Abstract
Background/Aim:
Recent interest in the liver stem cell field has led to the identification and characterization of several hepatic progenitor cell populations from fetal and adult tissues. We isolated a hepatic progenitor cell from naïve adult liver and the current studies focus on differentiation and growth.
Results:
A Sca-1+ hepatic progenitor cell was identified within the liver parenchyma. This cell expresses numerous liver related genes and transcription found in the developing and/or adult liver. It is located in the peri-portal region and expresses markers associated with undifferentiated hepatic cell populations, mature hepatocytes and biliary cells which distinguish it from the Sca-1-fraction.
Conclusion:
This hepatic progenitor cell from uninjured liver has features of both hepatocytic and biliary populations and demonstrates proliferative potential. Further studies will focus on sca-HPC subsets and conditions that regulate differentiation towards hepatic or biliary lineages.
Keywords: hepatic, liver, progenitor cell, stem cell, bipotential, differentiation
Introduction
The breadth of liver-related stem cell research has been focused on fetal-derived hepatic stem cell or oval cell populations [1-7]. Alternative approaches include isolating proliferative populations from the liver using hepatic injury models that inhibit mature hepatocyte replication [8, 9]. In addition, bipotent progenitor cells capable of multiple rounds of cell division have been identified without a preceding injury to the liver [10-12]. The population of “small hepatocytes” from adult rat liver described by Mitaka et al. is mononuclear with a less differentiated morphologic appearance [10]. Fujikawa et al isolated α-fetoprotein (AFP) positive cells from adult liver with immature endodermal characteristics that were capable of differentiating into both hepatic and biliary cell lineages [13]. More recently Fougere-Deschatrette et al isolated bipotential clonal cell lines from adult murine liver. They demonstrated the persistence of gene transcription of several liver related factors during extended passage along with overlap of select oval cell and biliary markers [12]. In contrast a select human hepatic stem cell population expressed numerous hepatic markers but no hematopoietic or biliary markers [14]. The summary of these works has generated multiple descriptions of potential hepatic progenitor cells with the possibility that overlap exists among them.
Several stem cell associated markers have generated interest in non-hematopoietic stem cell research. The murine stem cell antigen, SCA-1, (Ly6A/E) is a molecule that belongs to the Ly-6 multigene family [15, 16] and is expressed on hematopoietic stem cells (HSC) [15]. Petersen et al demonstrated Sca-1 expression on murine oval cells using an experimental injury model to induce oval cell proliferation [17]. Nierhoff et al demonstrated that a purified fetal liver epithelial cell population expressed Sca-1 and could give rise to functional mature hepatocytes [18]. Several groups have shown that liver-derived endothelial cells, and their precursors also express Sca-1 on the cell surface [15, 19].
We initially identified a tissue-derived hepatic progenitor cell population from adult liver contained within a mixed cell fraction of non-parenchymal cells [11]. The aim of this study was to enrich the hepatic progenitor cell compartment of the adult liver by identifying a marker that would aid in the purification of the hepatic progenitor cell population.
Materials and Methods
Reagents in these experiments were from Sigma, St. Louis, MO unless otherwise stated.
Mice
C57BL/6 mice (6-8 weeks old) were purchased from the Jackson Laboratory (Bar Harbor, ME) and used for all of the experiments. All care and use of animals was approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill in accordance with the principles and procedures outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Hepatic Progenitor Cell Isolation
Hepatic progenitor cell containing fraction was isolated as previously described [11].
Enrichment of Sca-1+-Hepatic Progenitor Cells
Sorting of liver cells was performed using a Sca-1 antibody conjugated to mini-magnetic beads (Miltenyi Biotec, Inc.; Auburn, CA, http://www.miltenyibiotec.com) according to the manufacturer's instructions. Sca1+ cells were eluted with a purity of >99% by flow cytometry and >80% viability.
In-vitro cell culture
The Sca-1+-HPCs (sca-HPCs) were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco; Carlsbad, California ) plus 10% fetal bovine serum (FBS), 20mM hepes, 10mM nicotinamide, 1 mM ascorbic acid 2-phosphate, 1 μM dexamethasone, 0.5 mg/L ITS (insulin-transferrin-selenium) solution, 30 mg/L proline, 100 mg/L antibiotic solution (Gibco), and 10 ng/mL epidermal growth factor (EGF) supplemented with conditioned media from previously unfractionated non-parenchymal cell population. The sca-HPCs were plated at a density of 8×104 cells/cm2. Tissue culture dishes were cultured in a 5% CO2 incubator at 37°C and the media was changed at regular time intervals.
Tissue Processing
Liver tissue was formaldehyde fixed and paraffin embedded. Five μm thick sections were cut and subjected to further immunohistochemical examination.
Histology and Immunohistochemical Characterization
Immunohistochemistry was performed by an indirect immunoperoxidase procedure for localization of select proteins. Primary antibody was against Sca-1 (BD Pharmingen™; San Jose, CA). The secondary antibody was from Vector Laboratories (Burlingame, CA) and the signal was detected using the ABC Elite and DAB kits (Vector Laboratories). All stained slides were viewed on the Nikon Microphot-FXA microscope equipped with an Optronics DEI 750 3-chip CCD camera and a Q Imaging Micropublisher CCD camera for digital image acquisition. Images were captured on an Apple Power Macintosh G3 computer utilizing Q Imaging software.
Fluroescent immunophenotyping and flow cytometry
Cells were stained for immunofluorescence using antibodies directly labeled with the relevant fluoroprobe. Unstained cells, cells stained with an irrelevant antibody and the same fluoroprobe or with the same antibody but no fluoroprobe were used as negative and isotype controls. Cells were suspended in room temperature HBSS containing 2% FBS and 2 mM HEPES buffer (SP buffer) at 5 × 106 cells per ml. Antibodies were primary conjugates against CD117, CD34 and CD45 (BD Biosciences; San Jose, CA). Cells were incubated for 90 minutes, centrifuged and resuspended in cold buffer at 5 × 106 per ml.
Isolation of Sca-1 negative cell fraction
The Sca-1 negative cell fraction was separated and confirmed using the Modular Flow (Mo Flo) cytometer from Cytomation Inc (Fort Collins, CO).
Reverse Transcription-Polymerase Chain Reaction
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Inc.; Valencia, CA) and quality confirmed on an Agilent 2100 BioAnalyzer (Agilent Technologies Inc.; Palo Alto, CA). Two micrograms of total RNA, were reverse-transcribed using the RETROscript® Reverse Transcription Kit (Ambion; Austin, TX). Polymerase chain reaction (PCR) was performed as follows: 94°C, 30 seconds; 55-60°C, 1 minute; and 72°C, 1 minute for 40 cycles. Primer sequences for the genes are in the supplemental data.
Western Blot Analysis
Cells were lysed in buffer containing 50mM Tris, 150mM NaCl, 1% Nonidet P40 (Roche; Indianapolis, IN) , 0.5% deoxycholate, 1mL protease inhibitor cocktail for every 100mL (5μg/mL aprotinin, leupeptin, pepstatin, and soybean trypsin inhibitor), and 1mL phosphatase inhibitor for every 100mL. Lysates were clarified by centrifugation at 14,000g × 2 min and stored at −20°C. Protein concentrations were determined by Bio-Rad protein assay. Total cellular proteins (50 μg/lane) were dissolved by SDS-PAGE, transferred to nitrocellulose membrane incubated with blocking buffer (5% nonfat dry milk in 1x Tris Buffered Saline with 0.05% Tween 20, pH 7.5), and probed with primary antibodies. After incubation with secondary antibodies, peroxidase activity was detected by enhanced chemiluminescence. Densitometric signals from Western blots were analyzed with NIH-ImageJ software (http://rsb.info.nih.gov/ij/) [20]. Protein levels were calculated in arbitrary units (AU) normalized with β-actin protein levels.
Cell Proliferation Assays
The 3-[4,5-dimethylthiazol-2-yl]-2,5- diphenyl tetrazolium bromide (MTT) assay quantifies cell number (Sigma; CGD-1) based on the MTT-Formazan assay [21, 22]. Cells were cultured on 96-well flat bottom plates in 150 μL of media. MTT equal to 10% culture volume in RPMI 1640 medium and the plates were kept at 37°C for 4 hrs in a humidified incubator. Media was removed and the cells were incubated with 150μL MTT solvent (0.1 N HCl in anhydrous isopropanol) until the purple formazan crystals were dissolved. The absorbance was read at 570 nm and 690nm using a microplate reader. The cell number was calculated from a standard absorbance curve [23].
RESULTS
Enrichment and Expansion of the Sca-1+ HPC (sca-HPC)
Having isolated a mixed cell population containing hepatic progenitor cells (HPC) from naïve adult murine liver tissue [11], our focus has been directed towards purification of the HPC from the non-parenchymal cell fraction. With our mechanical separation technique, there is an average of 29.3 ± 6.2 × 106 cells (HPC + NPC) in the supernatant fraction/ liver. This NPC fraction contains a heterogeneous mix of stellate cells, Kupffer cells and biliary cells in addition to the HPC population [11]. Other investigators demonstrated Sca-1 expression on fetal liver stem cell or select oval cell populations generated in specific injury models [17, 18, 24]. We analyzed the NPC fraction by flow cytometric analysis and found that 15.4% ± 3.7% of the cells in this fraction express Sca-1. [Figure 1]
Figure 1.
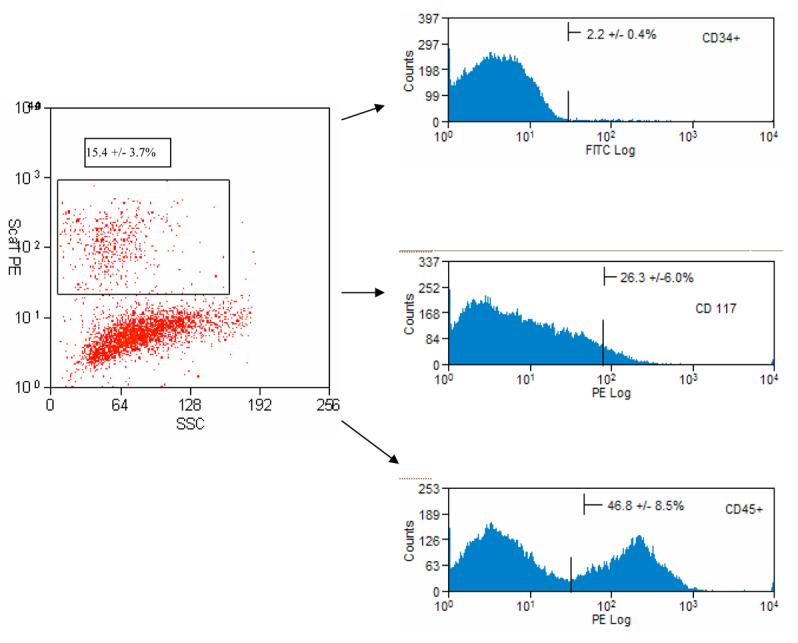
Representative flow cytometric analysis of freshly isolated hepatic progenitor cell (HPC)-containing cell fraction from liver and stained for Sca-1 antibody. The highlighted box (Sca-1+ HPC) represents 15.4 + 3.7 % of the total cell fraction (after depleting mature hepatocytes). The sca-HPC cell population was subsequently analyzed for expression of CD34, CD 117 or CD45 immediately after isolation. Analysis represents the average from three individual isolations.
To separate the HPCs from the NPC we incorporated a magnetic activated cell sorting (MACS®; Miltenyi, Biotec, Inc.) procedure. The MACS® process purifies our cell population giving us >98% Sca-1+ cells from the total cell fraction with >80% viability. (Data not shown) Sca-1+ HPC (sca-HPC) obtained with this protocol are used for all subsequent experiments.
Analysis for expression of additional stem cell related markers was performed on the immediately isolated sca-HPC population. The sca-HPC demonstrates co-expression of CD45, CD117 and CD34 (46.8±8.5%, 26.3±6.0% and 2.2±0.4% respectively) on subsets of this enriched population. [Figure 1]
The subsequent experiments were directed at expanding the sca-HPC population. In our initial experiments the purified sca-HPC population was cultured using routine culture conditions [11] but the cells did not proliferate or form colonies. We hypothesized that the missing NPC populations were requisite to support proliferation of the sca-HPC either via direct cell-cell interactions or cytokine production. We generated conditioned media by culturing the total NPC cell fraction (HPC + NPC) as had been used in our earlier work and media was collected from these dishes at 48 hour intervals. Fresh culture medium was then supplemented in a 3:1 ratio with the conditioned media to generate a 25% conditioned media for use in our experiments. Under these conditions there is reproducible sca-HPC colony formation. A phenotypic difference is demonstrated between the sca-HPC and the remaining Sca− (or NPC) fraction.[Data not shown]
Cellular Localization within Liver Parenchyma
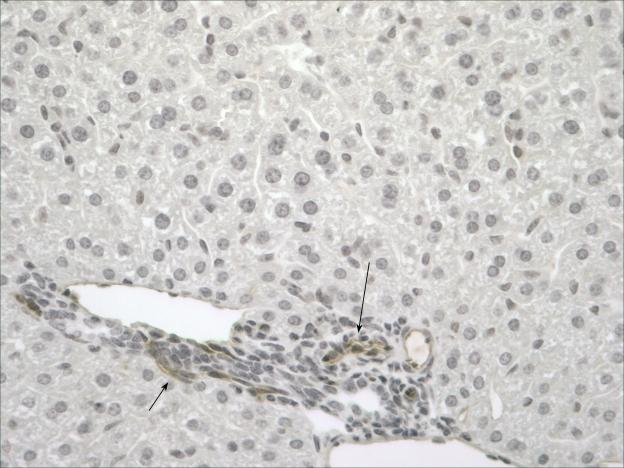
After selecting Sca-1 as a marker we wanted to identify where these cells arise within the liver. Murine liver was analyzed from two age groups; 15 day old mice and 6 week old mice to characterize the in situ location of these cells. In both age groups, cells are identified in the periportal region [Figure 2].
Figure 2.
Sca-1 staining was performed on liver at varying stages of development. Normal 6 week old liver; Sca+ cells are located in the portal region (black arrows) primarily around the bile ducts. [Original objective lens magnification: 20x].
Gene Expression of sca-HPCs
The next experiments characterize the sca-HPC's expression of genes associated with differentiated hepatocytes, biliary cells and transcription factors associated with the developing liver. Immediately after isolation the sca-HPC expresses numerous liver transcription factors. [Figure 3] The sca-HPC expresses a number of apolipoproteins along with genes associated with mature hepatocytes (i.e., tyrosine amino transferase [TAT], retinol binding protein, transferrin, albumin and tryptophan dioxygenase [TO]). The sca-HPC also expresses AFP (a marker of immature hepatic cells) and CK-19 (a marker of biliary epithelial cells). [Figure 3] For comparison, we analyzed the Sca-1 negative fraction of cells (the remaining NPC population). [Supplemental Material; Table 1]
Figure 3.

Reverse transcription-polymerase chain reaction (RT-PCR) analysis of select transcription factors and liver-related genes from Sca-HPC.
Differentiation and Bipotency of sca-HPCs
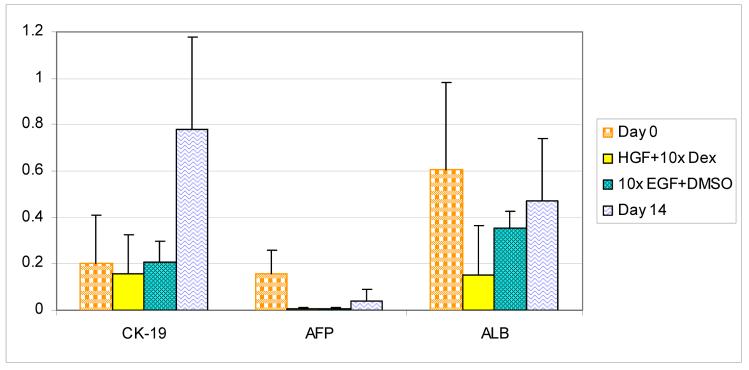
Sca-HPCs established in culture under experimental conditions are analyzed for their ability to differentiate into mature cell types. Using Western blot analysis the sca-HPCs are analyzed for albumin and AFP (hepatocytic markers) and CK-19 (a biliary marker) to define the cell's bipotent potential. Immediately after isolation the sca-HPCs express CK-19, AFP, and albumin which persists for two weeks of culture under our established growth conditions. [Figure 4 and Supplemental Data]
Figure 4.
Western blot analysis of sca-HPC. Sca-HPC were isolated and analyzed for expression of CK-19, AFP and albumin at day 14 in routine growth conditions; 50ng/mL EGF + 2% DMSO or 40ng/mL HGF + dexamethasone. Comparisons were made with primarily isolated Sca-HPC. Densitometric signals from Western blots were analyzed with NIH-ImageJ software (http://rsb.info.nih.gov/ij/). Protein levels were calculated in arbitrary units (AU) normalized to sample β-Actin protein levels.
Numerous culture conditions have been published with respect to supporting differentiation of liver cells towards biliary or hepatic phenotypes [25]. Based on this work the sca-HPCs are cultured under two distinct conditions to support differentiation. They are: 50ng/mL EGF + 2% DMSO and 40ng/mL HGF + dexamethasone without EGF in the media. For comparison sca-HPC in routine growth conditions plus 25% conditioned media were used as controls. In the control the cells demonstrate a decrease in albumin and AFP with a concomitant increase in CK-19 expression. Under both experimental conditions the cells demonstrate a decrease in expression of albumin, AFP and CK-19 compared with control cells.
Sca-HPC Viability and Proliferation
The formation of sca-HPC colonies begins at day 4-5 of primary cell culture. What remains unclear is whether this is an intrinsic delay in the pathways associated with cellular proliferation. We analyzed sca-HPC viability and proliferation in primary cell culture using thymidine incorporation during the first 4 days (or 96 hours) of primary culture. This time frame represents the period prior to visualization of colony formation. We also employed an MTT assay to evaluate viable sca-HPCs. In the [3H] thymidine assay there is a basal level of DNA synthesis during the first 48 hours of culture followed by a spike in activity during the subsequent 48 hour period. [Supplemental data] Using the MTT assay we observe a stable number of sca-HPCs through the first 4 days of culture followed by a >3 fold increase in sca-HPCs after day 4. [Supplemental data]
DISCUSSION
We previously isolated a heterogeneous cell population from naïve adult liver that contained a presumed hepatic progenitor cell population [11]. Unfortunately challenges in our initial work involved the heterogeneity of this cellular fraction as this population included the non-parenchymal cells (e.g. stellate cells, etc.). In these experiments we focused on enrichment and subsequent analysis of the sca-HPC separately from other NPC. Our strategy involved identifying one or more unique cell surface markers. Figure 1 demonstrates that a defined subset of the initial NPC express Sca-1 on their surface. Therefore using an antibody selection method we isolated these cells and established culture conditions that support their proliferation and ability to form colonies.
In-depth analysis of these cells demonstrates that a subset of the sca-HPC is positive for select stem-cell related markers (e.g. CD34, CD45 and CD 117) each of which has been shown to be important in various oval cell studies [6, 26-28]. [Figure 1] These findings are very interesting as they suggest that within the sca-HPC population there is diversity with multiple subpopulations. This finding could reflect variability in the state of cell differentiation and is important as several investigators have previously demonstrated heterogeneity of progenitor/stem cell populations with varying abilities to differentiate and/or proliferate [29, 30].
Investigating the in situ location of this population is critical to understanding the cell's origin and association with other cellular populations of the liver. Interestingly in the young mice the cells are primarily located in the periportal region [data not shown] while in the adult mice they are located in closer proximity to the bile ducts. [Figure 2] They also have the morphologic appearance of small periportal hepatocytes [31]. Sca+ cells were also demonstrated along the endothelium of the portal vein [29]. [Data not shown]
A series of experiments investigated transcription factor and liver related gene expression in the sca-HPC population suggest that the sca-HPC is similar to fetal liver cells[8]. [Figure 3 and Table 1] Interestingly, the Sca-negative fraction demonstrates a number of hepatocyte function markers which we believe this is secondary to cellular contamination with a number of mature hepatocytes. Using a culture system to distinguish the sca-HPC and Sca-negative cells we found that the latter population is compromised of a population of cuboidal cells that express very few of the developmental markers found in the Sca-HPC nor do they express AFP or CK-19. [Table 1]
We evaluated the sca-HPC for expression of hepatic and biliary markers during proliferation and found a decrease in albumin and AFP expression to levels that were approximately 50% of their expression immediately after isolation. Interestingly there was a more than 2-fold increase in the expression of CK-19 during this same time period. These findings are intriguing and we postulate that the sca-HPCs may be dedifferentiating in culture; a similar phenomenon had been demonstrated with mature hepatocytes in culture [25] and with other HPC populations [12]. Using a culture system that supports differentiation of hepatocytic cells along a biliary lineage the sca-HPCs did not differentiate towards a specific phenotype and while there was a loss of AFP expression with persistent expression of CK-19 there was also a decrease in albumin expression. [Figure 4]
In summary, these experiments are a step towards understanding the biology of hepatic progenitor cells from naïve adult liver. Our characterization is important as there are some similarities with previously described populations from fetal and adult tissues. In addition, the cell differentiation results are critical as we expand our knowledge of these unique populations.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institutes of Health DK059302 (DAG) and the Roche Organ Transplant Research Foundation (ROTRF) (DAG). We thank the Lineberger Cancer Center's Flow Cytometry and Histopathology Core Facilities for their assistance with experiments. We would also like to thank the Microscopy Services Laboratory and Hunter Best, PhD and Bill Coleman, PhD for their assistance with histology and imaging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kuai XL, Cong XQ, Li XL, Xiao SD. Generation of hepatocytes from cultured mouse embryonic stem cells. Liver Transpl. 2003;9:1094–1099. doi: 10.1053/jlts.2003.50207. [DOI] [PubMed] [Google Scholar]
- 2.Ishizaka S, Shiroi A, Kanda S, Yoshikawa M, Tsujinoue H, Kuriyama S, Hasuma T, Nakatani K, Takahashi K. Development of hepatocytes from ES cells after transfection with the HNF-3beta gene. Faseb J. 2002;16:1444–1446. doi: 10.1096/fj.01-0806fje. [DOI] [PubMed] [Google Scholar]
- 3.Choi D, Oh H-J, Chang U-J, Koo SK, Jiang JX, Hwang S-Y, Lee J-D, Yeoh GC, Shin H-S, Lee J-S, Oh B. In Vivo Differentiation of Mouse Embryonic Stem Cells Into Hepatocytes. Cell Transplantation. 2002;11:359–368. [PubMed] [Google Scholar]
- 4.Suzuki A, Zheng Y-W, Kondo R, Kusakabe M, Takada Y, Fukao K, Nakauchi H, Taniguchi H. Flow-cytometric separation and enrichment of hepatic progenitor cells in the developing mouse liver. Hepatology. 2000;32:1230–1239. doi: 10.1053/jhep.2000.20349. [DOI] [PubMed] [Google Scholar]
- 5.Kubota H, Reid LM. Clonogenic hepatoblasts, common precursors for hepatocytic and biliary lineage, are lacking classical major histocompatibility complex class I antigen. PNAS. 2000;97:12132–12137. doi: 10.1073/pnas.97.22.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujio K, Evarts RP, Hu Z, Marsden ER, Thorgeirsson SS. Expression of stem cell factor and its receptor, c-kit, during liver regeneration from putative stem cells in adult rat. Lab Invest. 1994;70:511–516. [PubMed] [Google Scholar]
- 7.Fiegel HC, Bruns H, Hoper C, Lioznov MV, Kluth D. Cell growth and differentiation of different hepatic cells isolated from fetal rat liver in vitro. Tissue Eng. 2006;12:123–130. doi: 10.1089/ten.2006.12.123. [DOI] [PubMed] [Google Scholar]
- 8.Li W-L, Su J, Yao Y-C, Tao X-R, Yan Y-B, Yu H-Y, Wang X-M, Li J-X, Yang Y-J, Lau JTY, Hu Y-P. Isolation and Characterization of Bipotent Liver Progenitor Cells from Adult Mouse 10.1634/stemcells.2005-0108. Stem Cells. 2006;24:322–332. doi: 10.1634/stemcells.2005-0108. [DOI] [PubMed] [Google Scholar]
- 9.Gordon GJ, Coleman WB, Hixson DC, Grisham JW. Liver regeneration in rats with retrorsine-induced hepatocellular injury proceeds through a novel cellular response. American Journal of Pathology. 2000;156:607–619. doi: 10.1016/S0002-9440(10)64765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitaka T, Kojima T, Mizuguchi T, Mochizuki Y. Growth and maturation of small hepatocytes isolated from adult rat liver. Biochemical and Biophysical Research Communications. 1995;214:310–317. doi: 10.1006/bbrc.1995.2289. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Clark JB, Rhee G-S, Fair JH, Reid LM, Gerber DA. Proliferation and Hepatic Differentiation of Adult-Derived Progenitor Cells. Cells, Tissues, Organs. 2003;173:193–203. doi: 10.1159/000070375. [DOI] [PubMed] [Google Scholar]
- 12.Fougere-Deschatrette C, Imaizumi-Scherrer T, Strick-Marchand H, Morosan S, Charneau P, Kremsdorf D, Faust DM, Weiss MC. Plasticity of hepatic cell differentiation: bipotential adult mouse liver clonal cell lines competent to differentiate in vitro and in vivo. Stem Cells. 2006;24:2098–2109. doi: 10.1634/stemcells.2006-0009. [DOI] [PubMed] [Google Scholar]
- 13.Fujikawa T, Hirose T, Fujii H, Oe S, Yasuchika K, Azuma H, Yamaoka Y. Purification of adult hepatic progenitor cells using green fluorescent protein (GFP)-transgenic mice and fluorescence-activated cell sorting. J Hepatol. 2003;39:162–170. doi: 10.1016/s0168-8278(03)00237-x. [DOI] [PubMed] [Google Scholar]
- 14.Herrera MB, Bruno S, Buttiglieri S, Tetta C, Gatti S, Deregibus MC, Bussolati B, Camussi G. Isolation and Characterization of a Stem Cell Population from Adult Human Liver. Stem Cells. 2006;24:2840–2850. doi: 10.1634/stemcells.2006-0114. [DOI] [PubMed] [Google Scholar]
- 15.Luna G, Paez J, Cardier JE. Expression of the hematopoietic stem cell antigen Sca-1 (LY-6A/E) in liver sinusoidal endothelial cells: possible function of Sca-1 in endothelial cells. Stem Cells Dev. 2004;13:528–535. doi: 10.1089/scd.2004.13.528. [DOI] [PubMed] [Google Scholar]
- 16.LeClair KP, Palfree RG, Flood PM, Hammerling U, Bothwell A. Isolation of a murine Ly-6 cDNA reveals a new multigene family. Embo J. 1986;5:3227–3234. doi: 10.1002/j.1460-2075.1986.tb04633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petersen BE, Grossbard B, Hatch H, Pi L, Deng J, Scott EW. Mouse A6-positive hepatic oval cells also express several hematopoietic stem cell markers. Hepatology. 2003;37:632–640. doi: 10.1053/jhep.2003.50104. [DOI] [PubMed] [Google Scholar]
- 18.Nierhoff D, Ogawa A, Oertel M, Chen YQ, Shafritz DA. Purification and characterization of mouse fetal liver epithelial cells with high in vivo repopulation capacity. Hepatology. 2005;42:130–139. doi: 10.1002/hep.20735. [DOI] [PubMed] [Google Scholar]
- 19.Cherqui S, Kurian SM, Schussler O, Hewel JA, Yates JR, 3rd, Salomon DR. Isolation and angiogenesis by endothelial progenitors in the fetal liver. Stem Cells. 2006;24:44–54. doi: 10.1634/stemcells.2005-0070. [DOI] [PubMed] [Google Scholar]
- 20.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 21.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 22.Dvir-Ginzberg M, Gamlieli-Bonshtein I, Agbaria R, Cohen S. Liver tissue engineering within alginate scaffolds: effects of cell-seeding density on hepatocyte viability, morphology, and function. Tissue Eng. 2003;9:757–766. doi: 10.1089/107632703768247430. [DOI] [PubMed] [Google Scholar]
- 23.Wagner WR, Muzzio DJ, Rilo HR, Deglau T, Ataai MM, Michalopoulos GK, Block GD. Effect of growth factos and defined medium on primary hepatocyte culture on polyester carriers wtih varying surface treatment. Tissue Eng. 1997;3:289. [Google Scholar]
- 24.Tsuchiya A, Heike T, Fujino H, Shiota M, Umeda K, Yoshimoto M, Matsuda Y, Ichida T, Aoyagi Y, Nakahata T. Long-term extensive expansion of mouse hepatic stem/progenitor cells in a novel serum-free culture system. Gastroenterology. 2005;128:2089–2104. doi: 10.1053/j.gastro.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 25.Block GD, Locker J, Bowen WC, Petersen BE, Katyal S, Strom SC, Riley T, Howard TA, Michalopoulos GK. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. Journal of Cell Biology. 1996;132:1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsusaka S, Tsujimura T, Toyosaka A, Nakasho K, Sugihara A, Okamoto E, Uematsu K, Terada N. Role of c-kit receptor tyrosine kinase in development of oval cells in the rat 2-acetylaminofluorene/partial hepatectomy model. Hepatology. 1999;29:670–676. doi: 10.1002/hep.510290304. [DOI] [PubMed] [Google Scholar]
- 27.Blakolmer K, Jaskiewicz K, Dunsford HA, Robson SC. Hematopoietic stem cell markers are expressed by ductal plate and bile duct cells in developing human liver. Hepatology. 1995;21:1510–1516. [PubMed] [Google Scholar]
- 28.Lemmer ER, Shepard EG, Blakolmer K, Kirsch RE, Robson SC. Isolation from human fetal liver of cells co-expressing CD34 haematopoietic stem cell and CAM 5.2 pancytokeratin markers. J Hepatol. 1998;29:450–454. doi: 10.1016/s0168-8278(98)80064-0. [DOI] [PubMed] [Google Scholar]
- 29.Tsuchiya A, Heike T, Baba S, Fujino H, Umeda K, Matsuda Y, Nomoto M, Ichida T, Aoyagi Y, Nakahata T. Long-term culture of postnatal mouse hepatic stem/progenitor cells and their relative developmental hierarchy. Stem Cells. 2007;25:895–902. doi: 10.1634/stemcells.2006-0558. [DOI] [PubMed] [Google Scholar]
- 30.Schmelzer E, Wauthier E, Reid LM. The phenotypes of pluripotent human hepatic progenitors. Stem Cells. 2006;24:1852–1858. doi: 10.1634/stemcells.2006-0036. [DOI] [PubMed] [Google Scholar]
- 31.Michalopoulos GK, Barua L, Bowen WC. Transdifferentiation of rat hepatocytes into biliary cells after bile duct ligation and toxic biliary injury. Hepatology. 2005;41:535–544. doi: 10.1002/hep.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.