Abstract
Wnts are secreted glycoproteins that regulate important cellular processes including proliferation, diiferentiation, and cell fate. In the β-catenin/canonical pathway, Wnt interacts with Fzd receptors to inhibit degradation of β-catenin and promote its translocation into the nucleus where it regulates transcription of a number of genes. Dysregulation of this pathway has been attributed to a host of diseases including cancer. As a result, components of the β-catenin/canonical pathway have been gaining recognition as promising targets for the discovery of novel therapeutic agents. Here we show, using an ELISA-based protein-protein binding assay that purified Wnt7a binds to the extracellular cysteine-rich domain of Fzd5 in the nanomolar range. We have developed a novel split eGFP complementation assay to visually detect Wnt7a-Fzd5 interactions and subsequent pathway activation in cells. These biological tools could help lead to a better understanding of Wnt-Fzd interactions and the identification of new modulators of Wnt signaling.
Keywords: Wnt signaling, Wnt7a, Fzd5, split eGFP, protein complementation
The β-catenin/canonical Wnt pathway defines a series of events that occur when Wnt proteins bind to cell-surface receptors of the Frizzled (Fzd) family. Currently, 19 Wnt proteins and 10 different Fzd members have been identified in the human genome[1; 2]. The Wnt-Fzd interaction promotes complex formation with a membrane-associated Lrp 5/6 co-receptor and activates signaling through the direct recruitment of a Dishevelled (Dsh) protein to the intracellular portion of Fzd[3; 4]. Dsh inhibits GSK-3 association with the Axin/APC complex of proteins that normally mark β-catenin for proteolytic degradation[5]. Intracellular pools of cytoplasmic β-catenin become stabilized, enter into the nucleus, and interact with the TCF/LEF family of transcription factors to promote specific gene expression[6; 7]. The Wnt-Fzd interaction has also been shown to activate two distinct non-canonical pathways; the planar cell polarity/JNK pathway and a Ca2+-mediated pathway[2; 8; 9].
Wnt was initially discovered as a proto-oncogene in mammary tumors activated by integration of the mouse mammary tumor virus[10]. More recently, studies have linked the β-catenin/canonical Wnt signaling pathway to disease onset and tumorogenesis [11-13]. The development of cancer has been attributed to mutations resulting in constitutive activation of β-catenin and either loss of expression or hypermethylation of extracellular Wnt antagonist proteins [14–16]. The involvement of Wnt signaling in human disease has raised significant interest to discover novel modulators of the β-catenin/canonical signaling pathway that may be considered for drug development. To date, most studies have been focused on identifying inhibitors that target downstream cytoplasmic and nuclear proteins of the pathway [17]. Our interest is to create a technique that will help elucidate specific Wnt-Fzd interactions and that may be employed to screen for inhibitors/activators of Wnt signaling that occur upstream at the receptor level.
Wnt7a regulates a variety of cell and developmental pathways that direct the prenatal growth of the female reproductive tract and maintain proper uterine function in the adult [18]. In the endometrium, Wnt7a functions in cell-cell communication and is responsive to changes in levels of sex steroid hormones [19]. Our lab has found that Wnt7a interacts with Fzd5 to activate β-catenin/canonical signaling and induce proliferation in endometrial cancer cells [Carmon and Loose, unpublished]. Additional studies have reported a critical role for Wnt7a in limb development and neuronal differentiation [20; 21]. Wnt7a was demonstrated to signal through the Fzd5/Lrp6 complex in neuronal PC12 cells as measured by increased β-catenin stability and activation of a TCF reporter construct[22]. In this study, we attempt to further characterize the binding of Wnt7a with Fzd5 and establish the proof of principle of an eGFP complementation assay for the detection of Wnt signaling in cells. This assay holds potential for the identification of novel modulators of the Wnt pathway that target Fzd receptors at the membrane.
Methods and Materials
Cell culture and transfection
Ishikawa cells were grown in RPMI 1640 medium with glutamine, without phenol red (Invitrogen) and supplemented with 10% FBS, 0.01 M HEPES, and penicillin/streptomyosin. Cells were split two days prior to transient transfection, and grown to 60–70% confluency. Transfections were conducted in using FuGene 6 (Roche).
Construction of the xDsh-NeGFP and Fzd5-CeGFP
Site-directed mutagenesis was performed by PCR using xDsh-eGFP. Forward primer 5’- ATACATCATGGCAGACAAACAATAGAATGGAATCAAAG -3’ and reverse primer 5’- CTTTGATTCCATTCTATTGTTTGTCTGCCATGATGTAT -3’ were used to convert the lysine residue 158 of eGFP to a stop codon, resulting in xDsh protein fused only to the N-terminal fragment of eGFP (1–158). A subcloning procedure was implemented to construct Fzd5-CeGFP. The single Xho1 restriction site downstream of the Fzd5 coding region of Fzd5-V5/His pcDNA3.1 was targeted for insertion of CeGFP. Briefly, cDNA encoding the C-terminal fragment of eGFP (158–238) was PCR amplified from xDsh-eGFP using forward primer ‘5–AGACTCGAGAAGAATGGAATCAAAGTTAACTTCA –3’ and reverse primer 5’-GAGACTCGAGTTATTTGTATAGTTCATCCATGCCA -3’. Primers were designed with an Xho1 restriction site flanking either end. Purified PCR product and Fzd5-V5/His were then digested with the Xho1 restriction enzyme. The CeGFP was subcloned into Fzd5-V5/His. Clones were screened for the presence of insert by PCR and sequenced.
Stable Expression of xDsh-NeGFP and Fzd5-CeGFP
To generate Ishikawa cell lines for stable expression of pcDNA3.1 vector, Wnt7a, or xDsh-NeGFP and Fzd5-CeGFP, cells were plated out in 1:40 dilution 24 hours post-transfection and cultured under G418 selection (600 ug/ml) for 10 to 14 days. At least 12 clones expanded from single cells were screened for expression via RT-PCR and/or Western blot analysis.
Co-immunoprecipitation and western blot analysis
Ishikawa cells were treated with 2 mM Dithiobis [succinimidylpropionate] (DSP) crosslinker (Pierce) at 24 hrs post-transfection for 30 minutes at room temperature and the reaction was terminated with 10mM Tris pH 7.5. Cells were harvested with TX-100 Buffer. Lysates from Ishikawa cells overexpressing xDsh-NeGFP alone or with Fzd5-CeGFP were immunoprecipitated with rabbit anti-NGFP (Sigma-Aldrich). Extracts overexpressing Fzd5-CeGFP were immunoprecipitated with goat anti-CGFP or (Santa Cruz Biotechnology). Western blot analysis was performed and immunoprecipitates were immunoblotted with rabbit anti-GFP (Abcam). To confirm stable expression of both split GFP fusion constructs in xDshN/Fzd5C cells, immunoprecipitation was performed with total lysate using rabbit anti-NGFP or goat anti-Fzd5 (Santa Cruz) prior to western blot analysis with anti-NGFP and anti-CGFP. Co-immunoprecipitation of Wnt7a was carried out with either 500 ng/ml Fzd5 cysteine-rich domain (CRD)-Fc IgG or Fc IgG using total lysate from wild type Ishikawa cells and Ishikawa cells stably overexpressing Wnt7a. Western analysis was performed using mouse anti-V5 (Invitrogen) and rabbit anti-human IgG (Santa Cruz Biotechnology)
Purification of recombinant Wnt7a 6xHis-tagged proteins
6xHis-tagged proteins were purified from stable cells expressing Wnt7a-V5/His by loading cleared lysate into Ni-NTA spin columns (Qiagen) and following protocol for mammalian cells under native conditions. Eluted samples were run through a second column and then dialyzed against PBS pH 7.4 with addition of 1.0% CHAPS. Protein expression and purity was verified via western analysis and Coomassie staining, respectively. Quantification was performed using a 6xHis-tag based ELISA protocol.
Wnt7a/Fzd5CRD-Fc ELISA binding assays
The 96-well plates were coated with 2µg/ml Fzd5CRD-Fc IgG (R&D Systems) or Fc IgG (Bethyl Laboratories) diluted in carbonate buffer pH 9.0 overnight at 4°C. The wells were washed with PBST (0.05% Tween-20, PBS pH 7.5) and blocked with PBSTM (0.05% Tween-20, 5% dried milk, PBS pH 7.5) for an additional hour at 25°C. Following four washes with PBST, 50-µl dilutions of Ni-NTA purified Wnt7a was incubated for 2 hours at 25°C. After four washes with PBST, 50 µl/well of mouse anti-V5 antibody (Invitrogen) diluted in PBSTM to a final concentration of 1 µg/ml was incubated in wells for 1 hour 30 minutes at 25°C. Another four washes in PBST were followed by incubation at 25°C with 1:500 dilution of HRP conjugated goat anti-mouse IgG (Santa Cruz). After a final set of four washes with PBS, 50 µl of TMB reagent (Sigma-Aldrich) was added. After 30 minutes the reaction was stopped with 50 µl 1M H2SO4. Absorbance at 450 nm was measured with a plate reader and OD values were converted to concentration based on a V5 standard curve. Non-linear regression analysis was performed using GraphPad Prism 5 Software.
TopFlash assays
The TopFlash firefly luciferase reporter and Renilla (pRL-TK) constructs were transfected at 0.5µg/well and 0.1µg/well of a 6-well plate, respectively. Ishikawa cells were treated with 300ng/ml Wnt7a or buffer 24 hours post-transfection and harvested 24 hours following treatment in 1 × passive lysis buffer (Promega ). Luciferase activity was measured by a Monolight 2010 luminometer (Analytical Luminescence Laboratory) using the Dual-Luciferase Reporter Assay (Promega) as indicated by manufacturer. Each experiment was repeated at least three times in duplicate to ensure reproducibility.
Fluorescence microscopy
For transient transfection experiments, Ishikawa cells were grown on cover-slips and transfected with xDsh-NeGFP (1µg), Fzd5-CeGFP (1µg), or co-transfected +/− Wnt7a-V5/His (1µg). Stable cells were treated with 300 ng/ml purified Wnt7a protein. After 24 hours cells were washed in PBS pH 7.4 and fixed in 10% formalin for 5 minutes. DAPI staining was used at 1 µg/ml. Cells were again washed in PBS pH 7.4 and mounted on slides with Fluoromount G (Southern Biotech). Images were collected via fluorescence microscopy at 5 s exposure and 400 × magnification.
Results and Discussion
Wnt7a Binds to the Fzd5 CRD in the Nanomolar Range
Previous reports have demonstrated that Wnts bind to the extracellular cysteine-rich domains (CRD) of frizzled receptors as means to activate Wnt signaling [23]. Given this knowledge, we tested whether the Wnt7a-Fzd5 interaction occurs through direct binding of Wnt7a to the CRD of Fzd5. Lysate from cells overexpressing Wnt7a was incubated with protein A/G agarose and a purified fusion protein consisting of the CRD of Fzd5 attached to the Fc portion of human IgG. Wnt7a lysate was incubated Fc IgG to serve as a negative control. Western analysis confirmed that Wnt7a specifically bound to the Fzd5CRD-Fc and not Fc (Figure 1A). These findings establish biochemically that Wnt7a associates with Fzd5 by binding to the CRD in order to activate canonical Wnt signaling.
Figure 1. Characterization of the Wnt7a-Fzd5 Interaction Using Purified Protein.
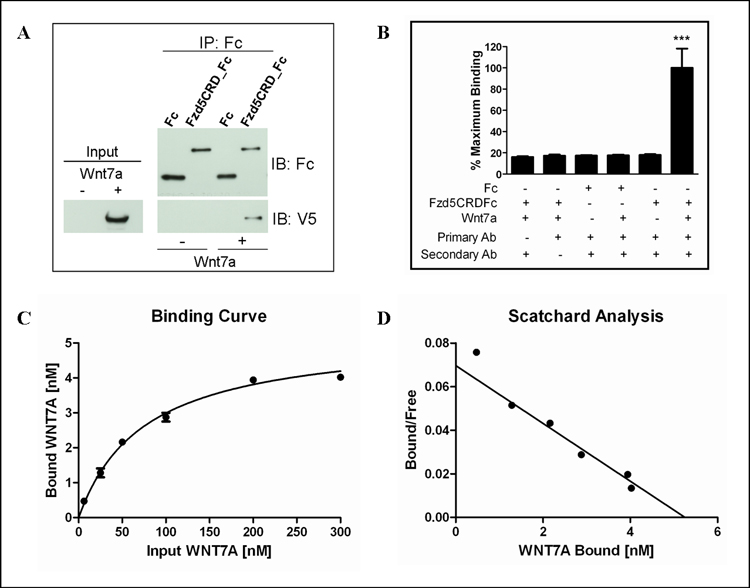
(A) Wnt7a directly interacts with the Fzd5 CRD. Ishikawa cell extracts from and extracts from cells stably overexpressing Wnt7a V5/His were immunoprecipitated with Fzd5CRD-Fc or Fc. (B) Negative controls for ELISA-based protein-protein binding assay indicating a minimal amount of background signal compared to maximum signal detected for Wnt7a and Fzd5CRD-Fc binding. ***p≤0.001 compared to all controls using ANOVA and Bonferroni post-hoc analysis (C) Wnt7a-Fzd5CRD-Fc binding curve and (D) scatchard plot obtained using non-linear regression analysis in GraphPad Prism. The calculated Bmax and Kd were 5.3 nM and 75.4 nM, respectively.
It is widely recognized that there are challenges associated with purifying large quantities of Wnts that retain biological activity. To date the only Wnts that have been successfully purified are Wnt1, Wnt3a, and most recently, Wnt5a [24; 25]. There is little information regarding the Wnt binding site and affinity of individual Wnts for different Fzd receptors. We have attempted to purify Wnt7a and show that it retains both its ability to bind Fzd and subsequently function to activate β-catenin/canonical signaling.
A unique ELISA-based protein-protein binding detection method was employed to determine relative affinity of Wnt7a binding to the CRD of Fzd5. The wells of a 96-well plate were coated with Fc or Fzd5CRD-Fc and incubated with increasing concentrations of purified Wnt7a. As shown in Figure 3B, when compared to wells containing Fzd5CRD_Fc plus the maximum concentration of Wnt7a, very little backround signal was measured from control wells that did not have the primary or secondary detection antibodies added. Likewise, no significant signal was detected from wells coated with Fc or in wells not containing Wnt7a (Figure 1B). However, when increasing amounts of Wnt7a were added to the Fzd5CRD coated wells, an increase in binding was detected at concentrations up to 200 nM (Figure 1C and 1D). Using non-linear regression analysis, the Bmax and Kd were calculated to be 5.3 nM and 75.4 nM, respectively. This data is consistent with previous reports of DWnt affinites for DFzds ranging from 16–70nM [26]. Our results strongly indicate that Wnt7a binds to Fzd5 with affinity in the nanomolar range.
Figure 3. Stable Expression of xDsh-NeGFP and Fzd5-CeGFP in Ishikawa Cells.
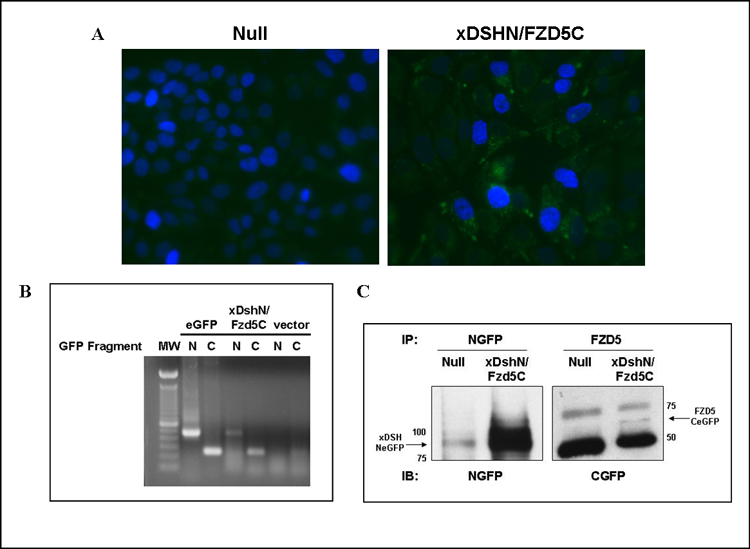
(A) Fluorescence microscopy of DAP1 stained vector versus xDshN/Fzd5C stable cells. (B) RT-PCR analysis of NeGFP and CeGFP expression in positive control eGFP-expressing cells, xDshN/Fzd5C stable cells, and vector (eGFP null) stable cells. (C) Immunoprecipitation and western blot analysis confirming expression of both split eGFP fusion proteins in the xDshN/Fzd5C stable cell line.
An eGFP Complementation Assay for Detection of Wnt Signaling
The split protein complementation assay technique has been previously described to detect a variety of different protein-protein interactions by using split proteins derived from dihydrofolate reductase, β-lactamase, Renilla and firefly luciferases, and the GFP and YFP reporters [27–34]. However, this technique has never been applied to proteins in the Wnt signaling pathway. We have developed a split enhanced green fluorescent protein (eGFP) complementation assay to investigate the interaction of Wnt7a with Fzd5 and subsequent activation of Wnt signaling in living cells. In our system, the eGFP reporter protein was split into two non-fluorescent fragments, the amino terminal fragment (NeGFP) and the carboxy terminal fragment (CeGFP), at lysine 158. The CeGFP fragment was fused to the C-terminal of Fzd5. Similarly, the NeGFP fragment was fused to the C-terminal of xDsh, a downstream component that directly associates with activated Fzd receptors and is universal to all three Wnt signaling pathways [9; 35]. The Ishikawa epithelial derived endometrial carcinoma cell line was then manipulated to express both xDsh-NeGFP and Fzd5-CeGFP fusion proteins. When a binding Wnt, such as Wnt7a, is introduced into our system and binds to Fzd5, xDsh is recruited to the membrane and initiates the reconstitution of the two eGFP fragments. As a result, the fluorescent output allows us to detect activation of Wnt signaling. (Figure 2A).
Figure 2. Visualization of Wnt7a-Fzd5 Pathway Activation Using a split eGFP Complementation Assay.
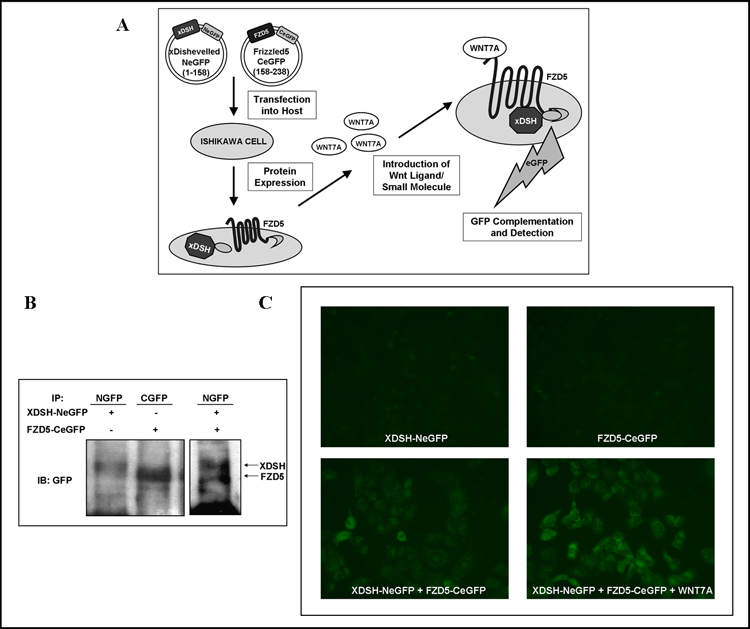
(A) Schematic depicts the basis of a split-eGFP complementation assay for Wnt signaling. The xDsh construct containing the N-terminous of eGFP and the Fzd5 construct with the eGFP C-terminous are transfected into Ishikawa cells for subsequent expression of fusion proteins. Wnt7a directly binds Fzd5-CeGFP and promotes the recruitment of xDsh-NeGFP to the intracellular membrane. This event results in the reconstitution of eGFP and associated fluorescent signal output. (B) Cells extract overexpressing xDsh-NeGFP with either empty vector or Fzd5-CeGFP were immunoprecipitated with anti-NGFP. Extracts overexpressing Fzd5-CeGFP were immunoprecipitated with anti-CGFP. Western blot was performed using anti-GFP. (C) Ishikawa cells grown on cover-slips were transiently transfected with xDsh-NeGFP, Fzd5-CeGFP, or co-transfected with both constructs +/−Wnt7a. Cells were fixed 24 hours post-transfection. Images were collected via fluorescence microscopy at 5s exposure and 400 x magnification. eGFP was detected only in the presence of both constructs and was intensified with the addition of Wnt7a ligand.
Overexpression of Wnt7a Induces the Fzd5 Interaction with xDsh and Reconstitution of eGFP
Co-immunoprecipitation was performed to confirm that the Fzd5-xDsh interaction was not disturbed by the fusion of the eGFP fragments to the C-terminal of both proteins. The xDsh-NeGFP and Fzd5-CeGFP expression constructs were individually transfected and immunoprecipitated with anti-NGFP and anti-CGFP, respectively. Western blot analysis was performed using an antibody against full length GFP (Figure 2B). xDsh-NeGFP and Fzd5-CeGFP co-immunoprecipitated with anti-NGFP after crosslinking with 2mM DSP and immunoblot was performed with anti-GFP (Figure 2B). This demonstrated a physical association between the two eGFP-fragment fusion proteins, presumably due to the presence of endogenous Wnt ligands.
To investigate Wnt7a binding and visualize the subsequent interaction of xDsh with the intracellular domain of the Fzd5 receptor, we co-transfected the expression vectors encoding xDsh-NeGFP and Fz5-CeGFP into Ishikawa cells. Cells were then examined 24 hours post-transfection using fluorescence microscopy. The expression of either xDsh-NeGFP or Fz5-CeGFP alone did not produce any significant fluorescence (Figure 2C, top left and right panels). There was observable fluorescence with the expression of both constructs together, in part due to endogenous Wnt ligand, including Wnt7a (Figure 2C, bottom left panel). Since we have already demonstrated that Wnt7a functions as a ligand for Fzd5, we wanted to explore whether co-transfection of Wnt7a with the xDsh-NeGFP and Fz5-CeGFP constructs would enhance eGFP complementation [Carmon and Loose, unpublished]. We found a marked increase in the fluorescence signal with co-transfection of Wnt7a (Figure 2C, bottom right panel). These findings demonstrate that eGFP fragment complementation is indicative of Wnt signaling activation due to Wnt7a binding to Fzd5.
Purified Wnt7a Activates β-catenin and eGFP Fluorescence
In order to monitor activation of Wnt signaling subsequent to treatment with purified recombinant Wnt7a using the split eGFP method, we initially modified Ishikawa cells for stable expression of xDsh-NeGFP and Fzd5-CeGFP. Ishikawa cells were transfected with either pcDNA3.1 vector or co-transfected with xDsh-NeGFP and Fzd5-CeGFP to establish the null and xDshN/Fzd5C stable cell line, respectively. Since Ishikawa cells natively express Wnt3, Wnt3a, and Wnt7a, the xDshN/Fzd5C cells were observed to elicit a diminutive amount of fluorescence due to GFP reconstitution (Figure 3A). The stable expression of both xDsh-NeGFP and Fzd5-CeGFP constructs was confirmed using RT-PCR and protein levels were assessed by immunoprecipitation and western blot (Figure 3B and 3C).
We next attempted to visualize alterations in fluorescence with the addition of purified Wnt7a protein. There was an observed increase in fluorescence intensity 24 hours after the addition of 300 ng/ml Wnt7a (Figure 4A, right bottom panel). This concentration was selected since it is in excess to previous reports of an EC50 = 100 ng/ml for Wnt3a and our finding of EC50 = 65 ng/ml for Wnt7a activation of β-catenin/canonical signaling [24]. No fluorescence was observed in eGFP null cells treated with or without Wnt7a (Figure 4A, left top and bottom panels). There was some fluorescence detected in the untreated xDshN/Fzd5C cells due to the presence of endogenous Wnts.
Figure 4. Purified Wnt7a Induces Reconstituion of split-eGFP and Activates βcatenin/Canonical Wnt Signaling.
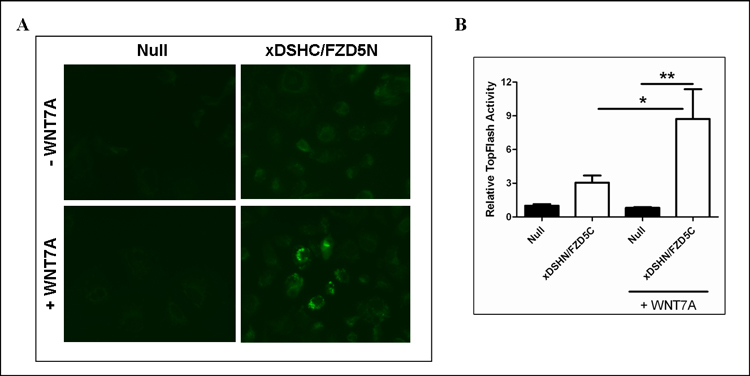
(A) Stable xDshN/Fzd5C cells and null cells were seeded on coverslips and treated with purified Wnt7a (300 ng/ml). Cells were fixed 24 hours post-treatment and images were collected via fluorescence microscopy at 400 x magnification. (B) Purified Wnt7a activates TopFlash in xDshN/Fzd5C cells. Luciferase activity was measured 24-h post-transfection. Results are representative of at least three separate experiments performed in duplicate ± S.E. * and **, indicate (p≤0.05) and (p≤0.01) based on ANOVA and Bonferroni post-hoc for multiple comparisons.
To determine if the Wnt7a treatment could successfully induce β-catenin/canonical Wnt signaling in our xDshN/Fzd5C cell line, we employed the luciferase reporter construct TopFlash. The TopFlash construct contains TCF-binding sites which are directly activated by the TCF/β-catenin complex. Both null and xDshN/Fzd5C cells were transiently co-transfected with TopFlash and a Renilla-TK luciferase reporter for normalization. TopFlash activity was 3-fold higher in xDshN/Fzd5C cells (Figure 4B). When xDshN/Fzd5C cells were treated with 300 ng/ml Wnt7a, β-catenin activity increased approximately 3-fold and 9-fold compared to treated eGFP null cells. No significant activation of canonical signaling was detected in treated versus untreated eGFP null cells. Since Ishikawa cells contain endogenous Wnts, including Wnt7a, it is likely the binding of endogenous Wnts to Fzd5 accounts for the increase in TopFlash activity in untreated xDshN/Fzd5C. Altogether, it is apparent that Wnt7a activation of β-catenin/canonical signaling is mediated through Fzd5 resulting in the reconstitution of both fragments of the split-eGFP.
In this study, we have demonstrated the utility of the split eGFP complementation approach for the visual detection of Wnt7a-Fzd5 interactions and subsequent Wnt signaling activation in endometrial cancer cells. This assay and our ELISA-based protein-protein binding technique may be employed as important biological tools for the identification of additional Wnts and/or other ligands that bind specific Fzds. The feasibility of high-throughput split eGFP complementation analysis could prove useful in screening for molecules that modulate Wnt signaling at the level of the cell membrane and to reveal novel compounds for drug discovery in the treatment of Wnt signaling-associated diseases.
Acknowledgements
We are grateful to the following individuals for providing reagents: J.B. Wallingford (U.T. Austin) for xDshGFP and O. Destree and H. Clevers (U. Utrecht) for TopFlash. This work was supported by a training fellowship from the Keck Center for Interdisciplinary Bioscience Training of the Gulf Coast Consortia, the American Legion Auxiliary Fellowship, and the NIH Uterine Cancer Specialized Programs of Research Excellence Grant # P50-CA098258.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu.Rev.Cell Dev.Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 2.Miller JR. The Wnts Genome Biol. 2002;3 doi: 10.1186/gb-2001-3-1-reviews3001. REVIEWS3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kishida S, Yamamoto H, Hino S, Ikeda S, Kishida M, Kikuchi A. DIX domains of Dvl and Axin are necessary for protein interactions and their ability to regulate beta-catenin stability. Mol.Cell.Biol. 1999;19:4414–4422. doi: 10.1128/mcb.19.6.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamai K, Zeng X, Liu CM, Zhang XJ, Harada Y, Chang ZJ, He X. A mechanism for Wnt coreceptor activation. Mol.Cell. 2004;13:149–156. doi: 10.1016/s1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- 5.Itoh K, Krupnik VE, Sokol SY. Axis determination in Xenopus involves biochemical interactions of axin, glycogen synthase kinase 3 and beta-catenin. Curr.Biol. 1998;8:591–594. doi: 10.1016/s0960-9822(98)70229-5. [DOI] [PubMed] [Google Scholar]
- 6.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 7.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc.Natl.Acad.Sci.U.S.A. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahumada A, Slusarski DC, Liu XX, Moon RT, Malbon CC, Wang HY. Signaling of rat Frizzled-2 through phosphodiesterase and cyclic GMP. Science. 2002;298:2006–2010. doi: 10.1126/science.1073776. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Yuan HD, Xie W, Mao JH, Caruso AM, McMahon A, Sussman DJ, Wu DQ. Dishevelled proteins lead to two signaling pathways - Regulation of LEF-1 and c-Jun N-terminal kinase in mammalian cells. J.Biol.Chem. 1999;274:129–134. doi: 10.1074/jbc.274.1.129. [DOI] [PubMed] [Google Scholar]
- 10.Nusse R, Varmus HE. Many Tumors Induced by the Mouse Mammary-Tumor Virus Contain A Provirus Integrated in the Same Region of the Host Genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 11.Morin PJ. beta-catenin signaling and cancer. Bioessays. 1999;21:1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura Y, Nishisho I, Kinzler KW, Vogelstein B, Miyoshi Y, Miki Y, Ando H, Horii A, Nagase H. Mutations of the adenomatous polyposis coli gene in familial polyposis coli patients and sporadic colorectal tumors. Princess Takamatsu Symp. 1991;22:285–292. [PubMed] [Google Scholar]
- 13.Prat J, Gallardo A, Cuatrecasas M, Catasus L. Endometrial carcinoma: pathology and genetics. Pathology (Phila) 2007;39:72–87. doi: 10.1080/00313020601136153. [DOI] [PubMed] [Google Scholar]
- 14.Aguilera O, Fraga MF, Ballestar E, Paz MF, Herranz M, Espada J, Garcia JM, Munoz A, Esteller M, Gonzalez-Sancho JM. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene. 2006;25:4116–4121. doi: 10.1038/sj.onc.1209439. [DOI] [PubMed] [Google Scholar]
- 15.Liu TH, Raval A, Chen SS, Matkovic JJ, Byrd JC, Plass C. CpG island methylation and expression of the secreted frizzled-related protein gene family in chronic lymphocytic leukemia. Cancer Res. 2006;66:653–658. doi: 10.1158/0008-5472.CAN-05-3712. [DOI] [PubMed] [Google Scholar]
- 16.Nojima M, Suzuki H, Toyota M, Watanabe Y, Maruyama R, Sasaki S, Sasaki Y, Mita H, Nishikawa N, Yamaguchi K, Hirata K, Itoh F, Tokino T, Mori M, Imai K, Shinomura Y. Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene. 2007;26:4699–4713. doi: 10.1038/sj.onc.1210259. [DOI] [PubMed] [Google Scholar]
- 17.Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat.Rev.Drug Discov. 2006;5:997–1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 18.Miller C, Sassoon DA. Wnt-7a maintains appropriate uterine patterning during the development of the mouse female reproductive tract. Development. 1998;125:3201–3211. doi: 10.1242/dev.125.16.3201. [DOI] [PubMed] [Google Scholar]
- 19.Miller C, Pavlova A, Sassoon DA. Differential expression patterns of Wnt genes in the murine female reproductive tract during development and the estrous cycle. Mech.Dev. 1998;76:91–99. doi: 10.1016/s0925-4773(98)00112-9. [DOI] [PubMed] [Google Scholar]
- 20.Bui TD, Lako M, Lejeune S, Curtis AR, Strachan T, Lindsay S, Harris AL. Isolation of a full-length human WNT7A gene implicated in limb development and cell transformation, and mapping to chromosome 3p25. Gene. 1997;189:25–29. doi: 10.1016/s0378-1119(96)00808-6. [DOI] [PubMed] [Google Scholar]
- 21.Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, Gotoh Y. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131:2791–2801. doi: 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- 22.Caricasole A, Ferraro T, Iacovelli L, Barletta E, Caruso A, Melchiorri D, Terstappen GC, Nicoletti F. Functional characterization of WNT7A signaling in PC12 cells - Interaction with a FZD5 center dot LRP6 receptor complex and modulation by dickkopf proteins. J.Biol.Chem. 2003;278:37024–37031. doi: 10.1074/jbc.M300191200. [DOI] [PubMed] [Google Scholar]
- 23.Dann CE, Hsieh JC, Rattner A, Sharma D, Nathans J, Leahy DJ. Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature. 2001;412:86–90. doi: 10.1038/35083601. [DOI] [PubMed] [Google Scholar]
- 24.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. Plos Biology. 2006;4:570–582. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 26.Wu CH, Nusse R. Ligand receptor interactions in the Wnt signaling pathway in Drosophila. J.Biol.Chem. 2002;277:41762–41769. doi: 10.1074/jbc.M207850200. [DOI] [PubMed] [Google Scholar]
- 27.Campbell-Valois FX, Michnick SW. Synthesis of degenerated libraries of the ras-binding domain of raf and rapid selection of fast-folding and stable clones with the dihydrofolate reductase protein fragment complementation assay Methods. Mol.Biol. 2007;352:249–274. doi: 10.1385/1-59745-187-8:249. [DOI] [PubMed] [Google Scholar]
- 28.Ding Z, Liang J, Lu Y, Yu Q, Songyang Z, Lin SY, Mills GB. A retrovirus-based protein complementation assay screen reveals functional AKT1-binding partners. Proc.Natl.Acad.Sci.U.S.A. 2006;103:15014–15019. doi: 10.1073/pnas.0606917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galarneau A, Primeau M, Trudeau LE, Michnick SW. beta-Lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein-protein interactions. Nat.Biotechnol. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh I, Hamilton AD, Regan L. Antiparallel leucine zipper-directed protein reassembly: Application to the green fluorescent protein. J.Am.Chem.Soc. 2000;122:5658–5659. [Google Scholar]
- 31.Luker KE, Smith MCP, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms DP. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc.Natl.Acad.Sci.U.S.A. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nyfeler B, Michnick SW, Hauri HP. Capturing protein interactions in the secretory pathway of living cells. Proc.Natl.Acad.Sci.U.S.A. 2005;102:6350–6355. doi: 10.1073/pnas.0501976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc.Natl.Acad.Sci.U.S.A. 2002;99:15608–15613. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang S, Ma C, Chalfie M. Combinatorial marking of cells and organelles with reconstituted fluorescent proteins. Cell. 2004;119:137–144. doi: 10.1016/j.cell.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 35.Sheldahl LC, Slusarski DC, Pandur P, Miller JR, Kuhl M, Moon RT. Dishevelled activates Ca2+ flux, PKC, and CamKII in vertebrate embryos. Journal of Cell Biology. 2003;161:769–777. doi: 10.1083/jcb.200211094. [DOI] [PMC free article] [PubMed] [Google Scholar]


