Abstract
The binding of a series of benzoxazole analogs with different amide- and ester-linked side chains to duplex DNA in the absence and presence of divalent metal cations is examined. All ligands were found to form complexes with Ni2+, Cu2+, and Zn2+, with 2:1 ligand/metal cation binding stoichiometries dominating for ligands containing shorter side chains (2, 6, 7, and 8), while 1:1 complexes were the most abundant for ligands with long side chains (9, 10 and 11). Ligand binding with duplex DNA in the absence of metal cations was assessed, and the long side-chain ligands were found to form low abundance complexes with 1:1 ligand/DNA binding stoichiometries. The ligands with the shorter side chains only formed DNA complexes in the presence of metal cations, most notably for 7 and 8 binding to DNA in the presence of Cu2+. The binding of long side-chain ligands was enhanced by Cu2+ and to a lesser degree by Ni2+ and Zn2+. The cytotoxicities of all of the ligands against the A549 lung cancer and MCF7 breast cancer cell lines were also examined. The ligands exhibiting the most dramatic metal-enhanced DNA binding also demonstrated the greatest cytotoxic activity. Both 7 and 8 were found to be the most cytotoxic against the A549 lung cancer cell line and 8 demonstrated moderate cytotoxicity against MCF7 breast cancer cells. Metal ions also enhanced the DNA binding of the ligands with the long side-chains, especially for 9, which also exhibited the highest level of cytotoxicity of the long side-chain compounds.
Introduction
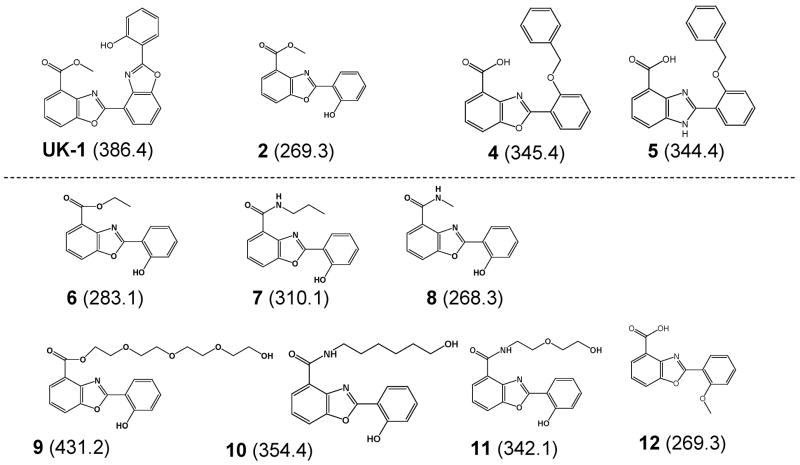
The discovery of the selective cytotoxic activity of UK-1 [1–3], a bis(benzoxazole) isolated as a secondary metabolite from Streptomyces, has stimulated the development of other benzoxazole and benzimidazole compounds with similar anticancer activities [4–6]. As a topoisomerase II inhibitor, one of the unique properties of UK-1 is its ability to bind biologically important divalent metal cations [3] and its metal-mediated DNA binding [3, 7]. While UK-1 has demonstrated cytotoxicity against a number of cell lines, it does not inhibit the growth of bacteria, yeast or fungi [1, 8], making the mechanism of this selective cytotoxic activity and the metal binding properties of UK-1 and new analogs of great interest.
In a recent study, we used electrospray ionization mass spectrometry (ESI-MS) in conjunction with cytotoxicity assays to examine several simple analogs of UK-1 to explore the metal ion binding requirements of these compounds, assess metal-mediated DNA binding and evaluate anticancer and antibacterial activity [6]. Interestingly, the only ligand that exhibited anticancer activity, 2 (Figure 1), was also the only metal-mediated DNA binder with a preference for Ni2+ as determined by ESI-MS experiments. Two other compounds, 4 and 5, formed complexes with DNA in a non-specific, non-metal mediated manner and showed antibacterial but not cytotoxic behavior. These results suggested a correlation between metal-mediated binding and anticancer activity of the compounds. To improve upon the solubility of the benzoxazoles in aqueous solutions and to allow further examination of the metal-mediated DNA binding and anticancer activity, a series of analogs of 2 have been synthesized with different ester and amide-linked side-chains (Figure 1) [9].
Figure 1.
Structures of the benzoxazole and benzimidazole ligands. Molecular weights are given (in Da) in the parentheses. UK-1, 2, 4, and 5 were examined in a previous study [6] and are only shown for comparison purposes.
In the present study, ESI-MS was used to screen the binding of the new ligands to a series of divalent metals including Mg2+, Ni2+, Cu2+, and Zn2+. Ligand binding to duplex DNA in the presence and absence of metal ions was also assessed. ESI-MS has been shown to be a useful tool for the analysis of non-covalent ligand/DNA complexes due to its sensitivity, low sample consumption, and fast analysis times that make it amenable to high throughput screening [10–12]. Early studies focused on examining well-studied, commercially available duplex DNA-binding compounds including both minor groove binders [13–16] and intercalators [17–19] to establish that the binding stoichiometries, selectivities, and specificities observed by ESI-MS correlate with known solution behavior. More recent studies have extended the use of the technique to novel compounds, including studies by our group examining the metal-dependent binding of UK-1 and related benzoxazoles and benzimidazole analogs [6, 7]. Our previous studies have demonstrated that one of the main advantages of a mass spectrometry-based analysis technique for metal-mediated DNA binding ligands over traditional spectroscopic techniques is that information about ligand/metal and ligand/metal/DNA binding stoichiometries is obtained. Moreover, typical solution methods to study the interaction of small molecules with DNA include absorption and fluorescence spectroscopy, yet the application of these methods to the types of metal ion ligands described here is made difficult by the complex equilibria involved and the effects of metal ions. The changes in UV/Vis absorbance spectra for UK-1 and analogs upon DNA binding are subtle compared to those due to metal ion binding. While UK-1 and related 2-hydroxyphenyl benzoxazoles are fluorescent, this fluorescence is quenched upon metal ion binding, which prohibits the use of the intrinsic fluorescence of these compounds in studies of their metal-mediated DNA binding. Displacement-based DNA binding assays employing DNA-binding dyes such as ethidium bromide are also precluded in the study of the metal-mediated DNA binding of UK-1 and analogs due to the quenching of the dye’s fluorescence by various transition metals of interest. Thus, a general and rapid alternative method, such as ESI-MS, for studying the metal-mediated DNA binding interactions of these compounds is needed.
For the present study, anticancer cytotoxicity assays were also done so that the activities of these analogs could be correlated to the metal binding behavior elucidated in the mass spectrometry studies and compared to other previously reported UK-1 analogs. Previous studies had identified 2 as a simplified analog of UK-1 that retains the selective cytotoxicity of the natural product [4]. The analogs of 2 examined here were designed to increase the potential for metal ion complexation and metal mediated DNA binding by modifying the ester side chain of 2, especially through the incorporation of additional sites for metal ion coordination.
Experimental
The syntheses of 2 and 12 have been previously reported [4, 20]. The synthesis of the benzoxazole analogs 6, 7, 8, 9, 10, and 11 will be reported separately [9]. The oligodeoxynucleotides (ODNs) used in this study, d(GCGGGGATGGGGCG), d(CGCCCCATCCCCGC), d(GCGGGAATTGGGCG), d(CGCCCAATTCCCGC), d(GCGGAAATTTGGCG), and d(CGCCAAATTTCCGC) were purchased from IDT Technologies (Coralville, IA) as ammonium salts. Duplex DNA was annealed by preparing equimolar (1 mM) concentrations of the non-self-complementary ODNs in 250 mM ammonium acetate. Concentrations were verified spectroscopically using Beer’s law and the extinction coefficients for the DNA strands provided by the manufacturer. The solutions were heated to 90°C and allowed to cool to room temperature over 7 hours. Analytical solutions containing a ligand and metal salt, a ligand and DNA duplex, or a ligand, metal salt and DNA duplex were prepared at equimolar (10 μM) concentrations unless noted otherwise, in 50 mM ammonium acetate solutions with 50% methanol.
The samples were directly infused at 3 μL/min into a Thermo Electron (San Jose, CA) LCQ mass spectrometer. For the DNA binding experiments, the instrument was operated in the negative ion mode with an electrospray voltage of 3.5 kV and a heated capillary temperature of 90 – 110 °C with sheath and auxiliary gas flows of 40 and 10 arbitrary units, respectively. Ligand/metal ion solutions were examined in the positive ion mode using an electrospray voltage of 4.5 kV and the same heated capillary and gas flow rates used for the solutions containing DNA.
Cytotoxicity was determined using the AlamarBlue cell viability assay as described previously [4]. Briefly, aliquots of 100 μl cell suspension (1–3 × 103 cells) were placed in microtiter plates in an atmosphere of 5% CO2 at 37 °C. After 24 h, 100 μL of culture media and 2 μl of the compound in DMSO were added to each well in duplicate, and the plates incubated an additional 72 h at 37 °C. Compounds, along with Mitomycin-C as a positive control, were evaluated at final concentrations ranging from 0.001 to 50 μM. Cell viability was determined by removing the culture media from each well, and adding 200 μL of fresh media and 20 μL of AlamarBlue reagent (Biosource), followed by an additional 6 h incubation prior to fluorescence measurement. Fluorescence was measured by a Beckman Coulter DTX880 plate reader with excitation at 530 nm and emission at 590 nm. The percent growth was calculated from the fluorescence data using the equation:
| (1) |
where:
Fo = the averaged measured fluorescent intensities of AlamarBlue reagent at the time just before the exposure of the cells to the test substance.
Ft = the averaged measured fluorescent intensities of AlamarBlue reagent after 72 h exposure of the cells to the test substance at a particular concentration.
Fc = the averaged measured fluorescent intensities of AlamarBlue reagent after 72 h exposure of the cells to the vehicle without the test substance.
The concentration of compound required to inhibit growth by 50% (IC50) was determined by nonlinear regression fitting the %Growth data to the equation:
| (2) |
where:
x = compound concentration.
y = % Growth.
Min = the minimum response plateau (0%Growth).
Max = the maximum response plateau (100% Growth).
H = the Hill slope co-efficient.
Cytotoxicity assays were carried out versus controls in which DMSO but no benzoxazole ligands were added to the cells. The concentration of DMSO employed in these assays does not affect the growth of the cells.
Results and Discussion
Ligand Binding to Metals
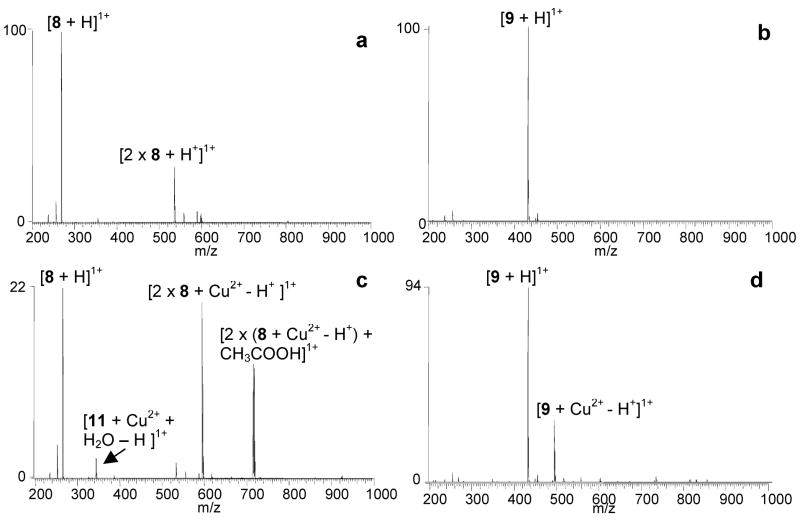
The binding of the benzoxazole ligands with metals including Mg2+, Ca2+, Ni2+, Cu2+, and Zn2+ was screened by ESI-MS to determine the binding preferences of the analogs and identify the metal ions that are most promising for enhancing ligand binding. Solutions containing a metal ion and a ligand were prepared at equimolar concentrations in the same ammonium acetate buffer used for subsequent DNA binding experiments and analyzed in the positive ion mode (Figure 2). No complexes were formed between any of the ligands and either Mg2+ and Ca2+. Figure 2 shows mass spectra for solutions containing Mg2+ with 8 (Figure 2a) and 9 (Figure 2b), demonstrating the absence of ligand/metal complexes. To ensure that the binding between the ligands and these metals was not concentration dependant, the metal ion concentration was increased 10 fold to 100 μM, however no complexation was observed.
Figure 2.
ESI-mass spectra for solutions containing benzoxazole ligands and metal ions: (a) 8 and Mg2+, (b) 9 and Mg2+, (c) 8 and Cu2+, and (d) 9 and Cu2+. Solutions contained a ligand and metal salt at equimolar (10 μM) concentrations in 50 mM ammonium acetate solutions with 50% methanol.
These results appear at first to contradict the results of previous studies in which the analogs of UK-1, including 2, were found to form complexes with Mg2+ [4]. However the prior binding studies were undertaken in a solvent containing 100% methanol due to the low solubility of the early analogs in aqueous buffers [4]. When methanol was used as a solvent in the current ESI-MS experiments, 2, 6, 7, 8, 9, 10, and 11 formed abundant complexes with Mg2+ (spectra not shown). These results demonstrate the importance of the solvent in the metal ion binding of the benzoxazoles, presumably due to the differences in metal ion solvation in different solvents. To maintain consistency with the buffer used for the DNA binding experiments, the 50 mM ammonium acetate buffer with 50% methanol (v/v) was used for the remaining metal ion binding experiments.
Ligands 2, 6, 7, 8, 9, 10, and 11 formed complexes with the divalent transition metals Ni2+, Cu2+, and to a lesser degree, Zn2+. Typical ligand/metal ion binding stoichiometries ranged from 1:1 to 2:2, with the 2:2 complexes most likely being dimers of the 1:1 complexes. Ligands with the shorter side chains, 6, 7, and 8 formed abundant 2:1 complexes, as shown in Figure 2c for a solution containing 8 with Cu2+. The short side-chain ligands were also capable of forming 1:1 complexes, however a water adduct was always bound to the complex, conceivably to fill the coordination shell of the metal. The long side-chain compounds, 9, 10 and 11, were able to form abundant 1:1 complexes without a solvent adduct as demonstrated by the spectrum of 9 with Cu2+ shown in Figure 2d. It is likely that the long side chain is able to wrap partially around the metal ion upon binding, thereby filling the coordination shell without addition of solvent molecules.
The only benzoxazole ligand that did not form complexes with Ni2+, Cu2+, and Zn2+ was 12 (spectra not shown). This compound is the only analog examined in this study that contains a methyl ester group rather than a hydroxyl group on the phenyl moiety of the compound. The 2-(2′-hydroxylphenyl)benzoxazole moiety is also present in synthetic metal ion chelators [21, 22], and is believed to play a key role in the metal ion chelation of the compounds [3]. The lack of metal ion binding observed by 12 in the ESI-MS experiments further implicates the role of the hydroxyl phenyl group in the metal ion binding of the benzoxazoles and makes 12 a good negative control compound for ligand/metal binding examined by ESI-MS.
The collisionally activated dissociation (CAD) mass spectra of the ligand/metal complexes were also examined by ESI-MS. Upon collisional activation, the 2:1 complexes containing 6, 7 and 8 dissociate via the ejection of a neutral ligand with the rapid adduction of a water molecule (always present in the trap in trace amounts) to the resulting 1:1 complex (spectra not shown). This is the same fragmentation pattern reported for complexes containing the anti-cancer analog 2 with Cu2+ and Ni2+ [6]. When the resulting 1:1 complexes were subjected to a second stage of CAD (MS3), the complexes did not undergo further observable fragmentation. It is likely that the attached water molecule is dislodged during collisional activation, but then rapidly re-attaches prior to ion detection. This type of solvent adduction process has been commonly observed for transition metal complexes in a quadrupole ion trap [23–27]. This was the same fragmentation pattern observed for the dissociation of the water-solvated 1:1 complexes containing 6, 7 and 8 and a transition metal.
Collisional activation of the 2:1 benzoxazole:metal complexes that contained the ligands with longer side-chains, 9, 10, and 11, showed dissociation via the loss of one ligand, leaving 1:1 complexes (spectra not shown). Upon MS3, these 1:1 complexes produced different fragmentation pathways that were dependent on the ligand. Complexes containing 7 and 9, the ligands with a polyethylene glycol side chain, dissociated via the loss of small portions of the side chain, such as C2H4O, while the metal ion remained bound to the remainder of the ligand. These losses are not observed for 10, which contains a six carbon alkyl chain with a terminal hydroxyl group. Instead, the 1:1 complexes containing 10 dissociate via the loss of the metal ion with the spontaneous adduction of methanol or water. In general, these initial ESI-MS experiments not only revealed that Ni2+, Cu2+, and Zn2+ are apparently the favored metals for ligand binding, but also showed differences in the preferred binding stoichiometries and fragmentation patterns for complexes containing ligands with long versus short side-chains.
DNA binding of the ligands without metals
In previous ESI-MS studies, benzoxazole and benzimidazole ligands that exhibited antibacterial activity were found to bind to DNA in the absence of metal cations, while compounds exhibiting anticancer activity, notably UK-1 and 2, only formed complexes with duplex DNA in a metal-mediated manner. Therefore, the DNA binding of the new analogs of 2 (Figure 1) in the absence of metal cations was of great interest and was examined by ESI-MS in the present study. Three non-self-complementary DNA duplexes, d(GCGGGGATGGGGCG/CGCCCCATCCCCGC), d(GCGGGAATTGGGCG/CGCCCAATTCCCGC), and d(GCGGAAATTTGGCG/CGCCAAATTTCCGC), were used for the initial screening study. The duplex sequences were selected to have different degrees of G/C and A/T base pair composition to investigate possible sequence preferences of the ligands. Solutions containing one ligand and one duplex at equimolar (10 μM) concentration were prepared in 50 mM ammonium acetate with 50% methanol. The high methanol composition was necessary to ensure the ligands remained soluble in the analytical solutions. To allow for comparison of the DNA binding of the new analogs with 2, experiments involving 2 were included in the present study to maintain consistency with the earlier study [6].
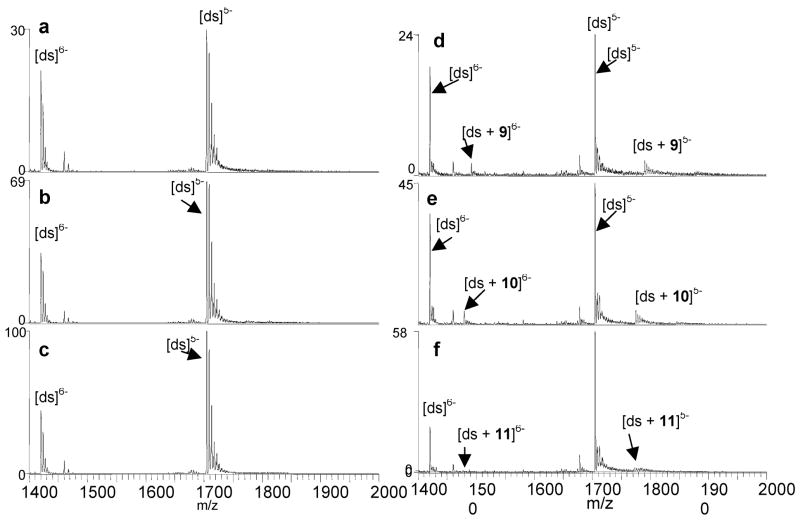
As demonstrated by the series of spectra shown in Figure 3 for solutions containing one of six ligands with d(GCGGGAATTGGGCG/CGCCCAATTCCCGC), three of the ligands, 9 (Figure 3d), 10 (Figure 3e), and 11 (Figure 3f) were found to form low abundance complexes with 1:1 ligand/DNA binding stoichiometries, while 6 (Figure 3a), 7 (Figure 3b) and 8 (Figure 3c) did not form any complexes detectable by ESI-MS. As expected, 2 likewise did not bind to DNA in the absence of metal ions (spectra not shown). The binding results of the ligands with the other two duplexes, d(GCGGGGATGGGGCG/CGCCCCATCCCCGC) and d(GCGGAAATTTGGCG/CGCCAAATTTCCGC), were similar to those shown in Figure 3, suggesting that these ligands do not have significant sequence selectivities. The ligands that do bind to the duplex all contain ester- or amide-linked side chains that are longer than those compounds that did not bind to duplex DNA, suggesting the side chains could play a role in promoting non-metal mediated DNA binding of the benzoxazoles.
Figure 3.
ESI-mass spectra of solutions containing duplex 3/4 and (a) 6, (b) 7, (c) 8, (d) 11, (e) 9, and (f) 10. Solutions contained a ligand, metal salt and DNA duplex at equimolar (10 μM) concentrations in 50 mM ammonium acetate solutions with 50% methanol.
The non-metal mediated binding of 9, 10 and 11 differs from that of the antibacterial ligands 4 and 5 examined in our previous study. The complexes formed by 4 and 5 had greater relative abundances, and the binding stoichiometries ranged from 1:1 to 3:1 and were highly dependent on ligand concentration [6]. The binding behavior of ligands 4 and 5 was similar to that of commercial intercalators examined in previous ESI-MS studies [17, 28]. While 9, 10 and 11 formed complexes with duplexes without metals, the binding stoichiometries never exceed 1:1, and their relative abundances are low.
DNA binding of the ligands with metals
Metal cations are documented to play a key role in the DNA binding of UK-1 and 2; the 1:1 binding of divalent metal ions by these ligands can result in cationic complexes with increased electrostatic attraction towards DNA. In analogy with the magnesium-dependent DNA binding by quinobenzoxazines [29], the metal ion-mediated binding of UK-1 and analogs with DNA may involve shared coordination of the ligand and phosphate groups on the DNA with the metal ion. This metal-mediated binding behavior is thought to be related to the anti-cancer activity of these ligands over other non-metal-mediated benzoxazole and benzimidazole compounds. Previous studies of the solution metal ion binding ability of UK-1 and analogs had indicated that Mg2+ and Zn2+ were implicated in the metal-mediated DNA binding by these compounds [4], while in ESI-MS studies Ni2+, Co2+, and Zn2+ promoted the greatest duplex binding for UK-1 [7], and the duplex binding of 2 was found to be mediated by Ni2+ [6].
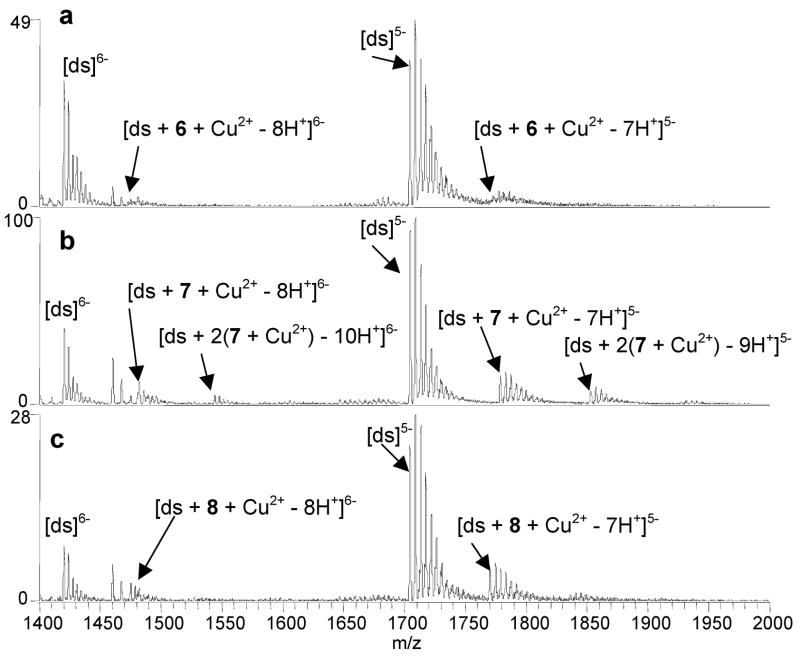
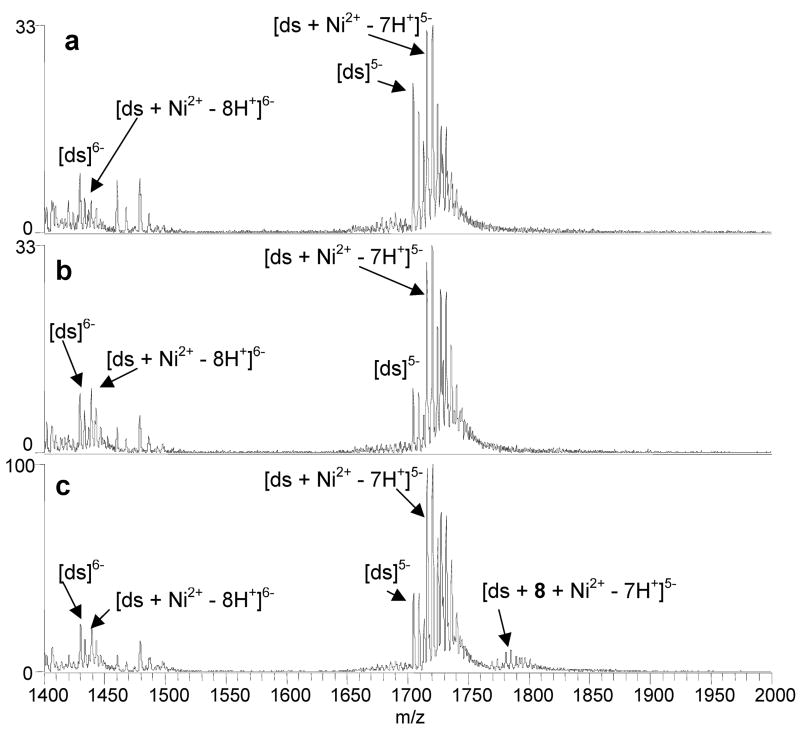
To determine if the binding of the new benzoxazole ligands is also metal-mediated, ESI-MS experiments were undertaken for solutions containing equimolar (10 μM) concentrations of a benzoxazole ligand, duplex d(GCGGGAATTGGGCG/CGCCCAATTCCCGC), and a metal salt. Based on the first section of results described above, only the metals that were found to form complexes with the ligands, Ni2+, Cu2+, and Zn2+, were used in this phase of the study. The most significant enhancement in duplex/ligand binding was observed with Cu2+ as demonstrated by the spectra shown in Figure 4, which correspond to solutions containing Cu2+, the duplex and either 6 (Figure 4a), 7 (Figure 4b), and 8 (Figure 4c). While none of these three ligands bound DNA in the absence of metal ions (i.e. Figure 3a, 3b, 3c), complexation is observed for all three in the presence of copper, with the most significant degree of binding observed for 7 and 8. Complexes containing ligand/metal/DNA binding stoichiometries of 1:1:1 and 2:2:1 were observed for 7 and 8, while only low abundance 1:1:1 were formed with 6. Similar results were obtained for 2 (spectra not shown). The ions that are not labeled in Figure 4 correspond to sodium adducts of the DNA and ligand/Cu2+/DNA complexes, with the source of sodium contamination likely being the benzoxazole solutions.
Figure 4.
ESI-mass spectra of solutions containing duplex 3/4 and Cu2+ with (a) 6, (b) 7, and (c) 8. Solutions contained a ligand, metal salt and DNA duplex at equimolar (10 μM) concentrations in 50 mM ammonium acetate solutions with 50% methanol.
While 9, 10 and 11 were found to bind to duplex DNA even in the absence of metal cations, their binding was enhanced in the presence of Cu2+, most significantly for 9 and 10 (spectra not shown). To compare the changes in the degree of ligand binding upon addition of the metal salt, the fraction of bound DNA values were calculated by expressing the sum of the abundances of all ions attributed to DNA/ligand complexes as a fraction of the total abundances of all DNA-containing ions (both free DNA and DNA/ligand complexes) as has been previously reported for other ligand/DNA complexes [30]. The abundances for all of the sodium adducts associated with each complex were included in the calculation, and only ions in the 5- charge state were used since this was the dominant charge state in the mass spectra. Inclusion of the abundances for the 6- charge state does not change the results. The increase in the extent of binding by 9, 10 and 11 is reflected in the fraction of bound DNA values summarized in Table 1. The results here and summarized in Table 1 demonstrate that Cu2+ has the greatest impact on ligand binding, enhancing the binding of 9, 10 and 11 and promoting binding by the other ligands with the most dramatic metal-mediated behavior seen for 7 and 8, and to a lesser degree, 6.
Table 1.
Fraction of bound DNA valuesa for the benzoxazole ligands and d(GCGGGAATTGGGCG/CGCCCAATTCCCGC) with Ni2+, Cu2+, or Zn2+ and in the absence of metals. Solutions contained equimolar (10 μM) concentration of ligand, DNA, and, where appropriate, metal ion.b
| Ligand | No metal | Ni2+ | Cu2+ | Zn2+ |
|---|---|---|---|---|
| 2 | 0 | 0.18 | 0.17 | 0 |
| 6 | 0 | 0 | 0.14 | 0 |
| 7 | 0 | 0 | 0.40 | 0 |
| 8 | 0 | 0.14 | 0.27 | 0 |
| 9 | 0.17 | 0.34 | 0.27 | 0.27 |
| 10 | 0.20 | 0.13 | 0.32 | 0.17 |
| 11 | 0.09 | 0.08 | 0.15 | 0.17 |
All values ± 0.05. This error represents the largest standard deviation for experiments that were repeated on three separate days.
All solutions were prepared in a solvent composed of 50 mM ammonium acetate with 50% methanol (v/v).
In general, Ni2+ had a small impact on ligand binding as demonstrated by the spectra shown in Figure 5. Extensive binding of Ni2+ to the duplex DNA is observed in Figure 5, but this does not translate into an enhancement of ligand binding. Ligand 8 was the only new analog that exhibited binding in the presence of Ni2+ yet did not bind to DNA without metal ions, as shown in Figure 5c. A low abundance 1:1:1 complex containing 8 is formed (Figure 5c), but with lower abundance relative to the complexes formed by 8 in the presence of Cu2+ (Figure 4c). The duplex binding of 9 was also enhanced in the presence of Ni2+ (spectra not shown). For 9, the fraction of bound DNA increased from 0.17 in the absence of metal ions to 0.34 in the presence of Ni2+ (Table 1). Experiments with 2 confirmed the results of our previous study that found the binding of the ligand to be Ni2+-mediated. The fraction of bound DNA values based on the 2/Ni2+/DNA complexes was 0.18 which is similar to that of the 8/Ni2+/DNA complexes, 0.14.
Figure 5.
ESI-mass spectra of solutions containing duplex 3/4 and Ni2+ with (A) 6, (B) 7, and (C) 8. Solutions contained a ligand, metal salt and DNA duplex at equimolar (10 μM) concentrations in 50 mM ammonium acetate solutions with 50% methanol.
Figures 5A and 5B show that neither 6 nor 7 bound to the DNA with Ni2+, nor did Ni2+ enhance the binding of 10 or 11 (spectra not shown). As summarized in Table 1, the fraction of bound DNA values for 10 and 11 in the absence of metal ions were 0.09 and 0.20, respectively (Figures 3e and 3f). While both ligands formed 1:1:1 ligand/Ni2+/DNA complexes, the relative abundances of these complexes were either the same as or lower than the abundances of the complexes formed in the absence of metal ions. This is reflected in the fraction of bound DNA values for solutions containing the ligands with Ni2+ and DNA which were calculated to be 0.13 for 10 and 0.08 for 11.
Zn2+ did not have a significant impact on the benzoxazole ligand binding to DNA. Zn2+ did not promote DNA binding by 6, 7, 8, or 2, the ligands that likewise did not bind to DNA in the absence of metals (spectra not shown). Ligand 10 formed 1:1:1 ligand/Zn2+/DNA complexes, however the fraction of bound DNA value for ligand binding in the presence of zinc was 0.17 which is not significantly different than it was in the absence of metal, 0.20 (spectra not shown). The DNA binding of 9 and 11 was enhanced by Zn2+, as indicated by an increase in their fraction of bound DNA values (spectra not shown). For 11, the fraction of bound DNA without metals was 0.09 and in the presence of Zn2+ it increased to 0.17, while the values increased from 0.17 to 0.27 for 9.
Aside from 2, the ligands displaying the most pronounced metal-mediated DNA binding based on the ESI-MS results are 7 and 8. The binding of 6 was also mediated by Cu2+, however the complexes formed by the ligand are lower in relative abundance than those formed by 7 and 8. This result is somewhat unsurprising due to the similarities in the structures of 2, 7 and 8. The ester linkage of 2 is changed to an amide-linkage in 8. While both 8 and 2 exhibited metal mediated binding, copper had a more positive impact on 8 binding than it did on 2. Likewise, Ni2+ promoted greater binding for 2 than it did for 8. This difference may result from the preferential coordination of Cu(II) to nitrogen atoms over oxygen atoms [1]. Similar differences can be seen for the binding of 7, which also has an amide-linked side chain that is extended by two carbon atoms compared to 8 and 2. Compared to 2, 7 demonstrates greater DNA binding with copper and no binding with Ni2+.
While the longer side-chains of 9, 10 and 11 enhance non-metal-mediated DNA binding, they tend to preclude the dramatic metal-mediated DNA binding behavior observed for the ligands with the shorter side-chains. The longer side-chains of the ligands are believed to enhance the metal binding of the ligands by wrapping around and coordinating to the metals. The ester-linked polyethylene glycol side chain of 9 produced the best enhancement of metal mediated binding, as all three metal ions, Ni2+, Cu2+, and Zn2+, increased the binding of the ligand to DNA relative to the binding in the absence of any metal. The results for 11, with the shorter, amide-linked polyethylene glycol side chain were not as favorable. While the binding was enhanced by Cu2+ and Zn2+, the overall fraction of bound DNA values were generally lower than the values for the other ligands in the presence of metals. With an amide-linked side chain that did not contain ethylene glycol groups, the binding of 10 was only enhanced by Cu2+. The binding results of the ligands with the longer side chains suggests that the longer, polyethylene glycol side chain of 9 is the most favorable for overall metal-mediated binding.
Cytotoxicity Assays
There are some interesting correlations between the degree of metal-mediated binding behavior of the benzoxazole ligands determined by ESI-MS and their cytotoxicity to two cancer cell lines. As summarized by the IC50 values shown in Table 2, the new ligands with the greatest cytotoxicity against the A549 lung cancer cell line were 7, 8, and 9, all having IC50 values that were similar to 2. Both 7 and 8 exhibited the most dramatic degree of metal-mediated DNA binding as their fraction of bound DNA values increased from 0 in the absence of metal ions to 0.40 and 0.27, respectively, in the presence of Cu2+. Ligand 8 was also found to form complexes in the presence of Ni2+. While 9 was not a truly metal-mediated DNA binding ligand, it exhibit the most consistent and dramatic enhancement in DNA binding in the presence of metal ions compared to all the ligands that bound to DNA in the absence of metal ions.
Table 2.
IC50 values(in μM) for the benzoxazole compounds against the A549 (lung cancer) and MCF7 (breast cancer) cell lines.
| Ligand | A549 | MCF7 |
|---|---|---|
| 2 | 12 ± 3 | 4 ± 2 |
| 6 | 40 ± 10 | 15 ± 5 |
| 7 | 14 ± 4 | 30 ± 10 |
| 8 | 11 ± 1 | 10 ± 8 |
| 9 | 11 ± 1 | 13 ± 2 |
| 10 | 39 ± 11 | 31 ± 7 |
| 11 | 41 ± 17 | ➢50 |
| 12 | > 50 | > 50 |
Ligands 8 and 9 also showed cytotoxicity against the MCF7 breast cancer cell lines with IC50 values of 10 ± 8 μM and 13 ± 2 μM, respectively. Both of these values are on par with the IC50 value of 2 (4 ± 2 μM). Interestingly, 7, which showed activity against A549 cells comparable to that of 2 was relatively inactive against MCF7 cells (IC50 = 30 ± 10 μM), and 6, which was relatively inactive against A549 cells, retained some activity against MCF7 cells (IC50 = 15 ± 5 μM). At this point the reasons for these differences are unclear, but we note that of the two analogs of 2 that incorporate the most conservative structural changes, 6 and 8, only 8 demonstrates enhanced metal-mediated DNA binding relative to 2 and this analog also displays the most comparable cytotoxicity against both cancer cell lines, relative to 2. Ligand 9 also demonstrated good cytotoxic activity, and while it was not a truly metal-mediated DNA binder, its DNA complexation was consistently enhanced by the presence of Cu2+, Ni2+, and Zn2+. In contrast, compound 12, which does not bind metal ions, was the only compound in this series that did not display cytotoxicity towards either cell line.
Conclusions
The correlation between significant metal-mediated or metal-enhanced binding determined by ESI-MS and anticancer activity of the benzoxazoles ligands assessed by cytotoxicity assays is demonstrated in this study. ESI-MS experiments reveal that Cu2+ and Ni2+ form the most abundant complexes with 6, 7, 8, 9, 10, and 11, while less abundant complexes are formed with Zn2+. For the complexes containing short side-chains, 6, 7, and 8, 2:1 ligand/metal ion binding stoichiometries were predominant, whereas the compounds with longer side-chains, 9, 10, and 11, formed abundant 1:1 complexes. DNA binding experiments reveal that the analogs with longer side-chains formed complexes with duplex DNA in the absence of metal ions, while those with shorter side-chains did not.
Of the ligands that did not bind to duplex DNA in the absence of metal ions, the DNA binding by 7 and 8 was enhanced most dramatically by Cu2+. Ni2+ influenced duplex binding for 8 and 2, and enhanced the binding by 9. The metal ion with the least substantial effect was Zn2+ which only enhanced the binding of 9 and 11, two ligands that formed complexes with DNA regardless of the presence of metals.
Of the compounds examined in this study, both 7 and 8 were also found to be the most cytotoxic against the A549 lung cancer cell line and 8 demonstrated moderate cytotoxicity against MCF7 breast cancer cells. Metal ions also enhanced the DNA binding of the ligands with the long side-chains, most notably for 9, which also exhibited the highest level of cytotoxicity of the long side-chain compounds.
Acknowledgments
Funding from the Robert A. Welch Foundation (F1155 to JSB), the USDA (58-3148-5-106 to SMK) and the National Institutes of Health is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ueki M, Ueno K, Miyadoh S, Abe K, Shibata K, Taniguchi M, Oi SM. UK-1, A Novel Cytotoxic Metabolite from Streptomyces Sp 517-02.1. Taxonomy, Fermentation, Isolation, Physicochemical and Biological Properties. J Antibiot. 1993;46:1089–1094. doi: 10.7164/antibiotics.46.1089. [DOI] [PubMed] [Google Scholar]
- 2.Shibata K, Kashiwada M, Ueki M, Taniguchi M. UK-1, A Novel Cytotoxic Metabolite from Streptomyces Sp 517-02.2. Structural Elucidation. J Antibiot. 1993;46:1095–1100. doi: 10.7164/antibiotics.46.1095. [DOI] [PubMed] [Google Scholar]
- 3.Reynolds MB, DeLuca MR, Kerwin SM. The Novel Bis(benzoxazole) Cytotoxic Natural Product UK-1 is a Magnesium Ion-Dependent DNA Binding Agent and Inhibitor of Human Topoisomerase II. Bioorg Chem. 1999;27:326–337. [Google Scholar]
- 4.Kumar D, Jacob MR, Reynolds MB, Kerwin SM. Synthesis and Evaluation of Anticancer Benzoxazoles and Benzimidazoles Related to UK-1. Bioorg Med Chem. 2002;10:3997–4004. doi: 10.1016/s0968-0896(02)00327-9. [DOI] [PubMed] [Google Scholar]
- 5.Wang BB, Maghami N, Goodlin VL, Smith PJ. Critical Structural Motif for the Catalytic Inhibition of Human Topoisomerase II by UK-1 and Analogs. Bioorg Med Chem Lett. 2004;14:3221–3226. doi: 10.1016/j.bmcl.2004.03.095. [DOI] [PubMed] [Google Scholar]
- 6.Oehlers L, Mazzitelli CL, Brodbelt JS, Rodriguez M, Kerwin S. Evaluation of Complexes of DNA Duplexes and Novel Benzoxazoles or Benzimidazoles by Electrospray Ionization Mass Spectrometry. J Am Soc Mass Spectrom. 2004;15:1593–1603. doi: 10.1016/j.jasms.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 7.Reyzer ML, Brodbelt JS, Kerwin SM, Kumar D. Evaluation of Complexation of Metal-Mediated DNA-Binding Drugs to Oligonucleotides via Electrospray Ionization Mass Spectrometry. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.21.e103. art. no.-e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato S, Kajiura T, Noguchi M, Takehana K, Kobayasho T, Tsuji T. J Antibiot. 1997;54:102. doi: 10.7164/antibiotics.54.102. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez M, Kerwin S. 2007 in preparaion. [Google Scholar]
- 10.Hofstadler SA, Griffey RH. Analysis of Noncovalent Complexes of DNA and RNA by Mass Spectrometry. Chem Rev. 2001;101:377–390. doi: 10.1021/cr990105o. [DOI] [PubMed] [Google Scholar]
- 11.Beck JL, Colgrave ML, Ralph SF, Sheil MM. Electrospray Ionization Mass Spectrometry of Oligonucleotide Complexes with Drugs, Metals, and Proteins. Mass Spectrom Rev. 2001;20:61–87. doi: 10.1002/mas.1003. [DOI] [PubMed] [Google Scholar]
- 12.Hofstadler SA, Sannes-Lowery KA. Applications of ESI-MS in Drug Discovery: Interrogation of Noncovalent Complexes. Nat Rev Drug Dis. 2006;5:585–595. doi: 10.1038/nrd2083. [DOI] [PubMed] [Google Scholar]
- 13.Gale DC, Smith RD. Characterization of Noncovalent Complexes Formed Between Minor Groove Binding Molecules and Duplex DNA by Electrospray Ionization Mass Spectrometry. J Am Soc Mass Spectrom. 1995;6:1154–1164. doi: 10.1016/1044-0305(95)00530-7. [DOI] [PubMed] [Google Scholar]
- 14.Gabelica V, Galic N, Rosu F, Houssier C, De Pauw E. Influence of Response Factors on Determining Equilibrium Association Constants of Non-Covalent Complexes by Electrospray Ionization Mass Spectrometry. J Mass Spectrom. 2003;38:491–501. doi: 10.1002/jms.459. [DOI] [PubMed] [Google Scholar]
- 15.Gabelica V, Rosu F, Houssier C, De Pauw E. Gas Phase Thermal Denaturation of an Oligonucleotide Duplex and its Complexes with Minor Groove Binders. Rapid Commun Mass Spectrom. 2000;14:464–467. doi: 10.1002/(SICI)1097-0231(20000331)14:6<464::AID-RCM895>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 16.Rosu F, Gabelica V, Houssier C, De Pauw E. Determination of Affinity, Stoichiometry and Sequence Selectivity of Minor Groove Binder Complexes with Double-Dtranded Oligodeoxynucleotides by Electrospray Ionization Mass Spectrometry. Nucleic Acids Res. 2002;30:e82. doi: 10.1093/nar/gnf081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabelica V, De Pauw E, Rosu F. Interaction Between Antitumor Drugs and a Double-Stranded Oligonucleotide Studied by Electrospray Ionization Mass Spectrometry. J Mass Spectrom. 1999;34:1328–1337. doi: 10.1002/(SICI)1096-9888(199912)34:12<1328::AID-JMS889>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 18.Beck JL, Gupta R, Urathamakul T, Williamson NL, Sheil MM, Aldrich-Wright JR, Ralph SF. Probing DNA Selectivity of Ruthenium Metallointercalators Using ESI Mass Spectrometry. Chem Commun. 2003;5:626–627. doi: 10.1039/b212132h. [DOI] [PubMed] [Google Scholar]
- 19.Wan KX, Shibue T, Gross ML. Non-Covalent Complexes Between DNA-Binding Drugs and Double-Stranded Oligodeoxynucleotides: A Study by ESI Ion-Trap Mass Spectrometry. J Am Chem Soc. 2000;122:300–307. doi: 10.1016/S1044-0305(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 20.DeLuca MR, Kerwin SM. The Total Synthesis of UK-1. Tetrahedron Lett. 1997;38:199–202. [Google Scholar]
- 21.Hoveyda HR, Rettig SJ, Orvig C. Coordination Chemistry of 2-(2′-Hydroxyphenyl)-2-Benzoxazole with Gallium(III) and Aluminum(III) - 2 Uncommon Group-13 Metal Environments Stabilized by a Biologically Relevant Binding Group. Inorg Chem. 1993;32:4909–4913. [Google Scholar]
- 22.Tanaka K, Kumagai T, Aoki H, Deguchi M, Iwata S. Application of 2-(3,5,6-trifluoro-2-hydroxy-4-methoxyphenyl)benzoxazole and -benzothiazole to Fluorescent Probes Sensing pH and Metal Cations. J Org Chem. 2001;66:7328–7333. doi: 10.1021/jo010462a. [DOI] [PubMed] [Google Scholar]
- 23.Vachet RW, Callahan JH. Quadrupole Ion Trap studies of the Structure and Reactivity of Transition Metal Ion Pair Complexes. J Mass Spectrom. 2000;35:311–320. doi: 10.1002/(SICI)1096-9888(200003)35:3<311::AID-JMS918>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Williams SM, Brodbelt JS. MSn Characterization of Protonated Cyclic Peptides and Metal Complexes. J Am Soc Mass Spectrom. 2004;15:1039–1054. doi: 10.1016/j.jasms.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 25.Satterfield M, Brodbelt JS. Structural Characterization of Flavonoid Glycosides by Collisionally Activated Dissociation of Metal Complexes. J Am Soc Mass Spectrom. 2001;12:537–549. doi: 10.1016/S1044-0305(01)00236-7. [DOI] [PubMed] [Google Scholar]
- 26.Perera BA, Ince MP, Talaty ER, Van Stipdonk MJ. Gas Phase Attachment of Water and Methanol to Ag(I) Complexes with Alpha-Amino Acids in an Ion Trap Mass Spectrometer. Rapid Commun Mass Spectrom. 2001;15:615–622. doi: 10.1002/rcm.280. [DOI] [PubMed] [Google Scholar]
- 27.Vachet RW, Hartman JAR, Callahan JH. Ion-Molecule Reactions in a Quadrupole Ion Trap as a Probe of the Gas-Phase Structure of Metal Complexes. J Mass Spectrom. 1998;33:1209–1225. [Google Scholar]
- 28.Kapur A, Beck JL, Sheil MM. Observation of Daunomycin and Nogalamycin Complexes with Duplex DNA Using Electrospray Ionisation Mass Spectrometry. Rapid Commun Mass Spectrom. 1999;13:2489–2497. doi: 10.1002/(SICI)1097-0231(19991230)13:24<2489::AID-RCM816>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 29.Fan JY, Sun D, Yu H, Kerwin SM, Hurley LH. The Self-Assembly of a Quinobenzoxazine-Mg2+ Complex on DNA: A New Paradigm for the Structure of a Drug-DNA Complex and Implications for the Structure of the Quinolone Bacterial Gyrase-DNA Complex. J Med Chem. 1995;38:408–424. doi: 10.1021/jm00003a003. [DOI] [PubMed] [Google Scholar]
- 30.Mazzitelli CL, Kern JT, Rodriguez M, Brodbelt JS, Kerwin SM. Evaluation of Binding of Perylene Diimide and Benzannulated Perylene Diimide Ligands to DNA by Electrospray Ionization Mass Spectrometry. J Am Soc Mass Spectrom. 2006;17:593–604. doi: 10.1016/j.jasms.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 31.Douglas B, McDaniel D, Alexander J. Concepts and Models of Inorganic Chemistry. 3. John Wiley & Sons, Inc.; New York, NY: 1994. [Google Scholar]