Abstract
Epithelial Na+ channels are expressed widely in absorptive epithelia such as the renal collecting duct and the colon and play a critical role in fluid and electrolyte homeostasis. Recent studies have shown that these channels interact via PY motifs in the C terminals of their α, β, and γ subunits with the WW domains of the ubiquitin-protein ligase Nedd4. Mutation or deletion of these PY motifs (as occurs, for example, in the heritable form of hypertension known as Liddle’s syndrome) leads to increased Na+ channel activity. Thus, binding of Nedd4 by the PY motifs would appear to be part of a physiological control system for down-regulation of Na+ channel activity. The nature of this control system is, however, unknown. In the present paper, we show that Nedd4 mediates the ubiquitin-dependent down-regulation of Na+ channel activity in response to increased intracellular Na+. We further show that Nedd4 operates downstream of Go in this feedback pathway. We find, however, that Nedd4 is not involved in the feedback control of Na+ channels by intracellular anions. Finally, we show that Nedd4 has no influence on Na+ channel activity when the Na+ and anion feedback systems are inactive. We conclude that Nedd4 normally mediates feedback control of epithelial Na+ channels by intracellular Na+, and we suggest that the increased Na+ channel activity observed in Liddle’s syndrome is attributable to the loss of this regulatory feedback system.
Keywords: amiloride, salivary gland, sodium current, G protein, WW domains
Epithelial Na+ channels are expressed widely in absorptive epithelia such as the renal collecting duct (1), the colon (1), and salivary ducts (2, 3) and play a critical role in the homeostatic control of fluid and electrolyte transport (4). Recent studies have shown that these channels are composed of three homologous subunits: α, β, and γ (5), each of which contains a proline-rich PY motif in its C terminal (6, 7) that interacts with the WW domains in Nedd4 (6), a widely expressed ubiquitin-protein ligase (8–10) that is believed to regulate the rate of proteolysis and the stability in the plasma membrane of the Na+ channels by catalyzing the ubiquitination of the α and γ subunits (11). Mutation or deletion of these PY motifs prevents interaction of the Na+ channel with Nedd4, leading to increased Na+ channel activity (7, 12–17). This happens, for example, in Liddle’s syndrome, an autosomal dominant form of hypertension associated with low circulating aldosterone levels and hypokalemia (18), which has been found to be attributable to mutation or deletion of the PY motifs in the β or γ subunits of the Na+ channel (13, 14, 19, 20). Binding of Nedd4 to the PY motifs of the Na+ channel thus would appear to be part of a physiological control system down-regulating Na+ channel activity. The identity of this tonically inhibitory control system is, however, unknown.
Because this unknown control system acts to inhibit Na+ channel activity tonically, it is reasonable to limit the search for candidate control systems to those that down-regulate channel activity. These control systems mediate homocellular regulation; in other words, they regulate the rate of Na+ influx across the apical membranes so as to match the basolateral extrusion rate and thereby maintain a stable cell volume and cytosolic Na+ concentration (1, 21, 22).
The proposed mechanisms that underlie homocellular regulation of Na+ channels have been the subject of much controversy. Suggested mechanisms include (i) a direct inhibitory action of extracellular Na+ (23, 24), (ii) feedback inhibition by raised intracellular Na+ (25–27), (iii) feedback inhibition by decreased intracellular pH (28), (iv) feedback inhibition by increased intracellular free Ca2+ concentration (29, 30), and (v) feedback inhibition by raised intracellular Cl− concentration (31, 32). In mouse mandibular salivary duct cells, however, it has been possible to eliminate extracellular Na+ (33), intracellular pH, and intracellular Ca2+ (25) as important mediators of homocellular regulation and to establish that the Na+ channels in these cells are regulated by feedback systems involving both intracellular Na+ and intracellular Cl− (25) acting, respectively, via the G proteins Go and Gi2 (25, 34).
In this study, we used whole-cell patch-clamp techniques to investigate whether Nedd4 is involved in feedback inhibition of Na+ channels by intracellular Na+ or by intracellular Cl−. We found that Nedd4 mediates the ubiquitin-dependent down-regulation of Na+ channel activity in response to increased intracellular Na+ and that it operates downstream of Go. In contrast, we show that Nedd4 is not involved in the feedback control of Na+ channels by intracellular Cl−.
MATERIALS AND METHODS
Cell Preparation.
Isolated cells were prepared by collagenase digestion of mandibular glands from male mice (3, 32). The standard bath solution had the following composition (in mmol/l): NaCl (145), KCl (5.5), CaCl2 (1.0), MgCl2 (1.2), NaH2PO4 (1.2), Na-Hepes (7.5), H-Hepes (7.5), and glucose (10); the pH was adjusted to 7.4 with NaOH. After establishing the whole-cell configuration, we replaced the bath with a solution containing (in mmol/l) Na-glutamate (145), NaCl (5.0), MgCl2 (1.0), H-Hepes (10), glucose (10), and EGTA (1.0); the pH was adjusted to 7.4 with NaOH. The pipettes were filled with solutions containing (in mmol/l) N-methyl-d-glucamine (NMDG)-glutamate and NaCl (together totaling 150), MgCl2 (1.0), H-Hepes (10), glucose (10), and EGTA (5.0); the pH was adjusted to 7.2 with Tris base or NaOH (7–14 mmol/l) as appropriate.
Patch-Clamp Techniques.
Standard whole-cell patch-clamp methods were used as described (3, 32). Patch-clamp pipettes were pulled from borosilicate microhematocrit tubes (Modulohm, Hevik, Denmark) so as to have resistances of 1–3 MΩ. An Ag–AgCl pellet was used as the reference electrode, and all potential differences were corrected for liquid junction potentials as appropriate (32). An Axopatch-200A patch-clamp amplifier (Axon Instruments, Foster City, CA) was used to measure whole-cell currents. To determine current–voltage relations, a MacLab-4 data acquisition interface (ADInstruments, Sydney, Australia) attached to a Macintosh-IIci computer was used to generate command voltages and to sample whole-cell currents. Amiloride-sensitive currents were measured as the differences between the whole-cell currents before and after the addition of 100 μmol/l amiloride to the bath solution. Whole-cell current–voltage relations were obtained by applying voltage pulses of 250-ms duration from a resting potential of 0 mV. Steady-state currents were calculated as the average current between 150 and 250 ms after the start of the voltage pulse.
Nedd4 Antibodies.
A polyclonal serum was raised against the carboxyl region of Nedd4 as described (9). IgG then was purified from pre-immune and anti-Nedd4 serum by standard protein A chromatography.
Glutathione-S-transferase (GST)–WW Fusion Protein.
To generate the GST–WW fusion protein, the region of Nedd4 cDNA containing the three WW domains was amplified by PCR by using the primers 5′-GGATCCCAACCAGATGCTGCCACT and 5′-GAATTCTCTTGTAACTTCTGGAGTA. The PCR product was cloned into the BamHI/EcoRI sites of pGEX-2TK (Pharmacia) and transformed into the Escherichia coli strain DH5α. Overnight cultures of E. coli harboring the GST–WW expression plasmid were diluted 1 in 50, grown for 2 hr at 37°C, induced with 1 mmol/l isopropyl β-d-thiogalactoside and grown for an additional 4 hr at 37°C. Bacterial cell pellets were resuspended in NETN (100 mmol/l NaCl/1 mmol/l EDTA/20 mmol/l Tris·HCl, pH 8.0/0.5% Nonidet P-40), were lysed by sonication, and were clarified by centrifugation at 10,000 × g for 10 min. Glutathione Sepharose (Pharmacia) was incubated with the cleared lysate for 60 min at room temperature, and then the beads were washed three times in NETN. Fusion protein was eluted five times with glutathione elution buffer. Protein concentration was measured by using a BCA kit (Pierce).
Wild-Type and Mutant GST-Ubiquitin Proteins.
A pGEX-2TK-ubiquitin construct was provided kindly by J. M. Huibregtse (Rutgers University). The ubiquitin K48R mutant was generated from pGEX-2TK-ubiquitin by using the Stratagene Quickchange method and was verified by sequencing. Wild-type and mutant GST-ubiquitin plasmids were transformed into E. coli BL21 cells, and proteins were expressed and purified as described above for GST–WW protein, except that bacterial pellets were resuspended in PBS and glutathione Sepharose-bound proteins were washed in PBS.
Immunohistochemistry.
Formalin-fixed mouse mandibular gland tissue was sectioned at a thickness of 3 μm. Sections were incubated sequentially with 1:200 dilution of the primary antibody (rabbit preimmune or the anti-Nedd4 serum) followed by biotinylated anti-rabbit IgG and ABC reagent (both from Vector Laboratories) according to the manufacturer’s protocol. The specific immune complexes were detected by using an AEC substrate kit (Vector Laboratories). Sections were counterstained with haematoxylin and mounted in Aquamount (BDH).
Chemicals.
EGTA, Tris, and Hepes were obtained from Sigma, amiloride was obtained from Research Biochemicals, and collagenase (type IV) was obtained from Worthington. Recombinant rat α-subunits of Go and Gi2 were obtained from Calbiochem and were activated as described (34, 35).
RESULTS
Nedd4 Is Expressed in Mouse Mandibular Duct Cells.
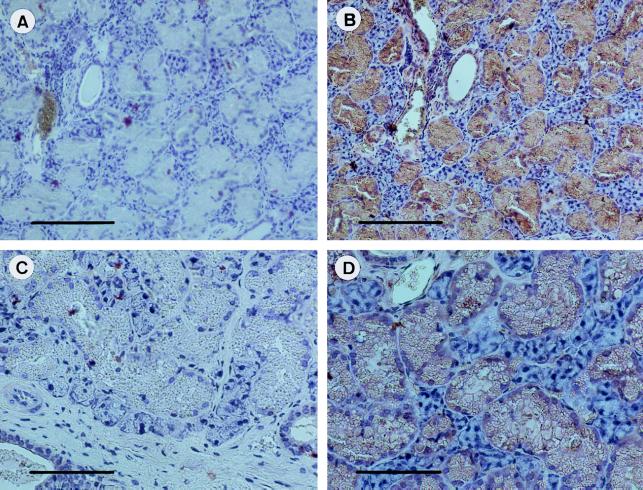
Immunohistochemistry showed marked staining of the granular ducts and the intralobular ducts of the mouse mandibular gland for Nedd4 (Fig. 1) whereas there was little or no staining of the endpieces.
Figure 1.
Expression of Nedd4 protein in the mouse mandibular gland. Sections of the formalin-fixed, paraffin-embedded mandibular gland tissue were subjected to immunohistochemical analysis by using either the pre-immune serum (A and C), or a 1:200 dilution of anti-Nedd4 serum (B and D). Note the strong expression of Nedd4 (brown) in all granular duct cells. [Bars = 160 μm (A and B), or 80 μm (C and D).]
Nedd4 Does Not Influence Na+ Channel Activity When the Na+ and Cl− Feedback Loops Are Inactive.
As we have reported (3, 33), when mouse mandibular duct cells are studied in the whole-cell patch-clamp configuration with a Na+-rich bath solution and a Na+-free, low-Cl− pipette solution (containing 150 mmol/l NMDG-glutamate), the predominant conductance seen is an amiloride-sensitive Na+ conductance that is not voltage-activated and is permeable to Li+ but not to K+. The channel type underlying this conductance appears to be the epithelial Na+ channel, which is known to be expressed in these cells (2). With NMDG-glutamate solution in the pipette and 150 mmol/l Na+-glutamate solution in the bath, the Na+ channel activity we observed appears to be maximal because we have discovered no treatment, including application of known Na+ channel activators such as para-chloromercuriphenylsulfonate or benzimidazolylguanidinium (34), that further increases Na+ channel activity.
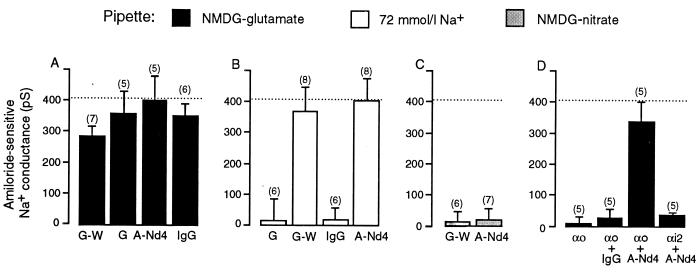
Because mutations of the PY domains of the Na+ channel that prevent Nedd4 from interacting with that channel lead to increased Na+ channel activity (see above), we first investigated whether preventing Nedd4 from interacting with the Na+ channel could further increase Na+ channel activity in mouse mandibular duct cells. We attempted to interfere with the interaction between Nedd4 and the Na+ channels in two ways: (i) by the inclusion in the pipette solution of a fusion protein constructed from GST and a segment of Nedd4, which included its three WW domains (GST–WW; final concentration 0.3 mg/ml) in the hope that it would compete with native Nedd4 for the PY domains in the Na+ channels and (ii) by the inclusion in the pipette solution of purified IgG raised against Nedd4 (anti-Nedd4; final concentration 1 μg/ml) in the hope of complexing all the native Nedd4 and rendering it unable to interact normally with the Na+ channels. We found that neither treatment increased the amiloride-sensitive Na+ conductance above the level observed with NMDG-glutamate in the pipette and 150 mmol/l Na-glutamate in the bath (Fig. 2A). Furthermore, neither the GST–WW fusion protein nor the anti-Nedd4 antibody caused any significant increase in the amiloride-sensitive Na+ conductance above that observed with the control GST protein or pre-immune IgG in the NMDG-glutamate pipette solution (Fig. 2A). Thus, we conclude that Nedd4 does not exert a tonic inhibitory effect on Na+ channels when the Na+ and Cl− feedback systems are inactive.
Figure 2.
(A) The effects of the inclusion in NMDG-glutamate pipette solution of the GST–WW fusion protein (G-W), GST control (G), anti-Nedd4 IgG (A-Nd4), or pre-immune IgG on the chord conductance measured at −80 mV of the amiloride-sensitive Na+ conductance. (B) The effects of the inclusion in 72 mmol/l Na+ pipette solution of the GST control, the GST–WW fusion protein, pre-immune IgG, or anti-Nedd4 IgG on the chord conductance measured at −80 mV of the amiloride-sensitive Na+ conductance. (C) The effects of the inclusion in NMDG–NO3 pipette solution of the GST–WW fusion protein or anti-Nedd4 antibody on the chord conductance measured at −80 mV of the amiloride-sensitive Na+ conductance. (D) The effects of the inclusion of activated α subunit of Go(αo), activated α subunit of Go plus pre-immune IgG, activated α subunit of Go plus anti-Nedd4 IgG, or activated α subunit of Gi2 plus anti-Nedd4 IgG in the NMDG-glutamate pipette solution on the chord conductance measured at −80 mV of the amiloride-sensitive Na+ conductance. The chord conductance observed by using the NMDG-glutamate pipette solution is shown in each panel as a dotted line.
Nedd4 Mediates the Na+ Feedback Loop.
In agreement with our previous studies (25, 34), we found that increasing the Na+ concentration in the pipette from 0 to 72 mmol/l effectively abolished the amiloride-sensitive Na+ conductance (Fig. 2B). We have shown (25) this inhibition to be caused by a reduction in the activity, rather than to a decrease in the single channel conductance of the Na+ channels (25). Here, we investigated whether inclusion of the GST–WW fusion protein in the 72 mmol/l Na+ pipette solution overcame the inhibitory effects of cytosolic Na+. We found that the inclusion of 0.3 mg/ml GST–WW in the 72 mmol/l Na+ pipette solution increased the amiloride-sensitive Na+ conductance to a level not significantly different from the conductance observed with the zero Na+ pipette solution (Fig. 2B), thus completely overcoming the inhibitory effect of cytosolic Na+. Inclusion of control GST protein in the 72 mmol/l Na+ pipette solution (0.3 mg/ml), on the other hand, failed to prevent inhibition of the amiloride-sensitive Na+ conductance by raised intracellular Na+ (Fig. 2B). Similarly, we found that inclusion of purified anti-Nedd4 IgG (1 μg/ml) in the pipette solution totally reversed the inhibitory effect of 72 mmol/l Na+ in the pipette solution (Fig. 2B) whereas inclusion of preimmune IgG in the 72 mmol/l Na+ pipette solution (1 μg/ml) was without effect (Fig. 2B).
Nedd4 Does Not Mediate the Cl− Feedback Loop.
We then examined whether Nedd4 mediated the Cl− feedback loop. We have reported (32) that the Na+ conductance in salivary duct cells is inhibited by the presence of anions such as Cl−, Br−, and NO3− in the cytosol (32) and have shown that this effect is mediated by the pertussis toxin-sensitive G protein (31) Gi2 (25, 34). We thus examined whether the effect of anions on the Na+ conductance is inhibited by the GST–WW fusion protein or by the anti-Nedd4 antibodies. As is our usual practice when studying control of Na+ channels by cytosolic anions (25, 31, 34), we used NO3− rather than Cl− in the pipette solution to inhibit the Na+ channels because NO3− reproduces the effects of Cl− on the Na+ channels (32) while having the additional benefit of eliminating the large Cl− currents (32, 36, 37) that would otherwise interfere with measurement of the Na+ current.
As shown in Fig. 2C, we found that neither inclusion of the GST–WW fusion protein (0.3 mg/ml) nor inclusion of the anti-Nedd4 antibody (1 μg/ml) in the NMDG–NO3− pipette solution reversed the inhibitory effects of NO3−, despite being used in concentrations that were adequate to reverse the inhibitory effects of cytosolic Na+ (compare Fig. 2B). We thus concluded that Nedd4 did not mediate the anion feedback control of the Na+ channels.
Nedd4 Mediates the Effects of Go on Na+ Channels.
As mentioned above, the inhibitory effect of intracellular Na+ on the Na+ channels in these cells is mediated by the α subunit of Go (25, 34). We have shown (34) that the inclusion of activated recombinant α subunit of Go in NMDG-glutamate pipette solution inhibits the amiloride-sensitive Na+ conductance (34). Consequently, in the present study, we investigated whether a blockade of Nedd4 would prevent inhibition of the amiloride-sensitive Na+ conductance by the activated α subunit of Go (0.2 μmol/l). As shown in Fig. 2D, we first reconfirmed our observation (25) that the activated α subunit of Go, when added to the NMDG-glutamate pipette solution, inhibited the amiloride-sensitive Na+ conductance. We then found that the addition of anti-Nedd4 antibody (1 μg/ml) totally overcame the inhibitory effect of adding activated α subunit of Go to NMDG-glutamate pipette solution (Fig. 2D). Finally, we confirmed that inclusion of purified pre-immune IgG in the pipette solution (1 μg/ml) was without effect (Fig. 2D). Thus, we concluded that Nedd4 mediates the effect of Go on Na+ channels. The inhibitory effect of the activated α subunit of Gi2 [which mediates the Cl− feedback system (25, 34)] was not inhibited by anti-Nedd4 antibody (Fig. 2D).
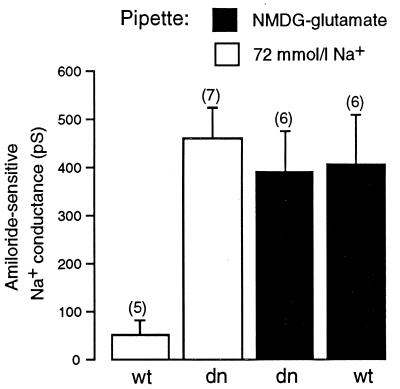
Down-Regulation of Na+ Channel Activity by Intracellular Na+ Requires Ubiquitin.
It recently has been reported (11) that the rate of proteolysis of epithelial Na+ channels and their stability in the plasma membrane are regulated by ubiquitination of the α and γ subunits of the channel (11), presumably catalyzed by the ubiquitin-protein ligase Nedd4. Because our data indicated that Nedd4 mediated the Na+ feedback pathway in mouse mandibular duct cells, we also investigated whether ubiquitination was involved in this control system. We did so by examining the effects of the inclusion of a dominant negative mutant of ubiquitin in the pipette solution on the Na+ feedback system (Fig. 3). This mutant (K48R) lacks the lysine that is required for the formation of multi-ubiquitin chains (38, 39). We found that inclusion of the ubiquitin K48R mutant (0.3 mg/ml) in the pipette solution overcame the inhibitory effects of intracellular Na+ but had no effect when the Na+ feedback system was inactive (Fig. 3). Inclusion of wild-type ubiquitin (0.3 mg/ml) in the Na+ containing pipette solution, on the other hand, did not affect the Na+ feedback system (Fig. 3).
Figure 3.
The effects of the inclusion in the 72 mmol/l Na+ pipette solution or in the NMDG-glutamate pipette solution of the GST–dn–ubiquitin (K48R) fusion protein (dn) and the GST–wt–ubiquitin fusion protein (wt) on the chord conductance measured at −80 mV of the amiloride-sensitive Na+ conductance.
DISCUSSION
In this paper, we have shown that Nedd4 is expressed in salivary duct cells (Fig. 1) and mediates the inhibitory effects of intracellular Na+ on the amiloride-sensitive Na+ channels (Fig. 2B) downstream of Go (Fig. 2D). In contrast, we have shown that Nedd4 does not mediate the corresponding anion feedback system (Fig. 2 C and D) that also regulates the amiloride-sensitive Na+ channel. Finally, we have shown that the effects of intracellular Na+ on the activity of the Na+ channels are blocked by a dominant negative mutant of ubiquitin (Fig. 3), indicating that Nedd4 influences channel activity as a consequence of its ubiquitin-protein ligase activity. Whether ubiquitination works by reducing the number of Na+ channels active in the cell membrane, as would be suggested by the findings that ubiquitination regulates the rate of Na+ channel turnover (11) and that deletion of the epithelial Na+ channel PY motifs inhibits endocytosis of Na+ channels (15, 16), or by decreasing open channel probability, as has been suggested on the basis of lipid bilayer studies (17), cannot be decided from our data.
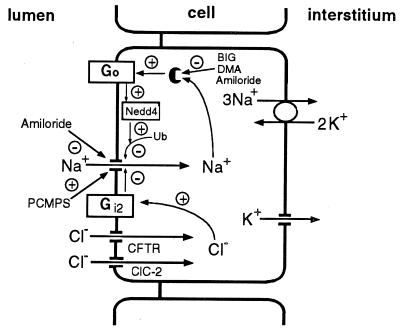
Our current knowledge of the Na+ and anion feedback systems that control the epithelial Na+ channels in mouse mandibular duct cells is summarized in Fig. 4. Intracellular Na+ binds an intracellular receptor site for Na+ (34), which can be blocked by amiloride and related compounds such as 5-N-dimethylamiloride and benzimidazolylguanidinium. The receptor, in turn, activates the G protein Go (25, 34), the α subunit of which then inactivates the Na+ channels by a mechanism that can be blocked by the GST–WW fusion protein (Fig. 2 B and D) and so must depend on the binding of Nedd4 to the PY motifs of Na+ channel. Nedd4 then ubiquitinates the Na+ channel (Fig. 3 and ref. 11), leading to its inactivation. From our data, it is not possible to determine whether Go acts on Nedd4, which in turn ubiquitinates the channels (as shown in Fig. 4), or whether Go puts Na+ channels into a conformation that is sensitive to ubiquitination by Nedd4.
Figure 4.
Proposed model for feedback regulation of Na+ channels in salivary duct cells by cytosolic Na+ and Cl− acting through G proteins and Nedd4.
In the anion feedback pathway, intracellular Cl− activates the G protein Gi2 (25, 34), which in turn inhibits the Na+ channels by a mechanism that does not involve Nedd4 (Fig. 2 C and D), implying that Go and Gi2 inhibit Na+ channel activity by different mechanisms. This inference is consistent with the ability of other effectors, such as Ca2+ channels, to discriminate between the Go and Gi classes of G protein (40), although it is perhaps surprising given the ability of the sulfhydryl reagent para-chloromercuriphenylsulfonate to override both the Na+ and the Cl− feedback systems (34).
Our finding that Nedd4 mediates the Na+ feedback pathway in mouse mandibular duct cells is important for two reasons. First, it identifies a physiological control pathway for Na+ channels, which, when interrupted, would lead to the increased Na+ channel activity that is observed in Liddle’s syndrome (13, 14, 19, 20) and in other circumstances when interaction between Nedd4 and Na+ channels has been prevented—for example, when Na+ channels with deleted PY motifs are expressed in Xenopus oocytes (15). The only other physiological control system that has been shown to be defective in Liddle’s syndrome, the loss of protein kinase A activation of Na+ channels (12), is a consequence of increased Na+ channel activity rather than an explanation for it. Second, our findings, when taken together with the occurrence in Liddle’s syndrome of abnormal Na+ channel activity in B lymphocytes (12) and in renal collecting ducts (18), suggest that the Na+ feedback pathway, which thus far has been characterized well only in mouse mandibular duct cells, may be of more general significance.
Acknowledgments
We thank Dr. J. M. Huibregtse (Rutgers University) for providing the pGEX-2TK-ubiquitin construct. This project was supported by the National Health and Medical Research Council of Australia and The Medical Foundation of the University of Sydney. D.I.C. is a Fellow of The Medical Foundation of the University of Sydney. S.K. is a Wellcome Senior Fellow in Medical Science. K.F.H. was supported by a Dawes Postgraduate Scholarship from the Royal Adelaide Hospital.
ABBREVIATIONS
- GST
glutathione-S-transferase
- NMDG
N-methyl-d-glucamine
References
- 1.Garty H, Palmer L G. Physiol Rev. 1997;77:359–396. doi: 10.1152/physrev.1997.77.2.359. [DOI] [PubMed] [Google Scholar]
- 2.Duc C, Farman N, Canessa C M, Bonvalet J P, Rossier B C. J Cell Biol. 1994;127:1907–1921. doi: 10.1083/jcb.127.6.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dinudom A, Young J A, Cook D I. Pflügers Arch. 1993;423:164–166. doi: 10.1007/BF00374977. [DOI] [PubMed] [Google Scholar]
- 4.Lifton R P. Science. 1996;272:676–680. doi: 10.1126/science.272.5262.676. [DOI] [PubMed] [Google Scholar]
- 5.Canessa C M, Schild I, Buell G, Thorens B, Gautschi I, Horisberger J D, Rossier B C. Nature (London) 1994;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 6.Staub O, Dho S, Henry P C, Correa J, Ishikawa T, McGlade J, Rotin D. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 7.Schild L, Lu Y, Gautschi I, Schneeberger E, Lifton R P, Rossier B C. EMBO J. 1996;15:2381–2387. [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar S, Tomooka Y, Noda M. Biochem Biophys Res Commun. 1992;185:1155–1161. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 9.Kumar S, Harvey K F, Kinoshita M, Copland N G, Noda M, Jenkins N A. Genomics. 1997;40:435–443. doi: 10.1006/geno.1996.4582. [DOI] [PubMed] [Google Scholar]
- 10.Staub O, Yeger H, Plant P J, Kim H, Ernst S A, Rotin D. Am J Physiol. 1997;272:C1871–C1880. doi: 10.1152/ajpcell.1997.272.6.C1871. [DOI] [PubMed] [Google Scholar]
- 11.Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciecanover A, Schild L, Rotin D. EMBO J. 1997;16:6325–6336. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bubien J K, Ismailov I I, Berdiev B K, Cornwell T, Lifton R P, Fuller C M, Achard J M, Benos D J, Warnock D G. Am J Physiol. 1996;270:C208–C213. doi: 10.1152/ajpcell.1996.270.1.C208. [DOI] [PubMed] [Google Scholar]
- 13.Snyder P M, Price M P, McDonald F J, Adams C M, Volk K A, Zeiher B G, Stokes J B, Welsh M J. Cell. 1995;83:969–978. doi: 10.1016/0092-8674(95)90212-0. [DOI] [PubMed] [Google Scholar]
- 14.Hansson J H, Schild L, Lu Y, Wilson T A, Gautschi I, Shimkets R A, Nelson-Williams C, Rossier B C, Lifton R P. Proc Natl Acad Sci USA. 1995;92:11495–11499. doi: 10.1073/pnas.92.25.11495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Firsov D, Schild L, Gautschi I, Merillat A M, Schneeberger E, Rossier B C. Proc Natl Acad Sci USA. 1996;93:15370–15375. doi: 10.1073/pnas.93.26.15370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimkets R A, Lifton R P, Canessa C M. J Biol Chem. 1997;272:25537–25541. doi: 10.1074/jbc.272.41.25537. [DOI] [PubMed] [Google Scholar]
- 17.Ismailov I I, Berdiev B K, Fuller C M, Bradford A L, Lifton R P, Warnock D G, Bubien J K, Benos D J. Am J Physiol. 1996;270:C214–C223. doi: 10.1152/ajpcell.1996.270.1.C214. [DOI] [PubMed] [Google Scholar]
- 18.Botero-Velez M, Curtis J J, Warnock D G. N Engl J Med. 1994;330:178–181. doi: 10.1056/NEJM199401203300305. [DOI] [PubMed] [Google Scholar]
- 19.Hansson J H, Nelson-Williams C, Suzuki H, Schild L, Shimkets R, Lu Y, Canessa C, Iwasaki T, Rossier B, Lifton R P. Nat Genet. 1995;11:76–82. doi: 10.1038/ng0995-76. [DOI] [PubMed] [Google Scholar]
- 20.Shimkets R A, Warnock D G, Bositis C M, Nelson-Williams C, Hansson J H, Scambelan M, Gill J R, Ulick S, Milora R V, Finling J W, et al. Cell. 1994;79:407–414. doi: 10.1016/0092-8674(94)90250-x. [DOI] [PubMed] [Google Scholar]
- 21.Palmer L G, Frindt G, Silver R B, Strieter J. In: Current Topics in Membranes and Transport, Vol. 34: Cellular and Molecular Biology of Sodium Transport. Schultz S G, editor. San Diego: Academic; 1989. pp. 45–60. [Google Scholar]
- 22.Turnheim K. Physiol Rev. 1991;71:429–445. doi: 10.1152/physrev.1991.71.2.429. [DOI] [PubMed] [Google Scholar]
- 23.Fuchs W, Larsen E H, Lindemann B. J Physiol (London) 1977;267:137–166. doi: 10.1113/jphysiol.1977.sp011805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Driessche W, Lindemann B. Nature (London) 1979;282:519–520. doi: 10.1038/282519a0. [DOI] [PubMed] [Google Scholar]
- 25.Komwatana P, Dinudom A, Young J A, Cook D I. Proc Natl Acad Sci USA. 1996;93:8107–8111. doi: 10.1073/pnas.93.15.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuthbert A W, Shum W K. Nature (London) 1977;266:468–469. doi: 10.1038/266468a0. [DOI] [PubMed] [Google Scholar]
- 27.Schultz S G. Am J Physiol. 1981;241:F579–F590. doi: 10.1152/ajprenal.1981.241.6.F579. [DOI] [PubMed] [Google Scholar]
- 28.Harvey B J, Thomas S R, Ehrenfeld J. J Gen Physiol. 1998;92:767–791. doi: 10.1085/jgp.92.6.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silver R B, Frindt G, Windhager E E, Palmer L G. Am J Physiol. 1993;264:F557–F564. doi: 10.1152/ajprenal.1993.264.3.F557. [DOI] [PubMed] [Google Scholar]
- 30.Frindt G, Silver R B, Windhager E E, Palmer L G. Am J Physiol. 1993;264:F565–F574. doi: 10.1152/ajprenal.1993.264.3.F565. [DOI] [PubMed] [Google Scholar]
- 31.Dinudom A, Komwatana P, Young J A, Cook D I. J Physiol (London) 1995;487:549–555. doi: 10.1113/jphysiol.1995.sp020899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinudom A, Young J A, Cook D I. J Membr Biol. 1993;135:289–295. doi: 10.1007/BF00211100. [DOI] [PubMed] [Google Scholar]
- 33.Komwatana P, Dinudom A, Young J A, Cook D I. J Membr Biol. 1996;150:133–141. doi: 10.1007/s002329900038. [DOI] [PubMed] [Google Scholar]
- 34.Komwatana P, Dinudom A, Young J A, Cook D I. J Membr Biol. 1998;162:225–232. doi: 10.1007/s002329900360. [DOI] [PubMed] [Google Scholar]
- 35.Lang J, Nishimoto I, Okamoto T, Regazzi R, Kiraly C, Weller U, Wollheim C B. EMBO J. 1995;14:3635–3644. doi: 10.1002/j.1460-2075.1995.tb00033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komwatana P, Dinudom A, Young J A, Cook D I. Pflügers Arch. 1994;428:641–647. doi: 10.1007/BF00374588. [DOI] [PubMed] [Google Scholar]
- 37.Dinudom A, Komwatana P, Young J A, Cook D I. Am J Physiol. 1995;268:G806–G812. doi: 10.1152/ajpgi.1995.268.5.G806. [DOI] [PubMed] [Google Scholar]
- 38.Chau V, Tobias J W, Bachmair A, Marriott D, Ecker D J, Gonda D K, Varshavsky A. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 39.Ward C L, Omura S, Kopito R R. Cell. 1995;3:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 40.Hescheler J, Rosenthal W, Trautwein W, Schultz G. Nature (London) 1987;325:445–447. doi: 10.1038/325445a0. [DOI] [PubMed] [Google Scholar]