Summary
Background
In the fission yeast Schizosaccharomyces pombe, cell growth takes place exclusively at both ends of the cylindrical cell. During this highly polarized growth, microtubules are responsible for the placement of the cell-end marker proteins, the Tea1-Tea4/Wsh3 complex, which recruits the Pom1 DYRK-family protein kinase. Pom1 is required for proper positioning of growth sites, and the Δpom1 mutation brings about monopolar cell growth.
Results
Pom1 kinase physically interacts with Rga4, which has a GAP (GTPase-activating protein) domain for Rho-family GTPase. Genetic and biochemical evidence indicates that Rga4 functions as GAP for the Cdc42 GTPase, an evolutionarily conserved regulator of F-actin. CRIB (Cdc42/Rac interactive binding)-GFP microscopy has revealed that GTP-bound, active Cdc42 is concentrated to growing cell ends accompanied by developed F-actin structures, where the Rga4 GAP is excluded. The monopolar Δpom1 mutant fails to eliminate Rga4 from the nongrowing cell end, resulting in monopolar distribution of GTP-Cdc42 to the growing cell end. However, mutational inactivation of Rga4 allows Cdc42 to be active at both ends of Δpom1 cells, suggesting that mislocalization of Rga4 in the Δpom1 mutant contributes to its monopolar phenotype.
Conclusions
Pom1 kinase recruited to cell ends by the Tea1-Tea4/Wsh3 complex is essential for proper localization of a GAP for Cdc42, Rga4, which ensures bipolar localization of GTP-bound, active Cdc42. Because of the established role of Cdc42 in F-actin formation, these observations provide a new insight into how the microtubule system achieves localized formation of F-actin to generate cell polarity.
Introduction
Microtubules and F-actin are two major cytoskeletal structures conserved among eukaryotic species. These polar filaments are the principal determinants of the cell polarity and morphology, which are intimately connected to the physiology and function of individual cell types. There has been emerging evidence in various organisms that microtubules provide an intrinsic cue to position cortical sites where F-actin structures are localized to generate cell polarity [1]. Currently, the fission yeast Schizosaccharomyces pombe is one of the best characterized organisms for the microtubule-dependent cell polarity [2, 3]. S. pombe is a unicellular organism of a cylindrical cell shape with microtubule bundles running along the long axis, providing the basis for polarized cell growth [4, 5]. The plus ends of microtubules growing toward cell tips carry a protein complex containing Tea1 and Tea4/Wsh3, which is then deposited to the cell-end cortex [6–8]. These proteins appear to serve as cell-end markers because they are localized to both cell ends throughout the cell cycle, marking regions for growth. Loss of the cell-end markers or factors required for their specific localization leads to frequent failure in initiation and positioning of growth sites [6–11].
Both cell tips of S. pombe are not constitutively active as growth sites during the cell cycle. Newly born daughter cells initiate growth only at the old end inherited from the mother. Later in G2 phase, the new end generated by the preceding division begins to grow, resulting in bipolar growth. This transition from monopolar to bipolar growth is known as NETO (New End Take Off) [12]. The transition also correlates with changes in the localization of F-actin patches, which are concentrated to actively growing cell ends [13, 14]. Disruption of the F-actin structures by the inhibitor drug latrunculin blocks cell tip growth [15], suggesting an essential role of F-actin in cell expansion. The localized formation of F-actin structures is likely to be brought about by Rho-family small GTPases, as is found in other eukaryotes [16]. The fission yeast genome encodes six Rho-family GTPases, Cdc42 and Rho1 to Rho5, among which Cdc42 is essential for polarized growth and F-actin organization [17]. Extensive studies of Cdc42 in diverse eukaryotes have found that the GTP-bound form of Cdc42 binds and activates a group of proteins with the CRIB (Cdc42/Rac-interactive binding) domain. The CRIB-domain proteins include p21-activated kinases (PAKs) and WASp (Wiskott-Aldrich Syndrome gene product), which induce actin polymerization and formation of F-actin structures [16].
Misplacement of the growth site and NETO defects in the tea1, tea3, and tea4/wsh3 mutants strongly suggest that the cell-end marker proteins placed by microtubules somehow contribute to formation of localized F-actin structures. Of particular interest in this regard is the protein kinase Pom1, mutations of which result in cell polarity phenotypes similar to those of the cell-end marker mutants [8, 18]. Pom1 is recruited to cell ends in a manner dependent on the cell-end markers, yet Pom1 appears to be neither a component of the cell-end marker complex nor required for the proper localization of the complex [7, 18, 19]. The simplest interpretation of these observations is that Pom1 may act as a downstream effector of the cell-end marker complex. Pom1 belongs to an evolutionarily conserved serine/threonine kinase family, DYRK (Dual-specificity tyrosine-phosphorylation-Regulated protein Kinase [20]). Both localization and activity of Pom1 kinase are important for its cellular function [18, 19], but very little is known as to how Pom1 contributes to the correct placement of F-actin structures and growth sites.
In this study, we have found that Pom1 kinase physically interacts with Rga4, which appears to function as GTPase-activating protein (GAP) for Cdc42. Rga4 is excluded from the cell tips where GTP-bound, active Cdc42 is concentrated to develop F-actin structures required for growth. Importantly, Pom1 is essential for the tip exclusion of Rga4, ensuring activation of Cdc42 at both cell tips for bipolar growth. We propose that Pom1 is a link between the microtubule-dependent cell-end markers and the Cdc42 GTPase, which organizes the actin cytoskeletons for polarized cell growth.
Results
Pom1 Kinase Interacts with Rga4, a GAP for Rho-Family GTPase
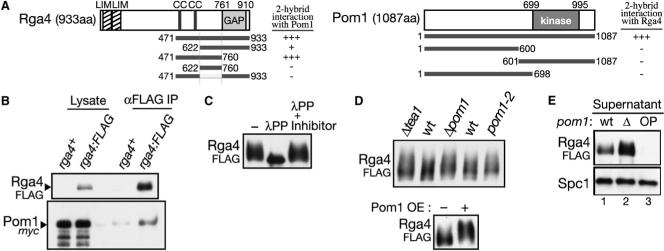
Aiming to identify effectors and regulators of Pom1 kinase, we conducted a yeast two-hybrid screen for proteins that physically interact with Pom1. The screen identified cDNA clones of the rga4+ gene (rho GTPase activating protein 4 [21]). Rga4 is a 933 amino acid protein with two LIM domains, two coiled-coil motifs, and a C-terminal GAP domain homologous to those that act on the Rho-family GTPases (RhoGAP domain; Figure 1A). Two cDNA clones isolated from the screen encode amino acid residues 471−933 and 622−933 of Rga4, respectively (Figure 1A), indicating that the N-terminal LIM domains and the coiled-coil motifs are not essential for interaction with Pom1. A further truncated clone for residues 471−760 of Rga4 demonstrated that its GAP domain also is not required for the interaction. Further deletion from its N- or C-termini of the Rga4[471−760] fragment yielded no additional positive fragments in the same assay (Figure 1A; data not shown). On the other hand, any partial Pom1 fragments examined did not interact with Rga4 (Figure 1A). Thus, both noncatalytic and catalytic domains of Pom1 kinase may be involved in the interaction with Rga4.
Figure 1. Pom1 Kinase Interacts with a RhoGAP, Rga4.
(A) Interaction between Pom1 and Rga4 in the yeast two-hybrid assay. Abbreviations: LIM, LIM domain; cc, coiled-coil; GAP, Rho-GAP domain; kinase, Ser/Thr-protein kinase domain. Domain prediction is based on the Pfam database. Numbers indicate positions in amino acid sequences. Interaction between Pom1 and Rga4 in the yeast two-hybrid assay was determined by growth on medium without histidine. “+++,” growth with 8 mM 3-AT; “+,” growth without 3-AT; “–,” no growth even without 3-AT. (B) Pom1 kinase is copurified with Rga4. pom1:myc and pom1:myc rga4:FLAG strains were grown in YES medium and their cell lysate was subjected to anti-FLAG immunoprecipitation, followed by anti-FLAG and anti-myc immunoblotting. (C) Rga4 is a phosphoprotein. Denatured crude lysate, prepared from a rga4:FLAG strain, was incubated with λ protein phophatase (λPP) with and without phosphatase inhibitors, and resolved by SDS-PAGE with reduced bis-acrylamide in the gel. (D) Pom1 affects the phosphorylation state of Rga4. Top, anti-FLAG immunoblotting was performed with the crude lysate of wild-type, Δpom1, Δtea1, and pom1−2 strains expressing Rga4FLAG. Bottom, anti-FLAG immunoblotting of the lysate prepared from the rga4:FLAG Δpom1 strain carrying the pREP1-pom1:myc before (−) and after (+) induction of Pom1myc by thiamine depletion. (E) Pom1 affects the solubility of the Rga4 protein. The supernatant was analyzed by immunoblotting. Lane 1, rga4:FLAG; lane 2, rga4:FLAG Δpom1; lane 3, rga4:FLAG Δpom1 overexpressing Pom1 from the pREP1-pom1:myc plasmid. Anti-Spc1 immunoblotting served as a loading control.
In order to detect the interaction between Pom1 and Rga4 in S. pombe, we constructed strains in which the chromosomal rga4+ and pom1+ open-reading frames (ORFs) are fused at their 3′ end with the FLAG or myc epitope sequences. The resultant strains, rga4:FLAG, rga4:myc, pom1:FLAG, and pom1:myc were indistinguishable from the wild-type strain, indicating that all the fusion proteins are functional. The Rga4 protein was insoluble in the cell lysate without nonionic detergent (data not shown), implying that Rga4 is a membrane-associated protein. Therefore, the following immunoprecipitation experiments were performed in the presence of the detergent Tween-20. From the cell lysate of the rga4:FLAG pom1:myc strain, immunoprecipitation of Rga4FLAG resulted in copurification of Pom1myc, significantly above the background level (Figure 1B). A reciprocal experiment with the rga4:myc pom1:FLAG strain further confirmed association of the two proteins (Figure S1A available online). In these coprecipitation experiments, only small fractions (<1%) of cellular Rga4 and Pom1 were detected as a complex (data not shown), implying that their interaction is transient in S. pombe. A similar experiment also detected association of Rga4 with the catalytically inactive Pom1−2 mutant protein [19], and therefore, the kinase activity of Pom1 is dispensable for its physical interaction with Rga4 (data not shown). Rga4 lacking residues 622−760, which seem to be involved in interaction with Pom1 (Figure 1A), also was coprecipitatable with Pom1, suggesting that binding of Pom1 is not limited to that region (data not shown). Interestingly, however, this internal deletion mutant of Rga4 was detected throughout the cytoplasm (Figure S1B), in contrast to wild-type Rga4 that is targeted to the cell cortex [22] (Figure 2A); residues 622−760 may contain a sequence required for the cortical localization of Rga4.
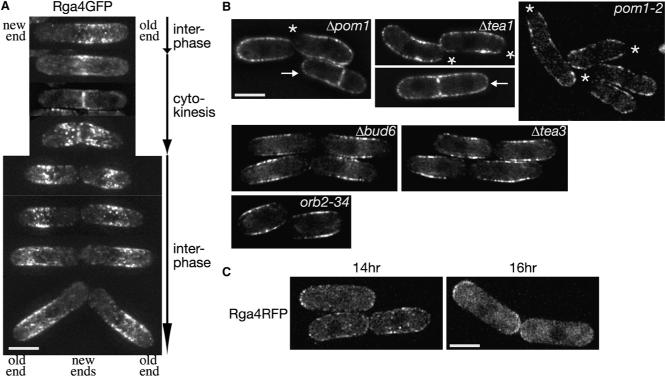
Figure 2. Cellular Localization of Rga4 Is Regulated by Pom1 Kinase.
(A) Localization of Rga4 during the cell cycle. rga4:GFP cells grown on EMM agarose were observed. Representative projection images are aligned in the order of cell cycle progression. (B) The Δpom1, pom1−2, and Δtea1 mutants show a sock-like localization pattern of Rga4. Rga4GFP was observed in Δpom1, pom1−2, Δtea1, Δbud6, Δtea3, and orb2−34 mutants. Midsection images are shown. Asterisks mark the cell ends from which Rga4GFP was excluded. Arrows indicate dividing cells. (C) Localization of Rga4 is altered by ectopic overexpression of Pom1 kinase. A rga4:RFP CRIB-GFP strain carrying the pREP1-pom1:myc plasmid was grown at 30°C in EMM medium without thiamine to overexpress Pom1 and observed at 14 hr and 16 hr after removal of thiamine. Projection images are shown. CRIB-GFP images of the same fields are presented in Figure S2A. Scale bars represent 5 μm. Images were taken at 0.4 μm steps and deconvolved.
Pom1 Kinase Affects the Phosphorylation State of Rga4
The Rga4 protein ran as a broad, smeared band in SDS-PAGE, especially in gels with reduced bis-acrylamide (Figure 1C). The smear collapsed into a sharp band when Rga4 was treated by protein phosphatase (Figure 1C), suggesting that Rga4 is phosphorylated at multiple sites. Because Pom1 kinase interacts with Rga4, we examined whether Pom1 affects the phosphorylation state of Rga4 and, thus, the mobility of Rga4 in electrophoresis. In both Δpom1 and pom1−2 mutants, Rga4FLAG still appeared as defused bands (Figure 1D), indicating that Rga4 phosphorylation is not largely dependent on Pom1 kinase. Rather, even slower migrating smears of Rga4FLAG were detectable in these strains as well as in the Δtea1 mutant, where Pom1 kinase is catalytically active but not localized to cell ends [18, 19]. The mobility of Rga4FLAG also was reduced when Pom1 was overexpressed from a plasmid (Figure 1D, bottom). Thus, the catalytic activity and localization of Pom1 affect the phosphorylation state of Rga4. However, Pom1 failed to phosphorylate Rga4 in vitro, and the rga4 mutations at the putative phosphorylation sites by DYRK (Rx1−3[S/T]P) [23] did not affect the electrophoretic mobility of Rga4 nor bring about any cellular phenotype (data not shown). Therefore, Rga4 does not seem to be a direct substrate of Pom1 kinase.
During these experiments, we noticed that the lysis buffer with the detergent Tween-20 failed to extract the Rga4 protein from cells overexpressing Pom1 (Figure 1E). On the contrary, an increased amount of Rga4 was recovered to the centrifugal supernatant from the Δpom1 cell lysate, compared to the wild-type control. The altered solubility of Rga4 may reflect Pom1-dependent changes in the subcellular localization of Rga4. Thus, we next examined whether Pom1 affects the Rga4 localization.
pom1 Mutants Fail to Exclude Rga4 from the Nongrowing Cell End
We constructed rga4:GFP and rga4:RFP alleles in which the chromosomal rga4+ ORF was fused to the GFP or RFP (Red Fluorescent Protein [24]) sequences. These fusion genes appeared to be functional because the constructed strains did not show any of the Δrga4 phenotypes [21, 22]. As has been reported [22], Rga4GFP and Rga4RFP showed punctate staining close to the cellular cortical regions, being largely excluded from both cell ends (Figure S1C). Figure 2A shows Rga4GFP localization in cells at different stages of the cell cycle. Late in G2, Rga4 was detected mostly in half of the cell with the old end inherited from the parental cell and absent from the vicinity of the new end (top). During cell division, Rga4 was concentrated toward the forming division septum (2nd and 3rd rows). Right after cytokinesis, Rga4 was repositioned from the division plane to the cell surface (4th row), moving away from the new end of the daughter cells (5th–8th rows).
Also in Δpom1 cells, Rga4GFP showed cortical staining (Figure 2B and Figure S1D), but, in contrast to wild-type cells, its signal was excluded only from one cell end (asterisk) during interphase, forming a “sock”-like localization pattern. F-actin staining (Figure S1E) showed that the Rga4-free end was the actin-rich, growing end of Δpom1 cells, which grow only in a monopolar manner [18], suggesting that exclusion of Rga4 from the nongrowing end is dependent on Pom1. In dividing Δpom1 cells, a significant amount of Rga4GFP remained throughout the cortex including cell tips (Figure 2B, arrow), in contrast to its concentrated localization to the division plate in wild-type cells (Figure 2A). Conversely, Pom1 appeared to be localized normally at both cell ends even in the Δrga4 mutant (Figure S1F). The “sock”-like localization of Rga4 also was observed in the kinase-inactive pom1−2 mutant as well as in mutants defective in Pom1 localization, Δtea1 [18] and Δtea4/wsh3 [8] mutants (Figure 2B and Figure S1D). On the contrary, other monopolar mutants, Δbud6 [25], Δtea3 [26], orb2−34 [27] and Δfor3 [28], showed Rga4 exclusion from both cell ends, as in wild-type cells (Figure 2B and Figure S1D). These results strongly suggest that both kinase activity and cell-end localization of Pom1 are important for exclusion of Rga4 from the nongrowing cell end to form the “corset”-like localization pattern. In the presence of functional Pom1, Rga4 is excluded from both growing and nongrowing ends of pre-NETO cells that have not switched to bipolar growth [22] (data not shown). Rga4 is also excluded from both ends of G1-arrested cdc10ts cells growing in a monopolar fashion (Figure S1G). Thus, it appears that the cell-end exclusion of Rga4 is not dependent on localized growth or F-actin patches at cell ends.
Consistent with Pom1-mediated regulation of the Rga4 localization, ectopic overexpression of Pom1 disturbs the cortical distribution pattern of Rga4. The characteristic corset localization of Rga4 was lost in 47% of cells (n = 247) after 14 hr induction of Pom1 from the strong nmt1 promoter [29]; Rga4RFP signals diffused throughout the cell cortex, including cell ends (Figure 2C). 16 hr after Pom1 induction, Rga4RFP often appeared more intense at cell ends (34.6% of interphase cells, n = 107). Prolonged Pom1 overexpression (>23 hr after thiamine depletion) led to aberrant cell morphology [19] with Rga4 diffused throughout the cytoplasm (data not shown).
Rga4 Is a GAP for Cdc42 GTPase
To determine which Rho GTPase is regulated by Rga4 among the six Rho-family members in S. pombe, we conducted a survey by the yeast two-hybrid assay, which detected interaction of Rga4 with the Cdc42-Q61L mutant protein, but not wild-type Cdc42 (Figure 3A). Cdc42-Q61L is a constitutively GTP-bound (“GTP-locked”) mutant [30], and its specific interaction with Rga4 suggests that Rga4 is a GAP for Cdc42 and binds only the GTP-bound form of Cdc42 to promote GTP hydrolysis. The interaction between Rga4 and GTP-Cdc42 was further confirmed biochemically; Rga4 was successfully precipitated from S. pombe cell lysate by the bacterially produced Cdc42-Q61L protein conjugated to agarose beads (Figure 3B).
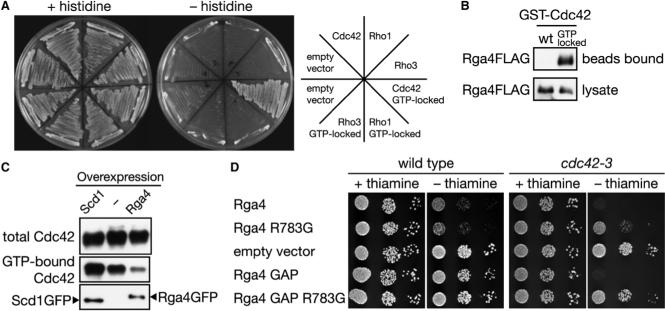
Figure 3. Rga4 Is a GAP for Cdc42.
(A) Rga4 interacts specifically with the GTP-bound form of Cdc42. Interaction of the Rga4 RhoGAP domain with the wild-type and GTP-locked mutants of Cdc42, Rho1, and Rho3 was assessed by histidine auxotrophy in the yeast two-hybrid assay. (B) Rga4 binds to the GTP-locked mutant of Cdc42. Bacterially expressed wild-type and GTP-locked mutant of GST-Cdc42 was immobilized to glutathione-beads and incubated with cell lysate of a rga4:FLAG strain, followed by anti-FLAG immunoblotting. (C) Overexpression of Rga4 reduces the cellular level of GTP-bound Cdc42. GFP-tagged Scd1 and Rga4 proteins were expressed from the nmt1 promoter in a strain carrying the HA:cdc42 allele. GTP-bound HA-Cdc42 in the cell lysate was collected by CRIB-beads, and detected by anti-HA immunoblotting. Overexpression of the GFP-tagged proteins was monitored by anti-GFP immunoblotting. (D) Overexpression of Rga4 or its GAP domain inhibits growth of the cdc42−3 mutant. Wild-type and cdc42−3 strains were transformed with the pREP1 vector (“empty vector”) and pREP1 carrying full-length rga4+, rga4R783G or their truncated fragments encoding the GAP domain (residues 681−933). Serial dilutions of the transformants were spotted onto EMM agar medium with or without thiamine and incubated at 30°C.
If Rga4 is a Cdc42 GAP, increased expression of Rga4 should reduce the cellular level of GTP-bound Cdc42. Therefore, we overproduced Rga4 in a strain expressing Cdc42 with a N-terminal HA epitope tag. The amount of GTP-bound HA-Cdc42 in cell lysate was determined by a pull-down assay using the CRIB domain immobilized on agarose beads, to which only GTP-bound, active Cdc42 can bind [31]. As shown in Figure 3C, overexpression of Rga4 significantly reduced the amount of GTP-bound HA-Cdc42 precipitatable by the CRIB-beads. The same assay detected an increase in the level of GTP-bound HA-Cdc42 after overexpression of Scd1/Ral1, a guanine nucleotide exchange factors (GEF) for Cdc42 [32, 33]. In Δrga4 cells, no significant change was detected in the cellular GTP-Cdc42 level (data not shown), probably because one or more of the other eight RhoGAPs in S. pombe [21] also function as Cdc42 GAP.
Genetic interaction between rga4+ and cdc42+ further suggested the Rga4 function as Cdc42 GAP. Through PCR-based mutagenesis, we isolated two hypomorphic alleles of cdc42, designated as cdc42−2 and cdc42−3. Overexpression of Rga4 or its GAP domain (residues 681−933) impaired the growth of these mutants more severely than that of wild-type cells (Figure 3D). This effect was reversed by mutating Rga4 Arg-783, the conserved “arginine finger” essential for GAP activity [34]. Together with the results described above, these observations indicate that Rga4 functions as a GAP for the Cdc42 GTPase.
Rga4 Regulates Polarized Distribution of GTP-Cdc42 and F-actin
Having found that Rga4 is a GAP for Cdc42, we delved into the role of Rga4 in the regulation of Cdc42 in vivo. To this aim, the GTP-bound form of Cdc42 was detected in living S. pombe cells by expressing GFP fused to a CRIB domain that specifically binds to GTP-Cdc42 [35]. Figure 4A shows representative CRIB-GFP images of a wild-type strain at different stages of the cell cycle. Late in G2 phase, CRIB-GFP was detected at both cell tips, where active cell expansion takes place (top). During M phase, CRIB-GFP became delocalized from cell tips and concentrated around the forming division septum (second and third rows). Immediately after completion of cytokinesis, CRIB-GFP was repositioned from the newly formed cell end to the old end, the initial growth site after cell division [12] (fourth and fifth rows). Later in G2 phase, CRIB-GFP reappeared also at the new cell end, showing bipolar localization, which is likely to represent NETO (sixth and seventh rows). Note that Cdc42 is not localized in the nucleus of fission yeast [36] and therefore, nuclear GFP signals are independent of Cdc42 but used as a convenient nuclear marker. These results strongly suggest that GTP-bound, active Cdc42 is concentrated at growing cell tips during interphase. We also examined CRIB-GFP localization in the Δscd1 mutant that lacks a Cdc42 GEF and is expected to have reduced GTP-Cdc42 levels [33]; cortical CRIB-GFP was hardly detectable in Δscd1 cells, which showed less polarized, spherical morphology (Figure 4B).
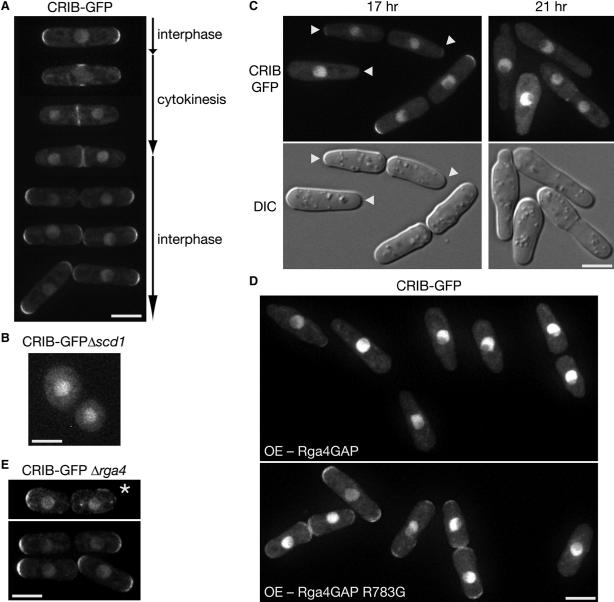
Figure 4. Rga4 Is Involved in Localization of Active Cdc42.
(A) Fluorescence microscopy of a wild-type strain expressing CRIB-GFP. Images of cells at different stages of the cell cycle are shown. (B) Fluorescence microscopy of a Δscd1 strain expressing CRIB-GFP. Cortical CRIB-GFP was hardly detectable. (C) Fluorescence (CRIB-GFP; top) and DIC (bottom) microscopy of a strain overexpressing Rga4. A wild-type strain expressing CRIB-GFP was transformed with the pREP1-rga4:FLAG plasmid and grown at 30°C for 17 hr and 21 hr in EMM without thiamine to induce Rga4FLAG expression. Arrowheads indicate the growing cell tip with reduced diameter. (D) Wild-type (Rga4GAP) or arginine-finger mutant (Rga4GAP R783G) GAP domains tagged with the FLAG epitope were overexpressed using the pREP1 vector in a CRIB-GFP strain. Cells were cultured in EMM medium without thiamine for 14 hr at 30°C to induce expression from the nmt1 promoter. Anti-FLAG immunoblotting confirmed that the wild-type and mutant Rga4 GAP domain fragments were expressed at similar levels (data not shown). (E) Fluorescence microscopy of a Δrga4 strain expressing CRIB-GFP. An asterisk marks a Δrga4 cell in which CRIB-GFP appears depolarized. Scale bars represent 5 μm. Images were taken at 0.4 μm steps and deconvolved for projection images.
Overexpression of Rga4 brings about a significant decrease in the cellular level of GTP-bound Cdc42 (Figure 3C). Consistently, 17 hr after induction of Rga4 from the nmt1 promoter, CRIB-GFP signals were almost completely lost from both ends in 36.7% of cells (n = 218) and the signals in other cells also appeared decreased (Figure 4C). The growing tip of these cells was reduced in diameter (Figure 4C, arrowheads) as reported previously [22], and residual CRIB-GFP signals occasionally were observed at the pointed tip. Prolonged Rga4 overexpression (21 hr) completely eliminated CRIB-GFP signals from the cell cortex, leading to growth inhibition (Figure 4C, right). The reduced CRIB-GFP signals in these experiments is likely to be brought about by the Cdc42 GAP activity of Rga4, because the GAP domain of Rga4, but not that of the arginine finger mutation (R783G), is sufficient to decrease cortical CRIB-GFP signals upon overproduction (Figure 4D).
In comparison with wild-type cells, Δrga4 cells are 20%−30% larger in diameter [21, 22], but CRIB-GFP staining also spanned the hemispherical ends of interphase Δrga4 cells (Figure 4E). The intensity of cortical CRIB-GFP signals in Δrga4 cells was comparable with that in wild-type cells (data not shown), suggesting that GTP-bound Cdc42 does not dramatically increase at the cell cortex in the Δrga4 mutant. However, a reduced percentage of interphase Δrga4 cells (35%, n = 60) showed evident CRIB-GFP signals at both cell tips in comparison with wild-type cells (69%, n = 97), reflecting the Δrga4 defect in bipolar cell growth [22] (Figures S2B–S2D). In some short Δrga4 cells immediately after cytokinesis (10% of interphase cells, n = 60), CRIB-GFP exhibited depolarized localization (Figure 4E, asterisk), which implies a transient delay in activation of Cdc42 at the old cell end. Thus, deletion of the rga4+ gene affects spatial control of Cdc42 activation in the cell cycle.
Polarized activation of Cdc42 is a key determinant of cellular F-actin organization and cell polarity [16]. Therefore, we examined F-actin structures in the Δrga4 mutant by fluorophore-conjugated phalloidin (Figure S2F). The percentage of Δrga4 cells with F-actin patches concentrated at both cell ends (35%, n ≥ 200) is significantly lower than that of wild-type cells (59%, n ≥ 200), consistent with the smaller population of Δrga4 cells with bipolar CRIB-GFP localization. In ∼10% of Δrga4 cells, depolarized actin patches were observed. On the other hand, F-actin cable structures appeared to be comparable between wild-type and Δrga4 cells. However, it was reported that in Δrga4 cells, the For3 formin was not properly localized to cell tips [22]; For3 is an essential nucleator of F-actin cables in interphase cells [28, 37]. Therefore, For3 localization in Δrga4 cells was reexamined by using the for3:GFP3 allele to express GFP-fused For3 [38]. We found that most interphase Δrga4 cells (92%, n = 100) showed polarized localization of For3GFP at either one end or both ends (Figure S2G). As was found with CRIB-GFP, depolarized For3GFP was observed only in a small percentage of short Δrga4 cells immediately after cell division (8% of interphase cells, n = 100) (Figure S2G, asterisk). The Δrga4 mutant seems to be delayed in re-polarizing GTP-Cdc42 at the old cell end right after division, and depolarized localization of GTP-Cdc42 in those cells may lead to temporarily dispersed For3 localization. Indeed, For3 interacts with GTP-bound Cdc42 [37], and its cell-tip localization is dependent on functional Cdc42 [39].
Cell-End Exclusion of Rga4 by Pom1 Is Important for Bipolar Activation of Cdc42
Simultaneous observation of Rga4RFP and CRIB-GFP (Figure 5A, left) demonstrated that in growing interphase cells, GTP-bound Cdc42 is confined to the cell-end cortex where Rga4, a Cdc42 GAP, is excluded. Complementary localization of Rga4 and GTP-Cdc42 also was apparent in monopolar Δpom1 cells; CRIB-GFP was concentrated exclusively to the growing tip, whereas Rga4RFP was localized over the rest of the cell (Figure 5A, right). This sock-like localization of the GAP appears to constrain GTP-Cdc42 to the single growing end of Δpom1 cells. We hypothesized that failure of the Δpom1 mutant in excluding Rga4 from the nongrowing end results in the monopolar accumulation of GTP-Cdc42, which coincides with monopolar growth. Therefore, we constructed the Δpom1 Δrga4 double mutant to examine whether deletion of rga4 allows activation of Cdc42 at both cell ends. Compared to the parental single mutants, the Δpom1 Δrga4 mutant grew 20%−30% slower, with severe flocculation. Some cells showed cytokinesis defects and significantly increased sizes due to polyploidy (data not shown). However, among the interphase cells growing more normally, 24% (n > 300) exhibited CRIB-GFP signals at both cell ends (Figure 5B, left), whereas less than 5% of Δpom1 single mutant cells (n > 200) showed bipolar distribution of CRIB-GFP. Similarly, the rga4R783G allele expressing catalytically inactive Rga4 allows bipolar CRIB-GFP distribution in interphase Δpom1 cells (30%, n = 250; Figure 5B, right). Altogether, these results suggest that Pom1 is required to eliminate Rga4 from the nongrowing cell end and that Rga4, a Cdc42 GAP, at the nongrowing end contributes to the monopolar phenotype of the Δpom1 mutant. However, detectable tip growth took place only at one end of the Δpom1 Δrga4 double mutant cells, indicating that Δrga4 cannot completely rescue the monopolar growth phenotype of Δpom1 cells (data not shown). This is probably because the Δrga4 mutation also impairs bipolar cell growth [22] (Figure S2B–D). It also is conceivable that Pom1 undertakes additional function for bipolar tip growth. Indeed, our yeast two-hybrid screens suggest that Pom1 interacts with multiple proteins (H.T., unpublished data).
Figure 5. Loss of Rga4 Allows Bipolar Distribution of Active Cdc42 in Δpom1 Cells.
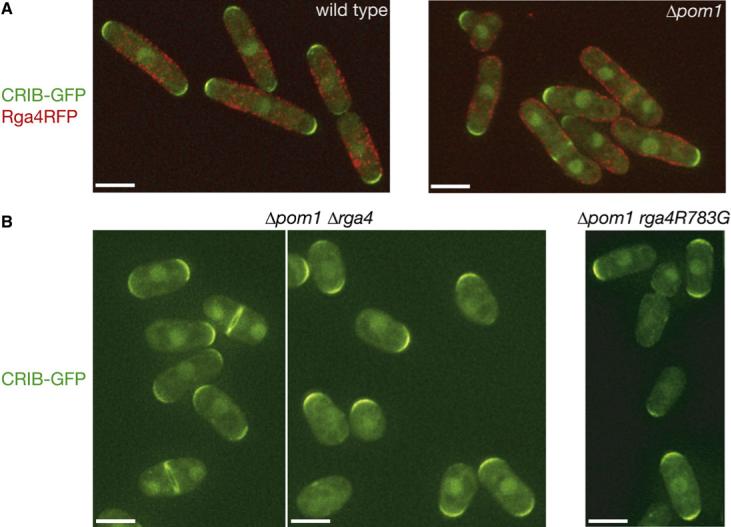
(A) Fluorescence microscopy of wild-type and Δpom1 strains expressing CRIB-GFP and Rga4RFP. (B) Fluorescence microscopy of Δpom1 Δrga4 and Δpom1 rga4R783G strains expressing CRIB-GFP. Immunoblotting confirmed that the Rga4R783G mutant protein was expressed at a level comparable to that of wild-type Rga4. Signals of CRIB-GFP and Rga4RFP were pseudo-colored green and red, respectively, and superimposed. Scale bars represent 5 μm. Images were taken at 0.4 μm steps and deconvolved for projection images.
Discussion
In fission yeast, concentrated F-actin patches continually attend growing cell tips, where active membrane expansion takes place [13, 40]. In this study, by using CRIB-GFP microscopy, we demonstrated for the first time that active growth sites in S. pombe is accompanied by the GTP-bound form of Cdc42, a conserved Rho-family GTPase essential for F-actin organization and polarized growth [17, 32, 33]. Furthermore, we determined that Rga4 [21, 22] functions as Cdc42 GAP, whose proper localization is dependent on Pom1 kinase at cell ends. It has been established that the polarized localization of Pom1 is governed by the microtubule-dependent cell-end marker proteins Tea1 and Wsh3/Tea4 [8, 18]. Therefore, the interaction of Pom1 with Rga4 appears to serve as a molecular link between the microtubule system and the polarized F-actin organization regulated by Cdc42.
Genetic, biochemical, and cell biological evidence presented in this paper consistently indicate that Rga4 functions as GAP for Cdc42. Rga4 is not likely to be the only Cdc42 GAP in S. pombe because the cellular level of GTP-bound Cdc42 does not markedly increase in the Δrga4 mutant. However, the following observations strongly suggest that Rga4 plays important roles in polarized distribution of GTP-Cdc42 within the cell. First, the complementary localizations of Rga4 and GTP-Cdc42 detected by CRIB-GFP are consistent with the idea that Rga4 defines the cortical region where Cdc42 is kept inactive. Second, without the Rga4 “corset,” active Cdc42 is expected to be more diffused and form an expanded growth zone at the cell end, which explains the increased cell diameter observed in the Δrga4 mutant [21, 22]. Third, in the Δrga4 mutant, depolarized localization of GTP-Cdc42 was often observed immediately after cell division, when cells need to re-establish Cdc42 activation at the old end. The frequent failure of Δrga4 cells in NETO [22] also suggests that this mutant is compromised in the re-organization of Cdc42 from monopolar to bipolar activation later in G2. Thus, during the transitions of the growth sites along the cell cycle, Rga4 becomes particularly important to attain the spatial reorganization of GTP-Cdc42. Impaired polarization of GTP-Cdc42 in Δrga4 cells may be responsible for the varying growth patterns under different temperatures [22] and other growth conditions (Figures S2B–S2D). However, cell polarity is not completely lost even in the absence of Rga4, probably because Cdc42 GEFs, Scd1/Ral1 and Gef1, still promote local activation of Cdc42 at cell ends [32].
We identified Rga4 as a protein that physically interacts with Pom1 kinase, and found that the phosphorylation state and solubility of Rga4 is modulated by the dosage of Pom1. Furthermore, Δpom1, pom1−2 and the cell-end marker mutants defective in Pom1 localization cannot exclude Rga4 from the nongrowing cell end, resulting in the sock-like Rga4 localization. Such mislocalization of Rga4 is not observed in other monopolar mutants, consistent with the idea that the proper corset localization of Rga4 is dependent on functional Pom1 at cell ends. Interestingly, the sock-like localization of Rga4 in pom1 mutants seems to be responsible for their monopolar activation of Cdc42 at the growing cell tip because inactivation of Rga4 allows bipolar distribution of GTP-Cdc42 in pom1 mutant cells. In addition to Rga4, Pom1 is also required for preventing cell-end distribution of Mid1/Dmf1, a key determinant of division plane positioning [41, 42]. In the Δpom1 mutant, Mid1 is dispersed over the cell cortex of the nongrowing end, leading to a division plane displaced from the cell center. Thus, a major cellular function of Pom1 kinase may be exclusion of certain cortical proteins, such as Rga4 and Mid1, from the nongrowing cell end. Consistently, in newly divided wild-type cells, Rga4 is promptly eliminated from the new end, where Pom1 is enriched [18] (Figure 2A). However, our data suggest that Rga4 is not likely to be a direct substrate of Pom1 kinase, and the exact molecular mechanism of Rga4 regulation by Pom1 remains to be elucidated. Even in Δpom1 cells, Rga4 is highly phosphorylated and detected as broad, smeared bands in SDS-PAGE. Recent studies in other yeast species identified cell cycle-dependent regulation of Cdc42 GAPs through multiple phosphorylation sites by cyclin-dependent kinases (CDKs) [43, 44]. As S. pombe Rga4 also contains 21 potential CDK phosphorylation sites, Rga4 may be subjected to intricate phosphorylation events for both CDK-dependent temporal regulation and Pom1-dependent spatial regulation described here.
Pom1 belongs to the DYRK family of Ser/Thr-protein kinases ubiquitously conserved among eukaryotes. Phylogenetic analysis suggests that DYRKs are classified into three subfamilies (Figures S3 and S4). The DYRK1 subfamily is animal specific and represented by the mammalian DYRK1, minibrain (mbk) in Drosophila, and C. elegans mbk-1. Budding yeast Yak1, Ppk15 in S. pombe, and their close orthologs form a fungal-specific subfamily. On the other hand, the DYRK2 subfamily is conserved among diverse eukaryotic organisms, including mammalian DYRK2, fly's smi35A, worm's mbk-2, and Pom1 in fission yeast. Interestingly, no member of the DYRK2 subfamily is found in budding yeast species, whose cell polarity is established by F-actin independently of microtubules [45]. Absence of a DYRK2 member in budding yeast might imply the function of the DYRK2 subfamily in the microtubule-driven cell polarity in other eukaryotes, as has been found with Pom1 [18]. Our results indicate that through a Cdc42 GAP, Pom1 plays a crucial role in translation of microtubule-dependent polarity cues into F-actin formation. Further studies of the DYRK2-family kinases in other cell systems will be of great interest to understand the conserved molecular function of DYRK2.
Experimental Procedures
S. pombe strains used in this study are listed in Table S1. Growth media and basic techniques for S. pombe have been described [46]. S. pombe cells were grown in yeast extract medium YES and synthetic medium EMM. Gene tagging with epitope and GFP sequences has been described previously [47, 48]. Live cell imaging was performed at room temperatures (23−28°C) [12]. F-actin staining with fluorescent-labeled phalloidin has been described previously [8, 14]. For the CRIB-GFP microscopy, a plasmid to express the CRIB domain (residues 1−208 of the S. cerevisiae Gic2 protein [35]) fused to three tandem copies of GFP under the control of the shk1 promoter was integrated at the ura4 locus. The procedure for yeast two-hybrid screens has been described previously [8]. Details of the screens are described in the Supplemental Data. The PCR-based method was used to isolate cdc42ts mutants, details of which are described in the Supplemental Data.
Acknowledgments
We are grateful to G. Lam, C. Lee, H. Nguyen, and C. Long for technical assistance and T. Matsumoto, E. Noguchi, P. Perez, T. Toda, R. Tsien, and the National BioResource Project for reagents. This work was supported by research grants from the Ministry of Education, Culture, Sports, Sciences and Technology of Japan (grant numbers. 18770163 and 19037004) awarded to K.N. and a National Institutes of Health grant (GM59788) awarded to K.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Supplemental Data Supplemental Experimental Procedures, four figures, and one table are available at http://www.current-biology.com/cgi/content/full/18/5/■■■/DC1/.
References
- 1.Siegrist SE, Doe CQ. Microtubule-induced cortical cell polarity. Genes Dev. 2007;21:483–496. doi: 10.1101/gad.1511207. [DOI] [PubMed] [Google Scholar]
- 2.Chang F. Establishment of a cellular axis in fission yeast. Trends Genet. 2001;17:273–278. doi: 10.1016/s0168-9525(01)02279-x. [DOI] [PubMed] [Google Scholar]
- 3.Hayles J, Nurse P. A journey into space. Nat. Rev. Mol. Cell Biol. 2001;2:647–656. doi: 10.1038/35089520. [DOI] [PubMed] [Google Scholar]
- 4.Umesono K, Toda T, Hayashi S, Yanagida M. Cell division cycle genes nda2 and nda3 of the fission yeast Schizosaccharomyces pombe control microtubular organization and sensitivity to anti-mitotic benzimidazole compounds. J. Mol. Biol. 1983;168:271–284. doi: 10.1016/s0022-2836(83)80018-7. [DOI] [PubMed] [Google Scholar]
- 5.Radcliffe P, Hirata D, Childs D, Vardy L, Toda T. Identification of novel temperature-sensitive lethal alleles in essential beta-tubulin and nonessential alpha 2-tubulin genes as fission yeast polarity mutants. Mol. Biol. Cell. 1998;9:1757–1771. doi: 10.1091/mbc.9.7.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mata J, Nurse P. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 7.Martin SG, McDonald WH, Yates JR, 3rd, Chang F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev. Cell. 2005;8:479–491. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Tatebe H, Shimada K, Uzawa S, Morigasaki S, Shiozaki K. Wsh3/Tea4 is a novel cell-end factor essential for bipolar distribution of Tea1 and protects cell polarity under environmental stress in S. pombe. Curr. Biol. 2005;15:1006–1015. doi: 10.1016/j.cub.2005.04.061. [DOI] [PubMed] [Google Scholar]
- 9.Browning H, Hayles J, Mata J, Aveline L, Nurse P, McIntosh JR. Tea2p is a kinesin-like protein required to generate polarized growth in fission yeast. J. Cell Biol. 2000;151:15–28. doi: 10.1083/jcb.151.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunner D, Nurse P. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell. 2000;102:695–704. doi: 10.1016/s0092-8674(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 11.Snaith HA, Sawin KE. Fission yeast mod5p regulates polarized growth through anchoring of tea1p at cell tips. Nature. 2003;423:647–651. doi: 10.1038/nature01672. [DOI] [PubMed] [Google Scholar]
- 12.Mitchison JM, Nurse P. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 1985;75:357–376. doi: 10.1242/jcs.75.1.357. [DOI] [PubMed] [Google Scholar]
- 13.Marks J, Hagan IM, Hyams JS. Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J. Cell Sci. Suppl. 1986;5:229–241. doi: 10.1242/jcs.1986.supplement_5.15. [DOI] [PubMed] [Google Scholar]
- 14.Arai R, Nakano K, Mabuchi I. Subcellular localization and possible function of actin, tropomyosin and actin-related protein 3 (Arp3) in the fission yeast Schizosaccharomyces pombe. Eur. J. Cell Biol. 1998;76:288–295. doi: 10.1016/S0171-9335(98)80007-1. [DOI] [PubMed] [Google Scholar]
- 15.Rupes I, Webb BA, Mak A, Young PG. G2/M arrest caused by actin disruption is a manifestation of the cell size checkpoint in fission yeast. Mol. Biol. Cell. 2001;12:3892–3903. doi: 10.1091/mbc.12.12.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 17.Miller PJ, Johnson DI. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol. Cell. Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bähler J, Pringle JR. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 1998;12:1356–1370. doi: 10.1101/gad.12.9.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bähler J, Nurse P. Fission yeast Pom1p kinase activity is cell cycle regulated and essential for cellular symmetry during growth and division. EMBO J. 2001;20:1064–1073. doi: 10.1093/emboj/20.5.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker W, Joost HG. Structural and functional characteristics of Dyrk, a novel subfamily of protein kinases with dual specificity. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:1–17. doi: 10.1016/s0079-6603(08)60503-6. [DOI] [PubMed] [Google Scholar]
- 21.Nakano K, Mutoh T, Mabuchi I. Characterization of GTPase-activating proteins for the function of the Rho-family small GTPases in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2001;6:1031–1042. doi: 10.1046/j.1365-2443.2001.00485.x. [DOI] [PubMed] [Google Scholar]
- 22.Das M, Wiley DJ, Medina S, Vincent HA, Larrea M, Oriolo A, Verde F. Regulation of cell diameter, For3p localization, and cell symmetry by fission yeast Rho-GAP Rga4p. Mol. Biol. Cell. 2007;18:2090–2101. doi: 10.1091/mbc.E06-09-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Himpel S, Tegge W, Frank R, Leder S, Joost HG, Becker W. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J. Biol. Chem. 2000;275:2431–2438. doi: 10.1074/jbc.275.4.2431. [DOI] [PubMed] [Google Scholar]
- 24.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glynn JM, Lustig RJ, Berlin A, Chang F. Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr. Biol. 2001;11:836–845. doi: 10.1016/s0960-9822(01)00235-4. [DOI] [PubMed] [Google Scholar]
- 26.Arellano M, Niccoli T, Nurse P. Tea3p is a cell end marker activating polarized growth in Schizosaccharomyces pombe. Curr. Biol. 2002;12:751–756. doi: 10.1016/s0960-9822(02)00821-7. [DOI] [PubMed] [Google Scholar]
- 27.Verde F, Mata J, Nurse P. Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J. Cell Biol. 1995;131:1529–1538. doi: 10.1083/jcb.131.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feierbach B, Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 2001;11:1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- 29.Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 30.Garcia-Mata R, Wennerberg K, Arthur WT, Noren NK, Ellerbroek SM, Burridge K. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 2006;406:425–437. doi: 10.1016/S0076-6879(06)06031-9. [DOI] [PubMed] [Google Scholar]
- 31.Benard V, Bohl BP, Bokoch GM. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J. Biol. Chem. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- 32.Garcia P, Tajadura V, Garcia I, Sanchez Y. Role of Rho GTPases and Rho-GEFs in the regulation of cell shape and integrity in fission yeast. Yeast. 2006;23:1031–1043. doi: 10.1002/yea.1409. [DOI] [PubMed] [Google Scholar]
- 33.Chang EC, Barr M, Wang Y, Jung V, Xu HP, Wigler MH. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 34.Bourne HR. G proteins. The arginine finger strikes again. Nature. 1997;389:673–674. doi: 10.1038/39470. [DOI] [PubMed] [Google Scholar]
- 35.Ozbudak EM, Becskei A, van Oudenaarden A. A system of counteracting feedback loops regulates Cdc42p activity during spontaneous cell polarization. Dev. Cell. 2005;9:565–571. doi: 10.1016/j.devcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Coll PM, Rincon SA, Izquierdo RA, Perez P. Hob3p, the fission yeast ortholog of human BIN3, localizes Cdc42p to the division site and regulates cytokinesis. Embo J. 2007;26:1865–1877. doi: 10.1038/sj.emboj.7601641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakano K, Imai J, Arai R, Toh EA, Matsui Y, Mabuchi I. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J. Cell Sci. 2002;115:4629–4639. doi: 10.1242/jcs.00150. [DOI] [PubMed] [Google Scholar]
- 38.Martin SG, Chang F. Dynamics of the formin for3p in actin cable assembly. Curr. Biol. 2006;16:1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 39.Martin SG, Rincon SA, Basu R, Perez P, Chang F. Regulation of the Formin for3p by cdc42p and bud6p. Mol. Biol. Cell. 2007;18:4155–4167. doi: 10.1091/mbc.E07-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gachet Y, Hyams JS. Endocytosis in fission yeast is spatially associated with the actin cytoskeleton during polarised cell growth and cytokinesis. J. Cell Sci. 2005;118:4231–4242. doi: 10.1242/jcs.02530. [DOI] [PubMed] [Google Scholar]
- 41.Celton-Morizur S, Racine V, Sibarita JB, Paoletti A. Pom1 kinase links division plane position to cell polarity by regulating Mid1p cortical distribution. J. Cell Sci. 2006;119:4710–4718. doi: 10.1242/jcs.03261. [DOI] [PubMed] [Google Scholar]
- 42.Padte NN, Martin SG, Howard M, Chang F. The cell-end factor pom1p inhibits mid1p in specification of the cell division plane in fission yeast. Curr. Biol. 2006;16:2480–2487. doi: 10.1016/j.cub.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 43.Sopko R, Huang D, Smith JC, Figeys D, Andrews BJ. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 2007;26:4487–4500. doi: 10.1038/sj.emboj.7601847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knaus M, Pelli-Gulli MP, van Drogen F, Springer S, Jaquenoud M, Peter M. Phosphorylation of Bem2p and Bem3p may contribute to local activation of Cdc42p at bud emergence. EMBO J. 2007;26:4501–4513. doi: 10.1038/sj.emboj.7601873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pruyne D, Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 2000;113:365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- 46.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. A laboratory course manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1993. Experiments with Fission Yeast. [Google Scholar]
- 47.Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 48.Shiozaki K, Russell P. Stress-activated protein kinase pathway in cell cycle control of fission yeast. Methods Enzymol. 1997;283:506–520. doi: 10.1016/s0076-6879(97)83040-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data Supplemental Experimental Procedures, four figures, and one table are available at http://www.current-biology.com/cgi/content/full/18/5/■■■/DC1/.