We tested this prediction first in larval eye imaginal discs of mosaics of the strong P allele Uba1s3484. A significant decrease in FK2 immunolabeling was detected in Uba1s3484 mutant clones (Fig. 3A). However, due to the apoptotic phenotype of strong Uba1 alleles (see below), clones were small and difficult to recover. The ts alleles Uba1D6/H33 provide a convenient alternative to obtain large clones for analysis. After induction of Uba1D6/H33 clones, these mosaics were incubated at 25°C. The animals were shifted to 29°C 12 hours before dissection and fixation of the discs. When Uba1D6/H33 mosaics were treated in this way, larger clones were obtained. FK2 labeling was significantly reduced in Uba1D6/H33 clones incubated at 29°C (Fig. 3B), suggesting that mono- and poly-ubiquitination was affected in strong Uba1 allele clones. Weak alleles also reduced FK2 immunoreactivity; however, this reduction was much more subtle compared to the strong alleles (Fig. 3C).
Fig. 3. Uba1 mutant clones affect ubiquitin conjugation.
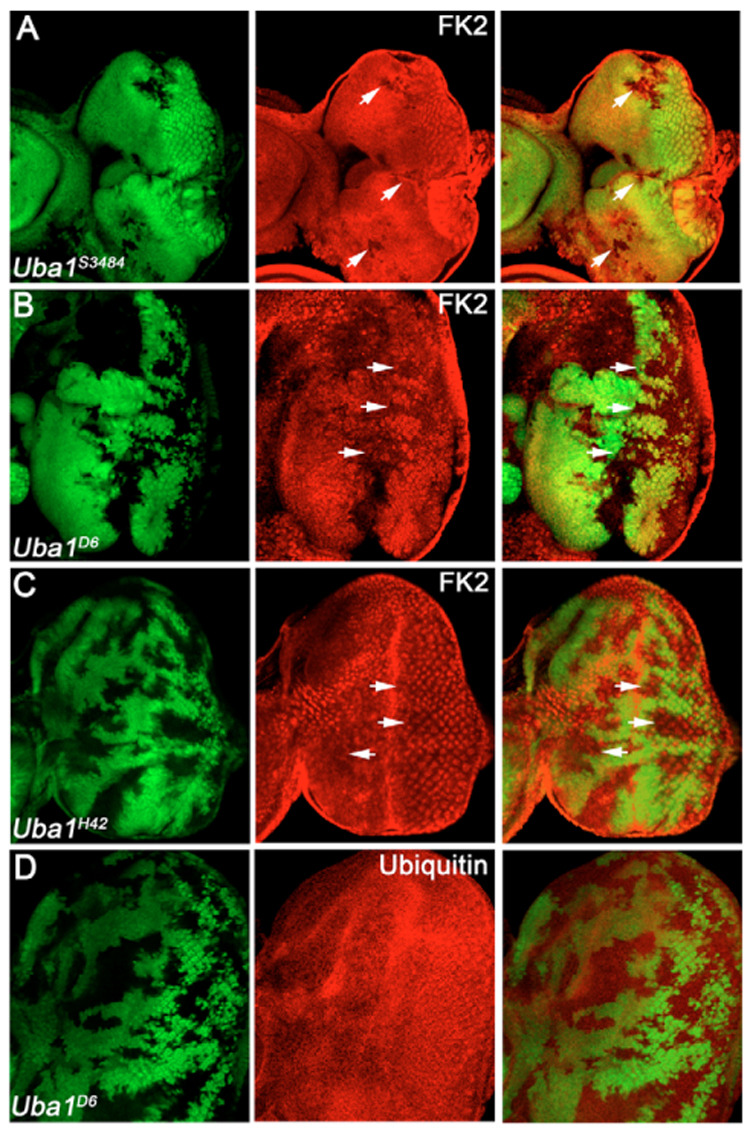
(A–D) The left panels indicate the positions of the Uba1 clones by absence of GFP, the middle panels show the experiment and the right panels are the merged images of left and middle panels. Genotype in all panels: ey-FLP; FRT42D Uba1/FRT42D P[ubi-GFP]. The allele is indicated in the panels. (A–C) Uba1 mutant discs were labeled with the FK2 antibody which specifically recognizes mono- and poly-ubiquitinated conjugates, but not unconjugated ubiquitin (Fujimuro et al., 1994). The strong alleles Uba1s3484 (A) and Uba1D6 (B, incubated at 29°C) significantly reduce FK2 immunoreactivity (see arrows). Clones of the weak allele Uba1H42 (C) mildly reduce FK2 immunoreactivity (arrows). (D) Uba1D6 mosaics incubated at 29°C. The overall protein levels of ubiquitin are unchanged in Uba1 mutant clones.
We considered that the absence of FK2 labeling in strong Uba1 clones may be due to an overall reduction of the protein levels of ubiquitin. Thus, as a control we tested the total ubiquitin protein levels. An antibody recognizing both conjugated and unconjugated ubiquitin did not reveal significant alterations of ubiquitin protein in Uba1D6/H33 clones compared with wild-type tissue at both 25°C and 29°C (Fig. 3D) suggesting that the loss of Uba1 has no effect on total protein levels of ubiquitin. Thus, the lack of FK2 immunolabeling in Uba1 clones is due to reduction in ubiquitin conjugation, and not to reduced levels of ubiquitin.
Taken together, these data illustrate that Uba1 is required to provide activated ubiquitin for protein conjugation, which is essential for ubiquitin-mediated protein degradation and other ubiquitin-dependent processes. With this knowledge in mind, we analyzed the observed apoptotic and growth phenotypes in more detail.
Weak Uba1 alleles protect from cell death, whereas clones of strong Uba1 alleles are apoptotic
We have identified weak Uba1 alleles as suppressors of GMR-hid-induced apoptosis (Fig. 1). Therefore, we analyzed the cause of the suppression of GMR-hid by Uba1. GMR-hid eye discs are characterized by two zones of apoptotic cell death, as shown by cleaved caspase 3 (Cas3*) labeling (Fig. 4A) (Srivastava et al., 2007). Consistent with the expectation, Cas3* activity induced by GMR-hid is significantly reduced in Uba1D6/H33 mutant clones in GMR-hid eye imaginal discs at 25°C (white arrows in Fig. 4B,B′). However, we also noticed that suppression of GMR-hid-induced cell death does not occur in all Uba1 clones. In some areas (red arrow in Fig. 4B,B′), Cas3*-positive cell death still occurs in Uba1 mutant clones. This region-specific behavior was consistently observed, even at lower temperature (22°C). It appears to be located at the midline, although we have not been able to pinpoint what distinguishes this region from others. Interestingly, this region-specific behavior suggests that there may be a form of hid-induced apoptosis that is independent of ubiquitin conjugation. This selective behavior may also explain why the suppression of GMR-hid by Uba1 is only moderately strong, and not as complete as observed for mutants of other components of the cell death pathway (Srivastava et al., 2007; Xu et al., 2005). Nevertheless, this analysis demonstrates that Uba1 directly or indirectly controls caspase activation.
Fig. 4. Weak Uba1 alleles suppress apoptosis.
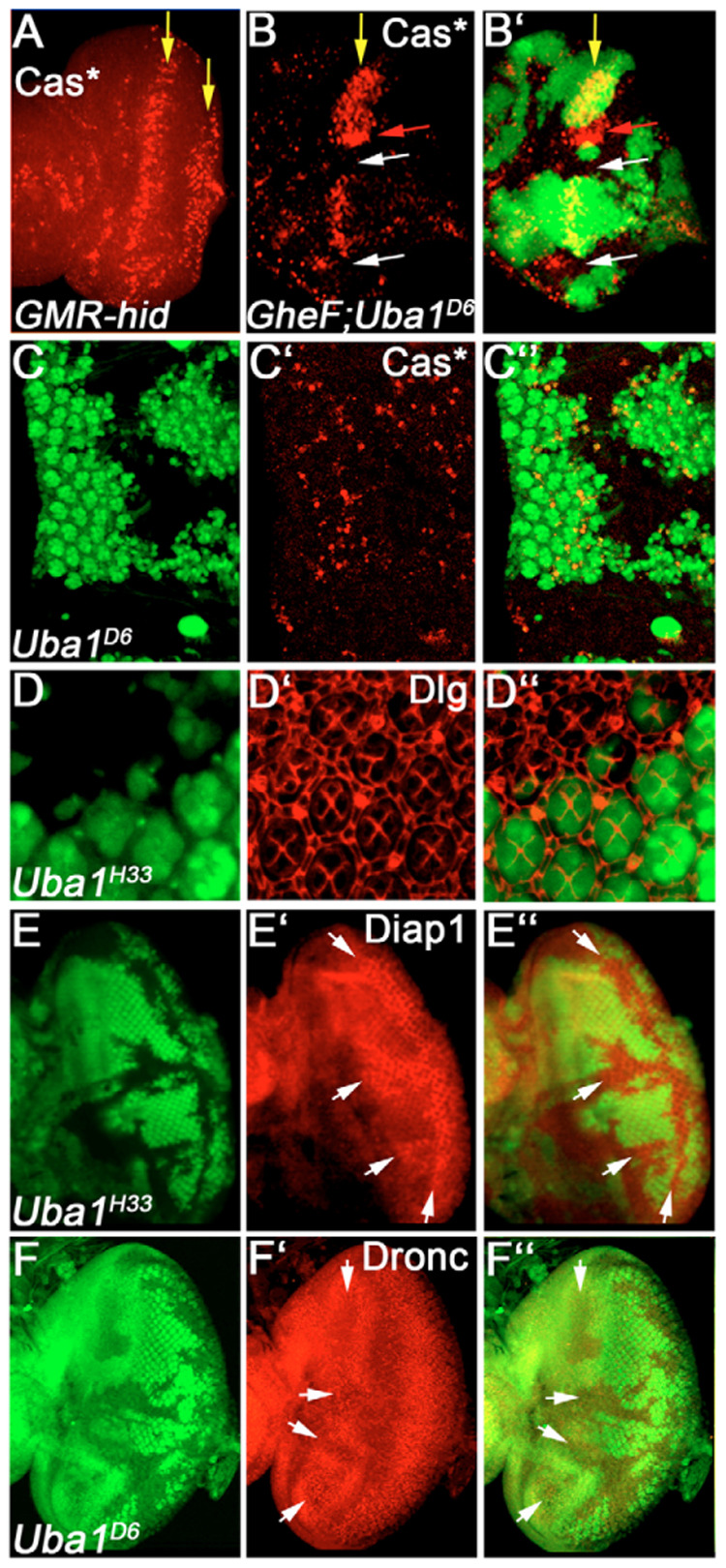
(A) GMR-hid eye disc labeled with cleaved Caspase-3 (Cas3*) antibodies. Yellow arrows indicate two zones of dying cells in GMR-hid. (B,B′) Suppression of Cas3*-positive cell death in GMR-hid discs. The yellow arrow indicates the position of the first zone of Cas3*-positive cell death in GMR-hid. Uba1D6 clones suppress Cas3*-positive cell death (white arrows). However, this suppression is not uniform and occurs in a region-specific manner. Clones in an area of the disc marked by the red arrow are still apoptotic. Genotype: GMR-hid ey-FLP; FRT42D Uba1D6/FRT42D P[ubi-GFP]; (C–C″) Uba1D6 clones suppress most normal developmental cell death in 28 hours APF pupal discs. Genotype as in B. (D–D″) Uba1H33 clones contain additional IOCs in 42 hours APF pupal discs. Genotype: ey-FLP; FRT42D Uba1H33/FRT42D P[ubi-GFP]. (E–E″) Diap1 protein levels are increased in Uba1H33 mutant clones (arrows). Genotype as in D. (F–F″) Dronc protein levels are decreased in Uba1D6 mutant clones (arrows). Genotype as in B.
We also analyzed whether Uba1 function is needed for normal physiologically occurring cell death. During normal eye development between 26 and 30 hours after puparium formation (APF), cell death is required to remove all cells that have not adopted a cell fate (Cagan and Ready, 1989; Cordero et al., 2004; Wolff and Ready, 1991). Uba1D6/H33 mutant clones raised at 25°C contain significantly less Cas3*-positive cells compared with wild-type tissue (Fig. 4C–C″), consistent with a genetic requirement of Uba1 for normal cell death. This lack of cell death results in increase in the number of interommatidial cells (IOCs). Wild-type ommatidia contain exactly nine IOCs. However, Uba1 clones contain up to 16 IOCs (Fig. 4D–D″). Thus, consistent with the Uba1 mutants being suppressors of GMR-hid, Uba1 is generally required for the control of normal cell death during Drosophila development.
Because activation of ubiquitin is the only known function of Uba1, these observations suggest that Uba1 is required for a ubiquitination-dependent event during cell death. Two components of the cell death pathway, the caspase inhibitor Diap1 [also known as Thread (Th) – FlyBase] and the initiator caspase Dronc [also known as Nedd2-like caspase (Nc) – FlyBase], are subject to ubiquitin-mediated degradation (Chai et al., 2003; Herman-Bachinsky et al., 2007; Holley et al., 2002; Ryoo et al., 2002; Wilson et al., 2002; Yoo et al., 2002). Whereas Dronc degradation occurs in surviving cells, Diap1 degradation is triggered in response to apoptotic stimuli. The degradation of both proteins depends on the E3-ligase activity of Diap1 (Herman-Bachinsky et al., 2007; Holley et al., 2002; Ryoo et al., 2002; Wilson et al., 2002; Yoo et al., 2002). Thus, we analyzed the protein levels of Diap1 and Dronc in Uba1 clones. Interestingly, whereas Diap1 protein levels are increased in Uba1 mutant clones (Fig. 4E–E″), Dronc protein levels are reduced (Fig. 4F–F″), consistent with previous reports showing that Diap1 controls the protein levels of Dronc (Chai et al., 2003; Ryoo et al., 2004; Wilson et al., 2002). Because increased levels of Diap1 do protect cells more efficiently from cell death (Hay et al., 1995), this observation suggests that the anti-apoptotic phenotype of Uba1 clones may be mediated by increased levels of Diap1. The fact that Dronc levels are reduced implies that ubiquitin conjugation still occurs in Uba1D6/H33 mutant clones at 25°C, providing further evidence that the Uba1 alleles isolated in our study are weak loss-of-function alleles.
We have shown above that higher temperature converts the alleles Uba1D6 and Uba1H33 to stronger loss-of-function alleles, which results in loss of Uba1 mutant clones and in non-autonomous phenotypes (see Fig. 2). Uba1D6/H33 clones in eye imaginal discs incubated at 29°C labeled with the Cas3* antibody are highly apoptotic (Fig. 5A) providing an explanation for the loss of Uba1 mutant clones in adult eyes. In control discs carrying clones of the non-ts Uba1H42 allele, Cas3* activity is low at 29°C (Fig. 5B). However, despite the fact that Uba1D6/H33 clones are highly apoptotic, Diap1 protein levels are still increased (Fig. 5C), yet they do not appear to be able to protect Uba1D6 mutant clones from apoptosis at 29°C. Interestingly, in contrast to incubation at 25°C, we found that the protein levels of Dronc are increased in Uba1D6/H33 clones at 29°C (Fig. 5D). Clones of the non-ts allele Uba1H42 do not contain increased levels of Dronc at 29°C (data not shown) suggesting that the increase of Dronc protein levels is caused by the transition of Uba1D6/H33 to stronger alleles at 29°C, and thus by loss of ubiquitin conjugation. Therefore, owing to the lack of activated ubiquitin, Diap1 can no longer efficiently act as an E3 ubiquitin-ligase for Diap1 and Dronc degradation. In conclusion, our study demonstrates that different proteins require different ubiquitination activity for efficient degradation. Diap1 requires much higher ubiquitination activity for degradation than Dronc (see also Discussion).
Fig. 5. The apoptotic phenotype of strong Uba1 alleles.
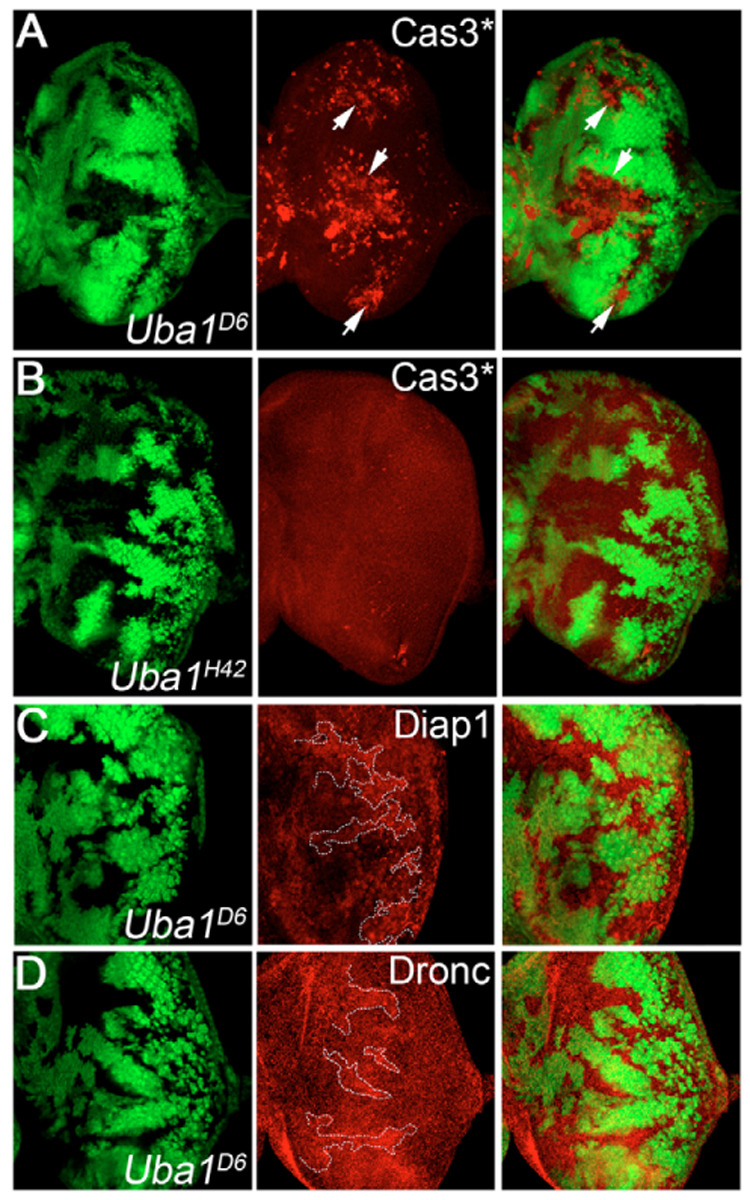
All discs were incubated at 29°C. (A,C,D) ey-FLP; FRT Uba1D6/FRT P[ubi-GFP]; (B) ey-FLP; FRT Uba1H42/FRT P[ubi-GFP]. (A) Uba1D6 clones are highly Cas3*-apoptotic (see arrows). (B) Uba1H42 clones lack apoptotic labeling. (C) Uba1D6 clones (outlined by dashed line) contain increased levels of Diap1 protein. (D) Uba1D6 clones (outlined by dashed line) contain increased levels of Dronc protein.
Strong Uba1 alleles cause autonomous cell cycle arrest and non-autonomous cell proliferation
Ubiquitin-induced degradation of cyclins is essential for propagation through the cell cycle (reviewed by Pines, 2006). Thus, we analyzed the effect of loss of ubiquitin conjugation on cell proliferation in strong Uba1 mutant clones. In Uba1D6/H33 clones at 29°C, cell proliferation, as determined by BrdU labeling, is strongly reduced (Fig. 6A). This cell cycle arrest phenotype correlates with increased Cyclin A and Cyclin B levels in Uba1D6/H33 mutant clones. This is particularly striking in the morphogenetic furrow (MF) in the eye disc, which does not contain cyclin proteins in wild-type tissue. However, Uba1D6/H33 clones spanning the MF fail to downregulate Cyclin A (Fig. 6C) and Cyclin B (not shown) at 29°C. The weak allele Uba1H42 and the ts alleles at 25°C do not affect cyclin levels in the MF, but do contain slightly increased cyclin levels in areas of the disc where cyclins are normally detected (Fig. 6D, arrow). However, these increased cyclin levels do not appear to significantly affect the cell cycle, as no obvious alterations in cell proliferation in Uba1H42 clones are detected (Fig. 6B).
Fig. 6. Cell cycle arrest and non-autonomous proliferation in Uba1 mosaics.
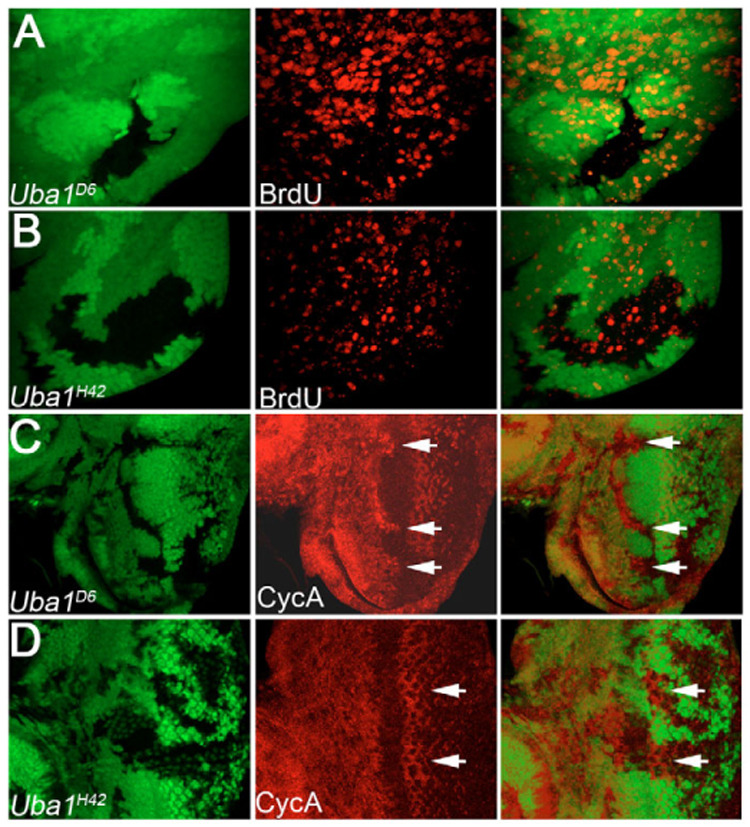
(A,B) Third instar mosaic Uba1D6 (A) and Uba1H42 (B) wing discs incubated at 29°C. Note lack of BrdU labeling in Uba1D6 mutant clones, and increased BrdU labeling in wild-type tissue surrounding Uba1D6 clones. Proliferation in Uba1H42 mosaics is homogenous within and outside the clones (B). (C,D) Cyclin A labeling of eye discs from third instar larval eye discs containing Uba1D6 (C) and Uba1H42 (D) mutant clones. Cyclin A levels are increased in Uba1D6 clones spanning the morphogenetic furrow (arrows in C). Clones of the weak allele Uba1H42 contain slightly increased Cyclin A levels (D, arrows). However, this increase does not affect cell proliferation (B).
Interestingly, cell proliferation in Uba1D6/H33 mosaics incubated at 29°C as demonstrated by BrdU incorporation is significantly increased in tissue adjacent to the mutant clones (Fig. 6A). This was also observed for the P allele Uba1s3484 (data not shown). Control discs (mosaic for the weak non-ts allele Uba1H42) show a homogenous distribution of proliferating cells both within and outside the clones (Fig. 6B). Thus, strong Uba1 clones appear to be the origin of increased proliferation in adjacent tissue, and the overgrowth phenotype in Uba1D6/H33 mosaics (Fig. 2) can most likely be explained by emission of signaling molecules from the mutant cells initiating non-autonomous proliferation in the adjacent wild-type tissue (see below).
In summary, consistent with previous reports, lack of ubiquitin conjugation causes cell cycle arrest, presumably through failure to proteolytically degrade cyclin proteins (Pines, 2006). Surprisingly, however, this study also reveals an unanticipated function of Uba1, the negative control of the cell cycle in neighboring cells.
Notch activity causes non-autonomous overgrowth in mosaics of strong Uba1 alleles
The non-autonomous overgrowth of mosaics of strong Uba1 alleles came as a surprise. However, we and others have recently reported a similar non-autonomous overgrowth phenotype caused by mutations in the ESCRT (endosomal sorting complex required for transport) components vps23 [also known as erupted (ept) and tsg101 – FlyBase] and vps25 (Herz et al., 2006; Moberg et al., 2005; Thompson et al., 2005; Vaccari and Bilder, 2005). ESCRT components are required at the early endosome for the sorting and targeting of activated cell surface receptors for lysosomal degradation (Babst, 2005; Gruenberg and Stenmark, 2004; Katzmann et al., 2002). Similar to Uba1 clones, vps23 and vps25 clones are highly apoptotic, but stimulate cell proliferation in neighboring tissue and non-autonomous overgrowth (Herz et al., 2006; Moberg et al., 2005; Thompson et al., 2005; Vaccari and Bilder, 2005). Interestingly, the signal for endocytosis and endosomal protein sorting is provided by ubiquitination of activated cell surface receptors (Haglund and Dikic, 2005); Gruenberg and Stenmark, 2004). Thus, in Uba1 mutants, endocytosis and endosomal protein sorting may be affected, too. In the case of vps23 and vps25, signaling by the Notch receptor is inappropriately increased, leading to secretion of the cytokine Unpaired which stimulates proliferation of neighboring cells by activation of the Jak/STAT pathway (Herz et al., 2006; Moberg et al., 2005; Thompson et al., 2005; Vaccari and Bilder, 2005). Therefore, we tested whether a similar mechanism causes non-autonomous proliferation and overgrowth in Uba1 mosaics.
In accord, we observe increased protein levels of Notch in Uba1D6/H33 mutant clones raised at 29°C (Fig. 7A). Using the Notch reporter E(spl)m8 2.61-lacZ, increased Notch activity was found in Uba1 mutant clones (Fig. 7B). Moreover, increased STAT activity, as judged by labeling with an antibody recognizing phosphorylated, i.e. activated STAT protein (pSTAT), is increased in a band of cells immediately adjacent to Uba1 clones, i.e. in a non-autonomous manner (Fig. 7D). Thus, similar to vps23 and vps25, Uba1 clones contain increased Notch activity resulting in activation of the Jak/STAT pathway in neighboring cells. Surprisingly, we also detected Notch activity in Uba1D6 clones raised at 25°C (Fig. 7C), although at this temperature non-autonomous activation of Jak/STAT signaling and overgrowth are not observed (data not shown; see Discussion).
Fig. 7. Notch signaling induces non-autonomous overgrowth in Uba1 mosaics.
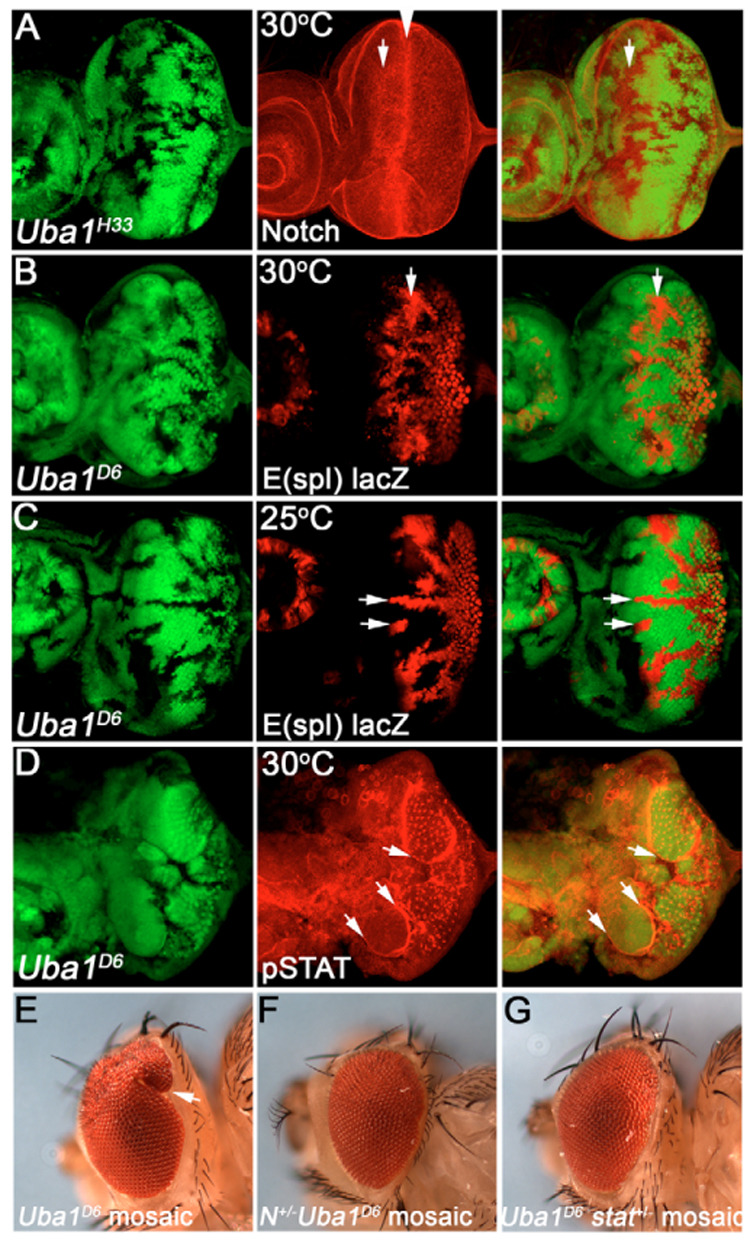
All discs were incubated at 30°C, except C which was incubated at 25°C. (A) Uba1H33 mosaic eye discs labeled with Notch antibody C17.9C6 which was raised against the intracellular domain of Notch. Arrows indicate increase of Notch protein in Uba1 clones. This is most prominent immediately anterior to the morphogenetic furrow (arrowhead). Genotype: ey-FLP; FRT Uba1D6/FRT P[ubi-GFP]. (B,C) Notch activity as visualized by the Notch reporter E(spl)m8 2.61-lacZ is increased in Uba1D6 clones at 30°C (B) and 25°C (C). Genotype: ey-FLP; E(spl)m8 2.61-lacZ FRT ubaD6/FRT P[ubi-GFP]. (D) pSTAT labeling is increased in a non-autonomous manner in wild-type tissue immediately adjacent to Uba1 clones (arrow). Genotype as in A. (E) Eyes of Uba1D6 mosaic animals are overgrown with bulges (arrow). Genotype as in A. (F,G) Heterozygosity of Notch (F) and stat92 (G) dominantly suppresses the Uba1D6 overgrowth phenotype. Genotype: Notch264-39/ey-FLP; FRT42 Uba1D6/FRT42 P[ubi-GFP] (F) and ey-FLP; FRT42 Uba1D6/FRT42 P[ubi-GFP]; stat9285c9 (G).
To test for a genetic requirement of Notch and Jak/STAT for the non-autonomous overgrowth phenotype, we analyzed Uba1 mosaics in a heterozygous Notch or stat92 mutant background. As a marker in this assay, we used the bulging phenotype of Uba1 mosaic eyes (Fig. 2D, Fig. 7E). Heterozygosity for Notch or stat92 dominantly suppressed the overgrowth phenotype of strong Uba1 mosaics (Uba1D6/H33 at 29°C; Fig. 7F,G). Together, these data strongly suggest that the overgrowth phenotype of strong Uba1 mosaics is caused by inappropriate Notch signaling, which triggers activation of the Jak/STAT signaling pathway in neighboring cells (see Discussion).
DISCUSSION
Before this study, E1 ubiquitin-activating enzymes have only been characterized in yeast and in mammalian cell lines (Ciechanover et al., 1984; Ciechanover et al., 1985; Finley et al., 1984; Ghaboosi and Deshaies, 2007; Kulka et al., 1988; McGrath et al., 1991; Salvat et al., 2000; Swanson and Hochstrasser, 2000). Here, we analyzed the only E1 gene in Drosophila, Uba1, and uncovered two unexpected mutant phenotypes. First, while partial loss of ubiquitin conjugation caused by weak Uba1 alleles inhibits cell death, strong Uba1 alleles are highly apoptotic. Second, while strong Uba1 clones are cell cycle arrested, they do stimulate neighboring wild-type cells to undergo cell proliferation and induce non-autonomous overgrowth. We also found that photoreceptor differentiation occurs both in clones of weak and strong Uba1 alleles. However, the onset of photoreceptor differentiation is slightly delayed in clones of strong alleles (see Fig. S1 in the supplementary material). Similar observations have been made in a different study (Pfleger et al., 2007).
Phenotypes affecting cell death
We identified Uba1 alleles as suppressors of the apoptotic phenotype caused by GMR-hid, and showed that Uba1 is also required for normal developmental cell death. This requirement is probably mediated through the control of Diap1 protein levels which in turn mediates ubiquitination of the caspase Dronc (Chai et al., 2003; Wilson et al., 2002). However, the GMR-hid-suppressing Uba1 alleles are weak. They affect overall ubiquitin conjugation only mildly (Fig. 3C) suggesting that ubiquitin-mediated degradation can still occur in an almost normal manner. In accord, the increased protein levels of Diap1 are even able to reduce the protein levels of Dronc in clones expressing weak Uba1 alleles.
It is interesting to note that whereas Diap1 protein levels are increased in clones expressing weak Uba1 alleles (Fig. 4E), other proteins such as Ci, Arm (not shown) or Dronc (Fig. 4F) are normal in abundance or even reduced, respectively. This suggests that some proteins such as Diap1 respond in a very sensitive manner to partial loss of activated ubiquitin, whereas other proteins do not. Because Diap1 has a fairly short half-life (~30–40 minutes) compared to Dronc (~3 hours) (Wilson et al., 2002; Yoo et al., 2002), the requirement of a fully functional ubiquitination machinery may be much stricter for Diap1, providing an explanation for why Diap1 responds so sensitively to a small reduction of activated ubiquitin for protein conjugation.
Alternatively, it is also possible that the Uba1 alleles isolated in this study specifically affect the interaction with UbcD1, the E2-conjugating-enzyme which targets Diap1 for ubiquitin-mediated degradation (Ryoo et al., 2002; Yoo, 2005). Thus, the interaction with other E2 enzymes may be normal, so that ubiquitin conjugation and degradation of other proteins may be normal. We have not tested which of these two possibilities applies.
Strong Uba1 alleles, which significantly reduce ubiquitin conjugation, affect the levels of all proteins we have analyzed. For example, although Diap1 levels are increased with strong Uba1 alleles, Dronc is no longer efficiently degraded. Instead, Dronc protein accumulates, suggesting that activated ubiquitin required for conjugation and degradation is no longer available. However, it is unclear why cells in Uba1 clones die. Dronc needs to be cleaved for activation, and Diap1 can directly bind to and inhibit caspases without degradation, at least in vitro (Chai et al., 2003; Meier et al., 2000; Zachariou et al., 2003). Thus, the increased Diap1 levels should still be able to inhibit the accumulated Dronc protein. Mutants in ark (also known as D-Apaf-1, hac-1 and dark), which encodes an adaptor protein required for Dronc activation (Kanuka et al., 1999; Mendes et al., 2006; Rodriguez et al., 1999; Srivastava et al., 2007; Zhou et al., 1999), block cell death in Uba1 (data not shown), suggesting that cell death in Uba1 mutants is indeed mediated via Dronc. Thus, simple binding of Diap1 to Dronc may not be sufficient to completely inhibit Dronc activity. Instead, ubiquitination may be required for full inactivation of Dronc.
Phenotypes affecting cell proliferation
Consistent with the expectation, loss of ubiquitin conjugation in strong Uba1 alleles causes cell cycle arrest. This correlates with increased protein levels of Cyclins A and B, the ubiquitin-dependent degradation of which is required for cell cycle progression (reviewed by Pines, 2006).
However, the non-autonomous overgrowth phenotype was unexpected. Strong Uba1 clones appear to be able to secrete a growth factor that promotes cell proliferation and overgrowth in adjacent wild-type tissue. In this capacity, Uba1 qualifies as a tumor suppressor gene.
It is interesting to note that the phenotypes observed for Uba1 are very similar to those of vps23 and vps25 (Herz et al., 2006; Moberg et al., 2005; Thompson et al., 2005; Vaccari and Bilder, 2005). In both cases, Notch signaling is inappropriately increased. Notch triggers Jak/STAT signaling in neighboring wild-type tissue, presumably through secretion of Unpaired, which encodes an Interleukin-like factor (Harrison et al., 1998) and acts as the ligand of the receptor of the Jak/STAT signaling pathway (Chao et al., 2004; Reynolds-Kenneally and Mlodzik, 2005; Tsai and Sun, 2004). However, the ultimate cause of Notch activation may be different. In the case of vps23 and vps25, Notch is internalized via endocytosis, however, endosomal protein sorting is impaired, thus turnover of Notch is affected. In the case of Uba1, it is not clear whether the lack of ubiquitination affects endocytosis of membrane-localized Notch or ubiquitin-mediated degradation of intracellular Notch in the nucleus. Failure of either may cause inappropriate signaling. The accumulation of Notch in Uba1 clones is not as striking as in vps25 clones, making it difficult to identify the subcellular localization of accumulated Notch.
Another interesting observation is the fact that we do observe increased Notch activity in clones of Uba1D6 at 25°C, at which temperature non-autonomous cell proliferation is not observed. Consistently, we do not detect increased STAT signaling under these conditions. If Notch signaling is increased at 25°C, why does this not induce non-autonomous proliferation? One potential reason may lie in the fact that Uba1 clones at 25°C are protected from apoptosis, whereas at 29°C they are apoptotic. Thus, an apoptotic environment may be necessary for the induction of non-autonomous proliferation. A similar phenomenon, referred to as apoptosis-induced compensatory proliferation, has recently been reported (Huh et al., 2004; Kondo et al., 2006; Perez-Garijo et al., 2004; Ryoo et al., 2004; Wells et al., 2006). In these studies, apoptotic cells trigger the secretion of Dpp and Wg which promote proliferation in neighboring cells. An involvement of Notch was not reported. However, in the aforementioned studies, apoptosis-induced compensatory proliferation is only detectable if cell death is simultaneously blocked. In the case of Uba1, vps23 and vps25, overgrowth occurs without inhibition of apoptosis (Herz et al., 2006; Moberg et al., 2005; Thompson et al., 2005; Vaccari and Bilder, 2005). Therefore, there may be different forms of compensatory proliferation in response to different apoptotic triggers.
In summary, we have largely focused on the effects of loss of ubiquitin conjugation for apoptosis and cell proliferation. Our analysis demonstrates that the loss of ubiquitin conjugation has significant consequences for the organism, and may implicate Uba1 as a tumor suppressor gene in Drosophila. The Uba1 alleles identified in this study will be of further use to analyze a general requirement of ubiquitination for other cellular processes as well.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/135/1/43/DC1
Supplementary Material
Acknowledgments
We would like to thank Erika Bach, Eric Baehrecke, Barry Dickson, Georg Halder, Pascal Meier, Hyung-Don Ryoo, Hermann Steller, The Bloomington Stock Center, Indiana, and The Developmental Studies Hybridoma Bank (DSHB), Iowa for fly stocks and reagents; Iswar Hariharan for sharing unpublished information; and the MD Anderson DNA Analysis Core Facility for sequencing (supported by Core Grant #CA16672 from the NCI). The project was supported by grant number R01 GM068016 form the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not represent the official views of the NIGMS or the NIH. We gratefully acknowledge support by the Robert A. Welch Foundation (G-1496).
References
- Babst M. A protein's final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Dev. Biol. 1989;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- Cashio P, Lee TV, Bergmann A. Genetic control of programmed cell death in Drosophila melanogaster. Semin. Cell Dev. Biol. 2005;16:225–235. doi: 10.1016/j.semcdb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, Shi Y. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat. Struct. Biol. 2003;10:892–898. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- Chao JL, Tsai YC, Chiu SJ, Sun YH. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development. 2004;131:3839–3847. doi: 10.1242/dev.01258. [DOI] [PubMed] [Google Scholar]
- Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Finley D, Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984;37:57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Finley D, Varshavsky A. Mammalian cell cycle mutant defective in intracellular protein degradation and ubiquitin-protein conjugation. Prog. Clin. Biol. Res. 1985;180:17–31. [PubMed] [Google Scholar]
- Cordero J, Jassim O, Bao S, Cagan R. A role for wingless in an early pupal cell death event that contributes to patterning the Drosophila eye. Mech. Dev. 2004;121:1523–1530. doi: 10.1016/j.mod.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Devoy A, Soane T, Welchman R, Mayer RJ. The ubiquitin-proteasome system and cancer. Essays Biochem. 2005;41:187–203. doi: 10.1042/EB0410187. [DOI] [PubMed] [Google Scholar]
- Fan Y, Soller M, Flister S, Hollmann M, Muller M, Bello B, Egger B, White K, Schafer MA, Reichert H. The egghead gene is required for compartmentalization in Drosophila optic lobe development. Dev. Biol. 2005;287:61–73. doi: 10.1016/j.ydbio.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Finley D, Ciechanover A, Varshavsky A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell. 1984;37:43–55. doi: 10.1016/0092-8674(84)90299-x. [DOI] [PubMed] [Google Scholar]
- Fujimuro M, Sawada H, Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- Fujimuro M, Sawada H, Yokosawa H. Dynamics of ubiquitin conjugation during heat-shock response revealed by using a monoclonal antibody specific to multi-ubiquitin chains. Eur. J. Biochem. 1997;249:427–433. doi: 10.1111/j.1432-1033.1997.00427.x. [DOI] [PubMed] [Google Scholar]
- Ghaboosi N, Deshaies RJ. A conditional yeast E1 mutant blocks the ubiquitin-proteasome pathway and reveals a role for ubiquitin conjugates in targeting Rad23 to the proteasome. Mol. Biol. Cell. 2007;18:1953–1963. doi: 10.1091/mbc.E06-10-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 2004;5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- Haas AL, Siepmann TJ. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Herman-Bachinsky Y, Ryoo HD, Ciechanover A, Gonen H. Regulation of the Drosophila ubiquitin ligase DIAP1 is mediated via several distinct ubiquitin system pathways. Cell Death Differ. 2007;14:861–871. doi: 10.1038/sj.cdd.4402079. [DOI] [PubMed] [Google Scholar]
- Herz HM, Chen Z, Scherr H, Lackey M, Bolduc C, Bergmann A. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133:1871–1880. doi: 10.1242/dev.02356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat. Cell Biol. 2002;4:439–444. doi: 10.1038/ncb798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Joazeiro CA, Bonfoco E, Kamada S, Leverson JD, Hunter T. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J. Biol. Chem. 2000;275:26661–26664. doi: 10.1074/jbc.C000199200. [DOI] [PubMed] [Google Scholar]
- Huh JR, Guo M, Hay BA. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr. Biol. 2004;14:1262–1266. doi: 10.1016/j.cub.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H, Miura M. Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Mol. Cell. 1999;4:757–769. doi: 10.1016/s1097-2765(00)80386-x. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Kondo S, Senoo-Matsuda N, Hiromi Y, Miura M. DRONC coordinates cell death and compensatory proliferation. Mol. Cell. Biol. 2006;26:7258–7268. doi: 10.1128/MCB.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulka RG, Raboy B, Schuster R, Parag HA, Diamond G, Ciechanover A, Marcus M. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J. Biol. Chem. 1988;263:15726–15731. [PubMed] [Google Scholar]
- Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51:283–290. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- McGrath JP, Jentsch S, Varshavsky A. UBA 1, an essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 2000;19:598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes CS, Arama E, Brown S, Scherr H, Srivastava M, Bergmann A, Steller H, Mollereau B. Cytochrome c-d regulates developmental apoptosis in the Drosophila retina. EMBO Rep. 2006;7:933–939. doi: 10.1038/sj.embor.7400773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg KH, Schelble S, Burdick SK, Hariharan IK. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev. Cell. 2005;9:699–710. doi: 10.1016/j.devcel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitinproteasome system. Nat. Rev. Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Perez-Garijo A, Martin FA, Morata G. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development. 2004;131:5591–5598. doi: 10.1242/dev.01432. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Harvey KF, Yan H, Hariharan IK. Mutation of the gene encoding the ubiquitin activating enzyme Uba1 causes tissue overgrowth in Drosophila. Fly. 2007;1:95–105. doi: 10.4161/fly.4285. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Reinstein E, Ciechanover A. Narrative review: protein degradation and human diseases: the ubiquitin connection. Ann. Intern. Med. 2006;145:676–684. doi: 10.7326/0003-4819-145-9-200611070-00010. [DOI] [PubMed] [Google Scholar]
- Reynolds-Kenneally J, Mlodzik M. Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye. Dev. Biol. 2005;285:38–48. doi: 10.1016/j.ydbio.2005.05.038. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat. Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat. Cell Biol. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev. Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Salvat C, Acquaviva C, Scheffner M, Robbins I, Piechaczyk M, Jariel-Encontre I. Molecular characterization of the thermosensitive E1 ubiquitin-activating enzyme cell mutant A31N-ts20. Requirements upon different levels of E1 for the ubiquitination/degradation of the various protein substrates in vivo. Eur. J. Biochem. 2000;267:3712–3722. doi: 10.1046/j.1432-1327.2000.01404.x. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Scherr H, Lackey M, Xu D, Chen Z, Lu J, Bergmann A. ARK, the Apaf-1 related killer in Drosophila, requires diverse domains for its apoptotic activity. Cell Death Differ. 2007;14:92–102. doi: 10.1038/sj.cdd.4401931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc. Natl. Acad. Sci. USA. 2001;98:8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Hochstrasser M. A viable ubiquitin-activating enzyme mutant for evaluating ubiquitin system function in Saccharomyces cerevisiae. FEBS Lett. 2000;477:193–198. doi: 10.1016/s0014-5793(00)01802-0. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, Mathieu J, Sung HH, Loeser E, Rorth P, Cohen SM. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev. Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Sun YH. Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis. 2004;39:141–153. doi: 10.1002/gene.20035. [DOI] [PubMed] [Google Scholar]
- Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev. Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38:871–885. doi: 10.1016/s0896-6273(03)00295-2. [DOI] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Wells BS, Yoshida E, Johnston LA. Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr. Biol. 2006;16:1606–1615. doi: 10.1016/j.cub2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werz C, Lee TV, Lee PL, Lackey M, Bolduc C, Stein DS, Bergmann A. Mis-specified cells die by an active gene-directed process, and inhibition of this death results in cell fate transformation in Drosophila. Development. 2005;132:5343–5352. doi: 10.1242/dev.02150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Steller H, Meier P. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat. Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Cell death in normal and rough eye mutants of Drosophila. Development. 1991;113:825–839. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 2005;132:2125–2134. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Wang Y, Willecke R, Chen Z, Ding T, Bergmann A. The effector caspases drICE and dcp-1 have partially overlapping functions in the apoptotic pathway in Drosophila. Cell Death Differ. 2006;13:1697–1706. doi: 10.1038/sj.cdd.4401920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- Yoo SJ. Grim stimulates Diap1 poly-ubiquitination by binding to UbcD1. Mol. Cells. 2005;20:446–451. [PubMed] [Google Scholar]
- Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, Feldman RM, Clem RJ, Muller HA, Hay BA. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Zachariou A, Tenev T, Goyal L, Agapite J, Steller H, Meier P. IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J. 2003;22:6642–6652. doi: 10.1093/emboj/cdg617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai RG, Hiesinger PR, Koh TW, Verstreken P, Schulze KL, Cao Y, Jafar-Nejad H, Norga KK, Pan H, Bayat V, et al. Mapping Drosophila mutations with molecularly defined P element insertions. Proc. Natl. Acad. Sci. USA. 2003;100:10860–10865. doi: 10.1073/pnas.1832753100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Song Z, Tittel J, Steller H. HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Mol. Cell. 1999;4:745–755. doi: 10.1016/s1097-2765(00)80385-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


