Abstract
The Gag polyproteins of gammaretroviruses contain a conserved p12 domain between MA and CA that plays critical roles in virus assembly, reverse transcription and nuclear integration. Here we show using nuclear magnetic resonance, that p12 is unstructured in a Moloney murine leukemia virus (MMLV) Gag fragment that includes the N-terminal domain of CA (p12-CAN). Furthermore, no long range interactions were observed between the domains, as has been previously predicted. Flexibility appears to be a common feature of Gag “late” domains required for virus release during budding. Residues near the N-terminus of CAN that form a β-hairpin in the mature CA protein are unfolded in p12-CAN, consistent with proposals that hairpin formation helps trigger capsid assembly.
Introduction
All retroviruses encode a polyprotein called Gag that serves as the major structural protein of the virus and is capable of assembling into virus-like particles in the absence of any other viral constituent. Gag proteins contain three major domains: an N-terminal matrix (MA) domain that regulates intracellular trafficking and membrane targeting, a capsid (CA) domain (that consists of N- and C-terminal subdomains) that promotes virus assembly and forms the capsid shell of the viral core during proteolytic maturation, and a nucleocapsid (NC) domain that is responsible for genome selection and encapsidation. Atomic level structures have been determined for several MA, CA and NC domains, and in general, the domain structures do not vary significantly among different genera of retroviruses [1], [2].
Gag proteins contain additional polypeptide elements and spacers that are often critical for proper virus assembly. In particular, short, proline-rich segments have been identified that facilitate viral budding at late stages of the viral lifecycle, and mutations within these segments typically give rise to elongated or partially formed virus-like buds that are not released from the plasma membrane [3]–[11]. These “late domains” are typically located in disparate regions of the Gag proteins of different retroviruses [9]. The late budding activity of the human immunodeficiency virus type-1 (HIV-1) is mediated by a conserved PT/SAP segment that resides within the C-terminal p6 region of Gag [3], [6]. In contrast, the late budding activity of the gamma-retroviruses is mediated by a conserved PPPY motif found within an 84 residue polypeptide called p12, which is located between the N-terminal MA and CA domains [12]. Disruption of the Moloney murine leukemia virus (MMLV) PPPY element gives rise to budding defects similar to those observed for HIV-1 PT/SAP mutants [12].
Proteolytic cleavage of the p12-CA junction is critical for viral infectivity and precedes cleavage of both the MA-p12 and CA-NC sites [13]. The N-terminal residues of mature N-tropic murine leukemia virus (N-MLV) CA (identical with MMLV CA except for five amino acid substitutions) adopt a β-hairpin that makes intermolecular CA-CA contacts upon hexamer formation [14]. It is likely that cleavage of the p12-CA junction is required for both hairpin formation and capsid assembly, as appears to be the case for HIV-1 capsid assembly [15], [16]. Cryo-electron microscopy (EM) images of immature MMLV virions revealed a low density zone of 25–60 Å between the MA and CA domains, suggesting that p12 is either unfolded or folded but highly mobile prior to proteolysis [17]. In addition to its role in virus assembly, the mature p12 peptide facilitates reverse transcription and the delivery of the pre-integration complex (PIC) to the nucleus for integration of the viral DNA into the host genome during the early phase of infection [18]–[23]. To further understand the structure and diverse roles of p12 as well as the cooperative nature of p12 and CA, we have characterized the dynamic properties and solution structure of a recombinant construct composed of p12 and the N-terminal domain of CA (CAN).
Methods
The pNCA MMLV proviral plasmid [24] was used to subclone MMLV p12CAN, with a C-terminal His-tag, into pET11a (Novagen) using NdeI and BamHI restriction sites, and subsequently transformed into BL21 codon plus RP (DE3) cells (Stratagene). Cells were grown in LB or M9 minimal media supplemented with 99.9 % enriched 15N-ammonium chloride and/or 99.8 % enriched 13C-glucose as the sole nitrogen and/or carbon sources. Protein expression was induced in shake flasks with 1 mM IPTG. The cells were harvested and lysed with a microfluidizer (Micorfluidics), clarified by centrifugation, and the target protein was purified to homogeneity using cobalt affinity (Talon), cation exchange and size exclusion chromatographies (Amersham). Fractions containing pure protein were concentrated using Centripreps (Amicon, MWCO = 3,500 Da) and Centricons (Amicon, MWCO = 3,500 Da). The mass of p12CAN was confirmed by mass spectroscopy.
NMR data were collected at 35°C with a Bruker AVANCE 600 MHz spectrometer equipped with a cryoprobe using samples of 0.8–1.0 mM p12CAN in buffer containing 50 mM sodium phosphate pH 5.5, 100 mM NaCl, 5 mM DTT and 10% D2O. Backbone assignments were obtained using 2D 1H,15N-HSQC, 3D 15N-edited TOCSY, and 3D HNCA and HN(CO)CA experiments [25]-[29]. 3D 15N-edited NOESY-HSQC, 4D 13C,15N-edited NOESY and 4D 13C,13C-edited NOESY experiments were used for side chain assignments and to identify long range intra- and interdomain [1H,1H] NOEs [30].
To gain insight into the dynamical properties of MMLV p12, {1H}–15N steady-state heteronuclear NOE (XNOE) data were obtained for the backbone 15N nuclei [31]. The XNOE experiment consists of a Reference and NOE experiment collected in an interleaved mode. The reference experiment contained a recovery delay of 8 sec and the NOE experiment applied proton saturation during the last 3 sec of the 8 sec recovery delay [31]. The XNOE value for a given residue is derived from the intensity ratio (I/I0) of its 15N/1H correlation peak in the presence of 3 sec proton saturation (I) and in the absence of proton saturation (I0). Peak intensities and the XNOE value were calculated using the HetNOE Analysis module in NMRVIEW. Errors were estimated from the baseline noise in the two spectra. All NMR data were processed using NMRPipe [32] and analyzed using NMRView [33]. Backbone chemical shifts were deposited in the Biological Magnetic Resonance Bank; Accession number 15672.
Results and Discussion
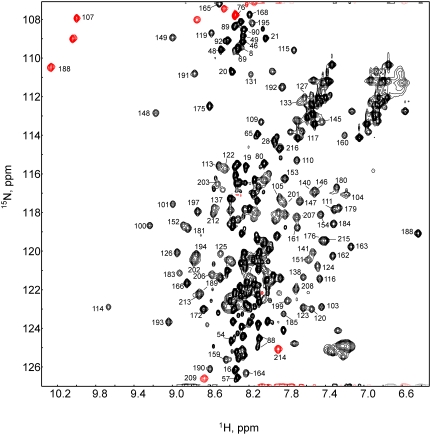
Initial examination of the 2D 1H,15N-HSQC spectra for p12CAN revealed a large subset of signals with poor chemical shift dispersion in the proton dimension, suggestive of a largely unstructured domain within the protein (Figure 1). Backbone resonance assignments established that these signals corresponded to residues in the p12 domain. The high degree of spectra overlap made backbone assignments difficult for this domain, yielding 60 % of the resonances assigned for p12. However, backbone assignments were made for 95 % of the non-proline residues of CAN.
Figure 1. 1H-15N correlation (HSQC) spectrum obtained for MMLV p12CAN.
Assignments are shown for signals in less-crowded regions of the spectrum. Red peaks represent signals folded in the 15N dimension.
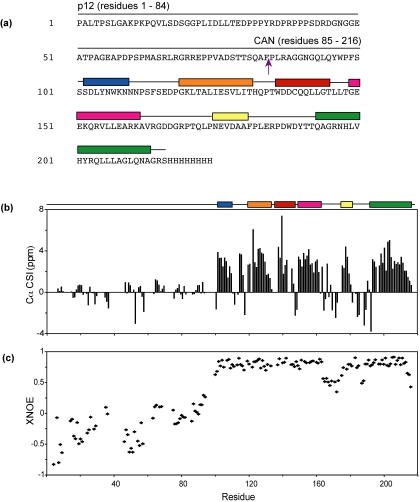
Backbone Cα NMR chemical shift indices (CSI, the difference between the observed chemical shift and shifts observed for random coil structures) provide information on local secondary structure [34], [35], with downfield shifts (positive deviations) reflecting α-helical conformations and upfield shifts reflecting extended (β-strand) conformations. Zero or near zero CSIs were observed for all assigned residues of the p12 domain of p12CAN, indicating that the p12 domain is unstructured, Figure 2. In contrast, most residues of the CAN domain of p12CAN exhibit positive deviations (Figure 2), in agreement with the crystal structure of MMLV CAN hexamer [14].
Figure 2. NMR chemical shift and relaxation data that identify regions of structure and mobility in p12CAN.
(A) Amino acid sequence of p12CAN (arrow denotes proteolytic cleavage site). Residues of CAN that adopt α-helical conformations in the N-MLV CAN crystal structure are denoted by colored rectangles. (B) NMR chemical shift indices for the backbone Cα atoms of p12CAN. Positive values denote helical regions, negative values denote regions of β-structure, and stretches of residues with near-zero values denote random coil conformations. For comparison, α-helical segments observed in the N-MLV CAN crystal structure are aligned at the top of the panel. (C) 15N{1H} heteronuclear NOE (XNOE) data obtained for p12CAN. Values near 1.0 reflect reduced molecular motion, and smaller or negative values reflect motion on a fast (ps-ns) timescale.
Atomic level structures have been determined for the mature CAN domains of several retroviruses, and in all cases, the first ∼15 residues form a β-hairpin stabilized by a salt bridge between Pro 1 and the carboxyl group of an Asp residue subsequent to proteolytic induced maturation [14], [15], [36]–[39]. These residues are unstructured in an HIV-1 MA-CA Gag construct, indicating that β-hairpin formation occurs subsequent to proteolytic cleavage of the MA-CA junction [40]. Near-zero CSIs observed here for assigned N-terminal residues of the CAN domain of p12CAN indicate that the β-hairpin observed in the X-ray structure of the mature MMLV CAN domain is similarly unfolded in the immature p12CAN construct.
To gain insight into the dynamical properties of N-MLV p12, {1H}–15N steady-state heteronuclear NOE (XNOE) data were obtained for the backbone 15N atom [31]. XNOEs provide information regarding high-frequency (psec-nsec) backbone motions and are therefore useful for identifying regions of the protein with high internal mobility. The maximum theoretical XNOE value of 0.86 reflects highly restricted internal motion, whereas values smaller than ∼0.7 are indicative of substantial internal motion [31]. XNOE values were measured for 44 residues of p12 and 106 of CAN that were well resolved in the 2D 1H,15N-HSQC spectra [31]. The residues of p12 exhibited primarily negative or near zero XNOEs, indicative of a high degree of internal flexibility and consistent with the random coil CSI analysis. The high degree of mobility extends through the first 16 residues of the CAN domain, confirming that the β-hairpin is unfolded in the immature p12CAN construct. In contrast, residues within the α-helices of the CAN domain exhibit relatively large XNOE values, consistent with a regularly folded tertiary structure.
Multi-dimensional 1H-1H NOE data (from 3D 15N-edited NOESY-HSQC, 4D 13C,15N-edited NOESY and 4D 13C,13C-edited NOESY experiments; data not shown) were also obtained to probe for intra- and potential inter-domain contacts. Except for the N-terminal residues of the CAN domain, which did not exhibit long-range interactions, the intra-CAN NOEs observed in all the NOE data were fully consistent with the X-ray structure of the mature CAN hexamer. No long-range intra- or inter-domain interactions were observed for residues of p12.
The combined CSI, XNOE and 1H-1H NOE NMR data indicate that the p12 domain of MMLV p12CAN is conformationally labile. The folded regions of the HIV-1 MA and CA domains are also separated by a stretch of flexible residues (∼20 in the MA-CAN NMR structure) [40], and flexibility between these domains may be important for allowing the CA-CA interactions to adjust during virus assembly and maturation. Flexibility in the p12 domain may also be required for interactions with host cell proteins involved in late assembly processes, including viral release. The PTAP element of HIV-1 Gag, which interacts directly with TSG101 [9], [41]–[45] (a component of the cellular protein sorting machinery), is also highly flexible in solution [9]. The inherent flexibility allows p6 to bind to a cleft on TSG101 UEV domain via an induced-fit mechanism [46], [47].
In summary, the MMLV p12 domain of p12CAN exhibits structural/dynamical properties similar to those observed for HIV-1 p6, despite that fact that p12 is twice as large as p6, is located in a very different region of Gag, and has additional functions during the early phase of viral replication.
Acknowledgments
We thank David King (U.C. Berkeley), Robert Edwards (UMBC, HHMI) and Chen Yu (UMBC, HHMI) for technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Funding was received from the NIH (AI30917 to MFS and GM076979 to SKK).
References
- 1.Turner BG, Summers MF. Structural Biology of HIV. J Mol Biology. 1999;285:1–32. doi: 10.1006/jmbi.1998.2354. [DOI] [PubMed] [Google Scholar]
- 2.Freed EO. HIV-1 Gag Proteins: Diverse Functions in the Virus Life Cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 3.Göttlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proceedings of the National Academy of Sciences USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Blanc I, Prevost MC, Dokhelar MC, Rosenberg AR. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J Virol. 2002;76:10024–10029. doi: 10.1128/JVI.76.19.10024-10029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wills JW, Cameron CE, Wilson CB, YXiang Y, Bennett RP, et al. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;96:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puffer BA, Parent LJ, Wills JW, Montelaro RC. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;72:10218–10221. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasuda J, Hunter E. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morita E, Sundquist WI. Retrovirus Budding. Annual Review of Cell and Develpomental Biology. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 10.Yuan B, Fassati A, Yueh A, Goff SP. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J Virol. 2000;74:7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HE, Norris KM, Mansky LM. Analysis of bovine leukemia virus gag membrane targeting and late domain function. J Virol. 2002;76:8485–8493. doi: 10.1128/JVI.76.16.8485-8493.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan B, Li X, Goff SP. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. Embo J. 1999;18:4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oshima M, Muriaux D, Mirro J, Nagashima K, Dryden K, et al. Effects of blocking individual maturation cleavages in murine leukemia virus Gag. J Virol. 2004;78:1411–1420. doi: 10.1128/JVI.78.3.1411-1420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mortuza G, Haire LF, Stevens A, Smerdon SJ, Stoye JP, et al. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. nature. 2004;431:481–485. doi: 10.1038/nature02915. [DOI] [PubMed] [Google Scholar]
- 15.Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, et al. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 16.von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, et al. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. Embo J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeager M, Wilson-Kubalek EM, Weiner SG, Brown PO, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan B, Fassati A, yueh A, Goff SP. Characterization of Moloney Leukemia Virus p12 Mutants Blocked during Early events of Infection. Journal of Virology. 2002;76:10801–10810. doi: 10.1128/JVI.76.21.10801-10810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auerbach MR, Shu C, Kaplan A, Singh IR. Functional characterization of a portion of the Moloney murine leukemia virus gag gene by genetic footprinting. Proc Natl Acad Sci USA. 2003;100:11678–11683. doi: 10.1073/pnas.2034020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes & Development. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 21.Yuan B, Li X, Goff SP. Mutations altering the Moloney murine leukemia virus p12 Gag affect virion production and early events of the virus life cycle. The EMBO journal. 1999;18:4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yueh A, Goff SP. Phosphorylated serine residues and an arginine-rich domain of the moloney murine leukemia virus p12 protein are required for early events of viral infection. J Virol. 2003;77:1820–1829. doi: 10.1128/JVI.77.3.1820-1829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SK, Nagashima K, Hu WS. Cooperative effect of gag proteins p12 and capsid during early events of murine leukemia virus replication. J Virol. 2005;79:4159–4169. doi: 10.1128/JVI.79.7.4159-4169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colicelli J, Goff SP. Sequence and spacing requirements of a retrovirus integration site. J Mol Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 25.Wüthrich K. New York: John Wiley & Sons; 1986. NMR of Proteins and Nucleic Acids. [Google Scholar]
- 26.Grzesiek S, Anglister J, Bax A. Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J Magn Reson. 1993;101:114–119. [Google Scholar]
- 27.Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. Journal of Magnetic Resonance. 1992;96:432–440. [Google Scholar]
- 28.Grzesiek S, Bax A. An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. Journal of Magnetic Resonance. 1992;99:201–207. [Google Scholar]
- 29.Grzesiek S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 30.Kay LE, Clore GM, Bax A, Gronenborn AM. Four-dimensional heteronuclear triple-resonance NMR spectroscopy of interleukin-1 beta in solution. Science. 1990;249:411–414. doi: 10.1126/science.2377896. [DOI] [PubMed] [Google Scholar]
- 31.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 32.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 33.Johnson BA, Blevins RA. NMRview: a Computer Program for the Visualization and Analysis For NMR Data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 34.Wishart DS, Bigam CG, Holm A, Hodges RS, Sykes BD. 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J Biomol NMR. 1995;5:67–81. doi: 10.1007/BF00227471. [DOI] [PubMed] [Google Scholar]
- 35.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 36.Gamble TR, Vajdos F, Yoo S, Worthylake DK, Houseweart SM, et al. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 37.Momany C, Kovari LC, Prongay AJ, Keller W, Gitti RK, et al. Crystal structure of dimeric HIV-1 capsid protein. Nature Struct Biol. 1996;9:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 38.Khorasanizadeh S, Campos-Olivas R, Summers MF. Solution structure of the capsid protein from the human T-cell leukemia virus Type-I. J Mol Biol. 1999;291:491–505. doi: 10.1006/jmbi.1999.2986. [DOI] [PubMed] [Google Scholar]
- 39.Campos-Olivas R, Newman JL, Summers MF. Solution structure and dynamics of the Rous Sarcoma Virus capsid protein and comparison with capsid proteins of other retroviruses. J Mol Biol. 2000;296:633–649. doi: 10.1006/jmbi.1999.3475. [DOI] [PubMed] [Google Scholar]
- 40.Tang C, Ndassa Y, Summers MF. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nature Struct Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 41.Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 43.Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 44.VerPlank L, Bouamr F, LaGrassa TJ, Agresta BE, Kikonyogo A, et al. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demirov D, Freed EO. Retrovirus budding. Virus Research. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Pornillos O, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Mol Biol. 2002;9:812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- 47.Sundquist WI, Schubert HL, Kelly BN, Hill G, Hill CP. Ubiquitin recognition by the human TSG101 protein. Mol Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]