Abstract
The human and shark Na–K–Cl cotransporters (NKCC), although 74% identical in amino acid sequence, exhibit marked differences in ion transport and bumetanide binding. We have utilized shark–human chimeras of NKCC1 to search for regions that confer the kinetic differences. Two chimeras (hs3.1 and its reverse sh3.1) with a junction point located at the beginning of the third transmembrane domain were examined after stable transfection in HEK-293 cells. Each carried out bumetanide-sensitive 86Rb influx with cation affinities intermediate between shark and human cotransporters. In conjunction with the previous finding that the N and C termini are not responsible for differences in ion transport, the current observations identify the second transmembrane domain as playing an important role. Site-specific mutagenesis of two pairs of residues in this domain revealed that one pair is indeed involved in the difference in Na affinity, and a second pair is involved in the difference in Rb affinity. Substitution of the same residues with corresponding residues from NKCC2 or the Na-Cl cotransporter resulted in cation affinity changes, consistent with the hypothesis that alternative splicing of transmembrane domain 2 endows different versions of NKCC2 with unique kinetic behaviors. None of the changes in transmembrane domain 2 was found to substantially affect Km(Cl), demonstrating that the affinity difference for Cl is specified by the region beyond predicted transmembrane domain 3. Finally, unlike Cl, bumetanide binding was strongly affected by shark–human replacement of transmembrane domain 2, indicating that the bumetanide-binding site is not the same as the Cl-binding site.
Keywords: kinetic, mutagenic analysis
Na–K–Cl cotransporters (NKCC) facilitate the diffusion of Na, K, and Cl ions across the plasmalemma of animal cells (1–5). In nonpolarized cells, Na–K–Cl cotransport plays a role in cell volume regulation and in maintenance of the electrolyte content (6), whereas in polarized cells the cotransport process is central to net transepithelial movement of salt and water. NKCC (also called BSC; ref. 7) is a member of the cation–Cl cotransporters (6, 8), a family that also includes the Na-Cl cotransporter (NCC or TSC; ref. 9) and the K–Cl cotransporter (8, 10). Two types of NKCC have been identified to date: NKCC1 is widely distributed and is most prominent in certain secretory epithelia, where it plays a pivotal role in electrolyte movement (1). NKCC2 is specific to the kidney, where it takes part in NaCl reabsorption across the apical membrane of epithelial cells; three splice variants (A, B, and F) are found in distinct regions of the thick ascending limb of the loop of Henle (4, 5).
Functionally, NKCC promotes the intracellular accumulation of ions by coupling the electroneutral uptake of 1 Na, 1 K, and 2 Cl ions. Transport is inhibited by loop diuretic drugs such as furosemide and bumetanide. For both NKCC1 and NKCC2, the binding of bumetanide requires the simultaneous presence of Na, K, and Cl (11–13). Regulation of cotransport activity is dependent on maneuvers that affect cell volume and intracellular concentration of Cl in a process that, at least for NKCC1, involves direct phosphorylation of the cotransport protein (14, 15).
Quantitative differences in functional properties have been found between the two NKCC isoforms, as well as between the same NKCC isoform in separate species. For example, we have recently shown that the Km(Na) and the inhibition constant for bumetanide [Ki(bumetanide)] of the rabbit NKCC2A (rNKCC2A; expressed as a chimera) are 10- to 12-fold lower than those of the shark NKCC1 (sNKCC1) and 40% lower than those of the human NKCC1 (hNKCC1) (16). Although hNKCC1 and sNKCC1 are 74% identical in amino acid sequence, the Km(ions) and Ki(bumetanide) are 4- to 6-fold higher in the shark cotransporter, compared with human (3). The Cl dependence of activation also differs between cotransporters: intracellular concentration of Cl is a crucial determinant in the activation of hNKCC1, whereas cell volume appears to play a relatively larger role in regulation of rNKCC2A (16).
Taking advantage of the species differences, we recently used chimeras of hNKCC1 and sNKCC1 to identify regions that are responsible for the differences in kinetic characteristics of the transporters (17). Using chimeras in which the N and the C termini were interchanged, we have shown that the differences in apparent affinities of ions and bumetanide are encoded by the large central transmembrane domain of the NKCC protein. We suggested that affinity-modifying residues are localized in the vicinity of ligand-binding residues and are able to modulate ligand-binding affinities through direct interactions or through allosteric effects.
In this study, we extended the chimera approach to the central domain and found that a region in transmembrane domain 2 of NKCC1 plays an important role in the species differences in cation and bumetanide affinities. Significantly, this domain corresponds to the alternatively spliced exon in NKCC2. In further analysis using site-specific mutagenesis, we found that substitution of a pair of residues in this region alters Na affinity, whereas substitution of another pair alters Rb affinity. In contrast, we found little effect of changes in this region on Cl affinity. Preliminary results were presented previously (18).
EXPERIMENTAL PROCEDURES
Chimeras and Point Mutations.
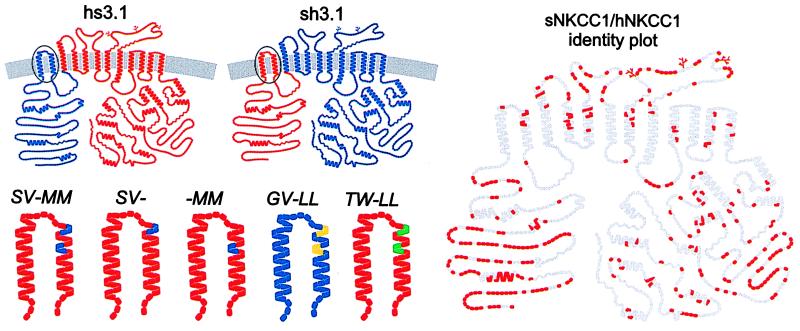
In our nomenclature (17) we use s, h, and r to designate domains from shark, human, and rabbit NKCC and numbers to indicate the position of the chimeric junctions relative to the 12 putative transmembrane domains. For example, hs3.1 represents a chimera with human sequence from the N terminus to the first residues of transmembrane domain 3, and with shark residues for the remainder. Models of hs3.1 and sh3.1 are shown in Fig. 1. In the case of the chimera between NKCC1 and NKCC2, the isoform designations are also included (h1r2A0.7).
Figure 1.
Models for sNKCC1, hNKCC1, two chimeras, and five point mutations in transmembrane domain 2 of sNKCC1 based on proposed structure (3). Amino acid residues are color coded according to their source: blue, hNKCC1; red, sNKCC1; yellow, rNKCC2A; green, hNCC (or TSC). Gray symbols in left model represent residues that are identical between sNKCC1 and hNKCC1.
The constructs used in this paper are listed in Table 1. All of the point mutations are in the second transmembrane domain of sNKCC1 or hNKCC1; these are designated by uppercase italicized letters to indicate the replacement residues. Unless noted otherwise, the mutations were generated in sNKCC1. For example, SV-MM represents a point mutation in which residues SV and MM from human replace the residues AL and GT of the shark sequence. In GV-LLhNKCC1, the residues GV and LL replace SV and MM in the human sequence. The location of these substitutions in the second transmembrane domain is shown in Fig. 1.
Table 1.
Wild-type and mutant NKCCs, highlighting the differences in transmembrane domain 2
| cDNAs and mutants | Transmembrane domain 2 | Backbone | Substitution from |
|---|---|---|---|
| sNKCC1 | G I G L A L L V I G T A T V V T T I T G | sNKCC1 | |
| hNKCC1 | G I G L S V L V I M M A T V V T T I T G | hNKCC1 | |
| rNKCC2A | G I G L G V V I I L L S T M V T S I T G | rNKCC2A | |
| hNCC | G I G L T W V I I L L S S F I T G I T G | hNCC | |
| SV-MM | G I G L S V L V I M M A T V V T T I T G | sNKCC1 | hNKCC1 |
| SV- | G I G L S V L V I G T A T V V T T I T G | sNKCC1 | hNKCC1 |
| -MM | G I G L A L L V I M M A T V V T T I T G | sNKCC1 | hNKCC1 |
| GV-LL | G I G L G V L V I L L A T V V T T I T G | hNKCC1 | rNKCC2A |
| TW-LL | G I G L T W L V I L L A T V V T T I T G | sNKCC1 | hNCC |
hs3.1 and sh3.1 were generated by fragment exchange at the common restriction site MunI. This site is in a conserved region of NKCC at the transition of transmembrane helix 3 and the intracellular connecting loop (base pair 1264 of hNKCC1 and base pair 1463 of sNKCC1). The junction corresponds to residues A367/I368 in hNKCC1 and A339/I340 in sNKCC1. MunI also occurs in the vector pJB20, in the ORF of sNKCC1 (base pair 2546), and in the 3′-untranslated region (base pair 4109). The other restriction sites used to generate the chimeras were KpnI and XhoI in the linker of both hNKCC1/pJB20 and sNKCC1/pJB20, XbaI and HindIII in the linker of a mutant Bluescript SK− (ref. 17), and BspEI in the ORF of sNKCC1/pJB20 (base pair 1579). An additional BspEI site exists at the 5′-linker junction of sNKCC1. hs3.1 was prepared in a four-step ligation. The BspEI fragment of sNKCC1/pJB20 (length 1584 bp) and the XbaI–HindIII fragment of the mutant Bluescript SK− (length 3000 bp) were ligated after the ends were treated with the Klenow fragment to generate sNKCC1del:1585–5260/Bluescript SK−. In the appropriate orientation, only the BspEI site at base pair 1579 remains intact. In sNKCC1del:1585–5260/Bluescript SK−, the KpnI–MunI fragment (length 1450 bp) was replaced by its counterpart KpnI–MunI fragment from hNKCC1/pJB20 (length 1270 bp) to generate hNKCC1del:1270–4115/Bluescript SK−. In this construct, the BspEI–KpnI fragment (length 3100 bp) was replaced with the BspEI–KpnI fragment of sNKCC1/pJB20 (length 7700 bp) to generate hs3.1/pJB20. sh3.1 was prepared in a single-step ligation by using the KpnI-MunI fragment of sNKCC1/pJB20, the MunI-XhoI fragment of hNKCC1/pJB20 (length 2800 bp), and the XhoI–KpnI fragment of sNKCC1/pJB20 (length 6200 bp), which corresponds to the vector.
The point mutations SV-, -MM, SV-MM, and TW-LL (Fig. 1 and Table 1) were generated by the method of Kunkel and coworkers (19): sNKCC1 was subcloned in the phagemid pTZ18U (Bio-Rad) to generate uracil-containing single-stranded DNA. In separate reactions, a synthetic oligonucleotide overlapping the region corresponding to helix 2 was hybridized to the template and extended with T7 polymerase. The mutations were reintroduced in sNKCC1/pJB20 by using the fragment flanked by the restriction sites BstEII and BspEI (length 1000 bp) located at base pairs 578 and 1579 of the sNKCC1 sequence. The oligonucleotides used to generate the mutations are shown in Table 2.
Table 2.
Oligonucleotides used to generate point mutations in NKCC1
| Mutation | Oligonucleotide sequence |
|---|---|
| SV-MM | GGG ATA GGT CTT TCT GTG CTA GTC ATT ATG ATG GCG ACT GTT G |
| SV- | GGG ATA GGT CTT TCT GTG CTA GTC ATT GG |
| -MM | GT CTT GCT CTA CTA GTC ATT ATG ATG GCG ACT GTT GT |
| GV-LL | GGA ATA GGT CTA GGA GTC CTT GTA ATA CTG CTG GCG ACT GTT G |
| TW-LL | GGG ATA GGT CTC ACG TGG CTA GTC ATT CTC CTG GCG ACT GTT G |
The point mutant GV-LLhNKCC1 (Fig. 1 and Table 1) was generated by PCR with two sets of synthetic oligonucleotides. The first set comprised a sense oligonucleotide overlapping the restriction site Bst1107I (base pair 986 in hNKCC1) and an antisense oligonucleotide containing the appropriate mutation in transmembrane 2 (Table 2). The second included a sense oligonucleotide corresponding to the reverse complement of the mutagenic oligonucleotide and an antisense oligonucleotide overlapping the restriction site AflII (base pair 1347 in hNKCC1). The two regions were amplified separately (25 cycles), and the PCR products were gel purified. The fragments from each reaction were then hybridized at equimolar concentration and extended with the Klenow fragment of DNA polymerase I. The final product was reamplified with the same external oligonucleotides, digested with Bst1107I and AflII (length 362 bp), and reintroduced in hNKCC1/pJB20.
Cell Lines.
Lines stably expressing hNKCC1, sNKCC1, and h1r2A0.7 were the same as in ref. 16. h1r2A0.7 is a chimera composed of the coding region of rNKCC2A with the 104 N-terminal amino acids replaced by the corresponding region in hNKCC1. The chimeras hs3.1 and sh3.1 and the five-point mutations were transfected into HEK-293 cells by calcium phosphate precipitation, and stable lines (generally 12–16) were isolated by G-418 resistance as described (17). Transfected HEK-293 cells were maintained in DMEM supplemented with 10% fetal bovine serum, penicillin, streptomycin, and G-418 (950 μg/ml), as described (15). By Western blot, the cell lines expressed comparable quantities of wild-type or mutant cotransporter with similar glycosylation patterns (results not shown); recognition by J3 (N-terminal epitope) or J7 (C-terminal epitope) antibodies was consistent with whether the corresponding terminus was derived from sNKCC1 (17). The level of functional expression achieved with the two chimeras (hs3.1 and sh3.1) and with four of the sNKCC1 point mutations (including -MM, SV-MM, GV-LLhNKCC1, and TW-LL) was comparable to the wild-type hNKCC1 and sNKCC1 (17), about 6- to 8-fold above the endogenous cotransport activity of mock-transfected HEK-293 cells. The mutant SV- was found to transport 86Rb at a lower rate, about 2.5-fold above the background. All point mutants except TW-LL were lost in a tissue culture disaster prior to completion of bumetanide inhibition studies.
Flux Studies.
As described (16, 17), HEK-293 cells were subcultured onto 96-well plates precoated with polylysine and grown to confluence (6–8 days). Ion transport rates and bumetanide binding were determined by 86Rb influx measurements. All experiments were done at room temperature (≈22°C) with all solutions at pH 7.4. Before each flux assay the cells were incubated in hypotonic low Cl (163 mosM, 2 mM) medium for 1 h (67.5 mM sodium gluconate/2.5 mM potassium gluconate/0.5 mM CaCl2/0.5 mM MgCl2/0.5 mM Na2HPO4/0.5 mM Na2SO4/7.5 mM Na Hepes) to activate the cotransporter (1). In experiments in which Ki for bumetanide inhibition was determined, the cells were reincubated for 15 min at various inhibitor concentrations in 20 mM Cl medium (1, 3). 86Rb influxes were performed for 1 min in a medium containing 135 mM NaCl, 5 mM RbCl (2 μCi/ml 86RbCl), 1 mM CaCl2, 1 mM MgCl2, 1 mM Na2HPO4, 1 mM Na2SO4, 15 mM Na Hepes, and 0.1 mM ouabain; when necessary, Na and Rb were replaced with equimolar amounts of N-methylglucamine, and Cl was replaced with gluconate. To determine bumetanide-sensitive influx, 250 μM bumetanide was added directly to the media. 86Rb uptake was terminated by addition of, and three rinses in, ice-cold stop solution (135 mM potassium gluconate/5 mM sodium gluconate/1 mM CaCl2/1 mM MgCl2/1 mM Na2HPO4/1 mM Na2SO4/15 mM Na Hepes/250 μM bumetanide/0.1 mM ouabain). Cells were solubilized in 2% SDS and assayed both for protein content and for 86Rb by counting Cerenkov radiation.
The two constructs (GV-LLhNKCC1 and h1r2A0.7) exhibited bumetanide-sensitive (not shown) 86Rb influx even in the absence of extracellular Na (Fig. 6). These are the two constructs with the highest Na affinity, raising the possibility that part of this “[Na] = 0” influx might have been related to carryover of Na from the preincubation solution. We estimate carryover to be <1% (<0.8 mM Na), which could account for a portion of the observed flux at “0 Na.”
Figure 6.
86Rb influx as a function of Na, Rb, and Cl concentration for point mutant TW-LL and sNKCC1. Flux procedures are described in Experimental Procedures. Data from 7 to 18 flux rows were averaged for TW-LL and compared with values for sNKCC1 from Fig. 3 (n = 34–55).
For each concentration curve, experiments were carried out in a single row of the 96-well plate. In a typical experiment there were two to six replicate rows. Data were handled on a per-row basis and counts were normalized to the value at the highest ligand concentration or to the value of uninhibited flux in inhibition studies. Normalized values were averaged and fit by using the Michaelis–Menten equation for binding at a single site for Na or bumetanide and for two sites for Cl (Hill coefficient = 2). Rows with obvious rogue values were omitted from averages, in each data set less than one row in eight. Data are expressed as means ± SEM among all rows in 4–15 experiments. Similarly, Km and Ki values were obtained on a per-row basis by nonlinear least-squares curve fitting using the Simplex algorithm (program plot, available from B.F.).
RESULTS
Chimeras hs3.1 and sh3.1.
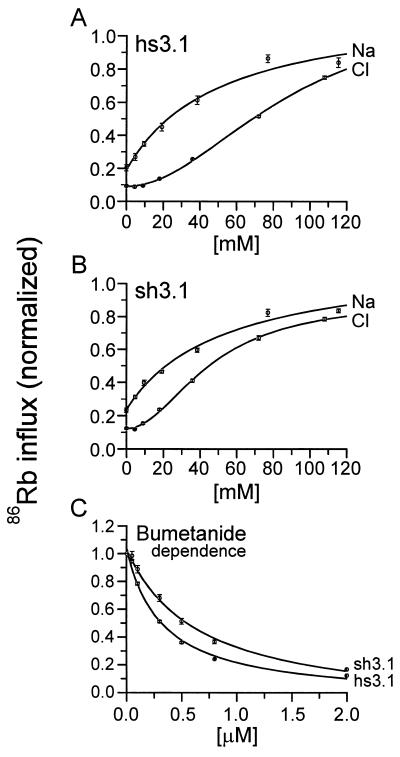
To identify a region within the central domain of the NKCC that affects ion and bumetanide binding, we characterized two human–shark chimeras with a junction at the beginning of the third transmembrane domain (see Fig. 1). Because neither the N nor the C terminus plays a role in determining the species differences in ion transport kinetics (17), and because transmembrane domains 1 and 3 are fully conserved between hNKCC1 and sNKCC1, chimera hs3.1 and its complement sh3.1 allow us to determine the relative contribution of transmembrane domain 2 to ion and bumetanide affinities. Data illustrating the kinetic behavior of these chimeras are presented in Fig. 2, and a comparison of kinetic constants with those of the sNKCC1 and hNKCC1 is given in Fig. 3.
Figure 2.
86Rb influx as a function of Na, Cl, and bumetanide concentration for hs3.1 or sh3.1. Flux procedures are described in Experimental Procedures. Data from 44 flux rows were averaged for sh3.1 and data from 17 rows were averaged for hs3.1.
Figure 3.
Kinetic constants for chimeras sh3.1 and hs3.1. Data are from experiments of Fig. 2 for the two chimeras. Data for hNKCC1 and sNKCC1 are from ref. 17 (n = 9–11) with the addition of data from recent experiments (n = 3–4); these were time-period matched to be in parallel with the chimera data and the derived constants are essentially identical with those reported (17).
As shown in Fig. 3, the behavior of two chimeras with regard to the cation dependence of transport is intermediate between human and shark. For Km(Na), each of the two chimeras are approximately half way between shark and human, i.e., the Km is 52 and 55 mM in the chimeras compared with 15 and 109 mM in hNKCC1 and sNKCC1, respectively. Because we previously found that interchanging N termini has no effect on kinetic behavior (17), these data suggest that variant residues in the second transmembrane domain confer part of the shark–human difference in Na affinity.
Similarly, the Km(Rb) of hs3.1 is between those of the wild-type transporters, 5.9 mM compared with 1.8 mM for hNKCC1 and 11.8 mM for sNKCC1. Thus, as for Na, much of the shark–human affinity difference in Rb binding appears to involve the second transmembrane domain. The behavior of the reverse chimera sh3.1 is closer to that of hNKCC1 (Km 2.6 mM) but is still intermediate between the behaviors of the two wild-type transporters.
The Cl dependence of 86Rb transport is also shown in Fig. 2. As previously shown, the sigmoidal relationship between 86Rb influx and the concentration of Cl is consistent with two binding and translocation sites for this ion. The Km(Cl) for each of the chimeras is close to one of the wild-type transporters: for hs3.1 the Km is 87 mM, compared with 107 mM in sNKCC1; and for sh3.1 the Km is 47 mM, compared with 31 mM for hNKCC1. The differences between chimeras and wild types are significant (P < 0.05) but are much smaller than observed for cations. Thus, it appears that changing the residues in the second transmembrane domain has only a small effect on anion affinity.
Loop diuretic drugs such as bumetanide have been found to be competitive with Cl in binding to the NKCC (refs. 11, 20, and 21; but see also ref. 22), raising the possibility that Cl and bumetanide bind to the same site. If so, one would expect that a change in bumetanide affinity would parallel a change in Cl affinity. Surprisingly, as illustrated in Figs. 2 and 3, the affinity for bumetanide is almost as low in sh3.1 as in sNKCC1 (0.76 vs. 1.04 μM) and it is almost as high in hs3.1 as in hNKCC1 (0.34 vs. 0.28 μM). Thus, in contrast to the result for Cl, the first two transmembrane domains appear to play an important role in determination of loop diuretic affinity.
Point Mutations in Transmembrane Domain 2.
Apart from the N terminus, the two chimeras discussed above differ from wild-type counterparts by only six residues in the first extracellular loop and in the second transmembrane domain (Fig. 1). To isolate the residues conferring the human-shark difference in cation binding and to test our hypothesis that the four residues in the second transmembrane segment are important, we examined three point mutations in which pairs of residues in sNKCC1 were replaced by residues from their human counterpart. Mutants “SV-” (ALsNKCC1 → SVhNKCC1), “-MM” (GTsNKCC1 → MMhNKCC1), and “SV-MM” are illustrated in Fig. 1.
The kinetic constants obtained for SV-, -MM, and SV-MM are compared with values for hs3.1 and sNKCC1 in Fig. 4. The point mutants are all seen to have a Km(Cl) very similar to that of sNKCC1; this is expected because the apparent Cl affinity of the hs3.1 chimera is similar to that of sNKCC1. With regard to Na and Rb affinities, it is shown that the SV-MM mutant is identical to the chimera hs3.1. This result confirms that the variant residues in transmembrane domain 2 account for the kinetic differences between hs3.1 and sNKCC1. When individual pairs of residues are examined, the results in Fig. 4 demonstrate that the AL → SV mutation is completely responsible for the change in Na affinity, whereas GT → MM is completely responsible for the change in Rb affinity.
Figure 4.
Kinetic constants for point mutants SV-, -MM, and SV-MM. Values from 12 to 30 flux rows were averaged and compared with values for sNKCC1 and hs3.1 from Fig. 3.
We previously proposed that transmembrane domain 2 may be important in determination of ion affinities in the renal cotransporter, NKCC2. This isoform occurs as three variants in which an alternatively spliced cassette spans putative transmembrane helix 2 (4). As reported elsewhere, in HEK-293 cells it has only been possible to study the behavior of NKCC2 using an N-terminal NKCC1 chimera (h1r2A0.7) to obtain expression (16). To investigate the role of transmembrane domain 2, we replaced this entire region in rNKCC2 with the sequence from the splice variants NKCC2A, B, and F. Unfortunately, we were unable to detect functional expression of these constructs (not shown).
We were able to measure expression of an hNKCC1 construct in which the four residues identified above were replaced with the corresponding residues from the rabbit kidney A variant (GV-LLhNKCC1, Fig. 1). The kinetic behavior of the GV-LL point mutant is compared with hNKCC1 and h1r2A0.7 in Fig. 5. The Km for Cl is the same for all three constructs, which is consistent with the above observation that changes in transmembrane domain 2 do not affect apparent Cl affinity. The GV-LL mutation in hNKCC1 was found to exhibit Km values for Na and Rb that are displaced toward the values obtained in the kidney construct. This further supports the idea that residues in transmembrane domain 2 play an important role in cation affinity and the prediction that the renal splice variants will exhibit different kinetic properties.
Figure 5.
86Rb influx as a function of Na, Rb, and Cl concentration for point mutant GV-LLhNKCC1 and chimera h1r2A0.7. Flux procedures are described in Experimental Procedures. Data from 18 to 24 flux rows were averaged for GV-LL and compared with values for hNKCC1 from Fig. 3 (n = 42–55) and h1r2A0.7 from ref. 16 (n = 18–35).
Similarly, we prepared a construct in which the two pairs of residues in sNKCC1 are replaced by the corresponding residues TW-LL from the thiazide-sensitive NCC (or TSC). As shown in Fig. 6, the Km(Na) of TW-LL is decreased relative to sNKCC1, comparable to the change seen with the SV-MM mutant. On the other hand, neither Km(Rb) nor Km(Cl) was substantially affected by the substitution.
DISCUSSION
The results reported here illustrate the importance of transmembrane domain 2 of the NKCC in cation transport. Evaluation of the kinetic behavior of two shark–human NKCC1 chimeras demonstrates that approximately half of the difference in cation affinities between sNKCC1 and hNKCC1 is determined by six residues in the second transmembrane domain and associated intracellular and extracellular loops. Further analysis utilizing point mutagenesis has confirmed that two pairs of residues in the second predicted transmembrane domain confer these differences in apparent ion affinities.
Although the second transmembrane domain appears to be of critical importance in affecting cation affinities, the changes we have made in this region do not appreciably alter the Km for Cl. Thus, it is clear that a part of the large central domain beyond the third transmembrane domain must regulate Cl affinity; as indicated in Fig. 1, there are candidate residues that are different between sNKCC1 and hNKCC1 in every putative transmembrane domain except the 6th and 10th. Some of these candidate residues must also confer the other half of the differences in cation affinities that are not encoded by the domain 2 residues studied here.
An important finding of these experiments is that one pair of residues appears to be involved with Na affinity and a second pair with Rb affinity. It is interesting to examine this result in the context of an ordered binding model of transporter function. From evaluation of ion movements under conditions of exchange and net transport in duck and ferret red blood cells, Lytle and McManus (23) concluded that ion binding is strictly ordered: the order of binding from the outside as well as of release from the inside is Na–Cl–K–Cl. The simplest structural correlate of this model would involve a deeply seated Na site and a more superficial K site relative to the extracellular surface. Our results suggest the opposite: the SV pair that confers higher Na affinity on the human isoform is localized near the extracellular end of transmembrane 2, and the MM pair that confers higher Rb affinity is localized further into the bilayer. This apparent discrepancy may be an argument against the structure depicted in Fig. 1 or against the ordered kinetic model, or it may indicate that more complex models are required. Alternatively, it is possible that the two pairs of residues are not directly involved in ion binding but that they confer affinity differences through conformational interactions.
In the current approach, we search for residues that confer species differences in the kinetics of ion translocation. These are possibly not the same residues that are essential for ion binding because many of those residues would be fully conserved through evolution and would not be identified by this differential technique. Nonetheless, it seems reasonable to propose that the regions and residues that confer differences in affinity are close to the ion-binding site and may, for instance, be involved in helix–helix interactions that are important for the integrity of the ion translocation pocket.
One strength of the chimera approach is illustrated by the reciprocal relationship between kinetic parameters in the two chimeras. The kinetic changes observed when the first three transmembrane domains in sNKCC1 are converted to the human counterpart (hs3.1) are mirrored when the same domain in hNKCC1 is changed to shark (sh3.1). Thus, the apparent affinities for Na and Rb are about half way between shark and human values in each of the chimeras, whereas the Cl affinities are only about 15% different from the parent species. There is some deviation from reciprocity with regard to Km(Rb), but, nonetheless, the directionality of the change is as expected. These observations reassure us that we have used systematic and informative mutations that are not acting simply by disrupting the normal structure of the protein.
In this paper we report Km values for ion transport and bumetanide inhibition. These parameters are related to actual binding site affinities in a complex manner that also depends on the rate constants in the transport cycle. Thus, it is theoretically possible that observed differences in Km values could be caused by changes in a translocation rate constant rather than to a change in a binding site affinity. Although the specific relationships are model dependent, in the general case one would expect to see parallel differences in apparent affinities for all three ions if this were a significant factor. In fact, we see substantial changes in Km(cations) but not in Km(Cl), and we see individual effects of specific residue pairs on Km(Na) or Km(Rb); these results are reconcilable with simple kinetic models only if individual ion affinities are being changed. We conclude that the differences between mutants in this study result from the exchange of affinity-modifying residues between human and shark.
The results of these studies provide a dramatic distinction between regions important in bumetanide binding and regions determining Cl affinity. Whereas Km(Cl) is changed <15% by modification of the residues in the first two transmembrane domains, about 80% of the species difference in Ki(bumetanide) is conferred by changes in that region. This is strong evidence against a hypothesis that the bumetanide-binding site is the same as the Cl-binding site, a popular idea (24) that had its origins in the competitive interaction of Cl in increasing Ki(bumetanide) (11, 20). This does not rule out the possibility that part of the bumetanide-binding site is shared with the Cl site.
Interestingly, although predicted transmembrane 2 does not contain any charged residues, it does include an unusually high concentration of serine and threonine residues (4)—8 of 14 residues in one section of this helix. Many of these are clustered on the same face of the predicted helix as three of the amino acids (SV- and -M) identified in this study. This suggests the possibility that this face of transmembrane 2 forms part of a hydrophilic channel involved in ion translocation. In future studies it should be possible to examine this hypothesis and to use the present approach to identify additional residues that participate in ion interactions.
Acknowledgments
We gratefully acknowledge the technical assistance of Mrs. Grace Jones. We also thank Rachel Behnke, Susan Brill, Deborah Lynn, Iñaki Giménez, and Andreas Flemmer for reading the manuscript. This study was supported by National Institutes of Health Grants DK-47661 and DK-17433. P.I. was supported by a Clinician Scientist Fellowship from the Medical Research Council of Canada.
ABBREVIATIONS
- NKCC
Na–K–Cl cotransporter(s)
- hNKCC
human NKCC
- sNKCC
shark NKCC
- rNKCC
rabbit NKCC
- NCC
Na–Cl cotransporter
References
- 1.Xu J, Lytle C, Zhu T T, Payne J A, Benz E, Jr, Forbush B., III Proc Natl Acad Sci USA. 1994;91:2201–2205. doi: 10.1073/pnas.91.6.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delpire E, Rauchman M I, Beier D R, Hebert S C, Gullans S R. J Biol Chem. 1994;269:25677–25683. [PubMed] [Google Scholar]
- 3.Payne J A, Xu J C, Haas M, Lytle C Y, Ward D, Forbush B., III J Biol Chem. 1995;270:17977–17985. doi: 10.1074/jbc.270.30.17977. [DOI] [PubMed] [Google Scholar]
- 4.Payne J A, Forbush B., III Proc Natl Acad Sci USA. 1994;91:4544–4548. doi: 10.1073/pnas.91.10.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Igarashi P, Vanden Heuvel G B, Payne J A, Forbush B., III Am J Physiol. 1995;269:F405–F418. doi: 10.1152/ajprenal.1995.269.3.F405. [DOI] [PubMed] [Google Scholar]
- 6.Haas M. Annu Rev Physiol. 1989;51:443–457. doi: 10.1146/annurev.ph.51.030189.002303. [DOI] [PubMed] [Google Scholar]
- 7.Gamba G, Miyanoshita A, Lombardi M, Lytton J, Lee W S, Hediger M A, Hebert S C. J Biol Chem. 1994;269:17713–17722. [PubMed] [Google Scholar]
- 8.Gillen C M, Brill S, Payne J A, Forbush B., III J Biol Chem. 1996;271:16237–16244. doi: 10.1074/jbc.271.27.16237. [DOI] [PubMed] [Google Scholar]
- 9.Gamba G, Saltzberg S N, Lombardi M, Miyanoshita A, Lytton J, Hediger M A, Brenner B M, Hebert S C. Proc Natl Acad Sci USA. 1993;90:2749–2753. doi: 10.1073/pnas.90.7.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payne J A, Stevenson T J, Donaldson L F. J Biol Chem. 1996;271:16245–16252. doi: 10.1074/jbc.271.27.16245. [DOI] [PubMed] [Google Scholar]
- 11.Forbush B, III, Palfrey H C. J Biol Chem. 1983;258:11787–11792. [PubMed] [Google Scholar]
- 12.Lytle C, Xu J-C, Biemesderfer D, Haas M, Forbush B., III J Biol Chem. 1992;267:25428–25437. [PubMed] [Google Scholar]
- 13.Moore M L, George J N, Turner R J. Biochem J. 1995;309:637–642. doi: 10.1042/bj3090637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lytle C, Forbush B., III J Biol Chem. 1992;267:25438–25443. [PubMed] [Google Scholar]
- 15.Lytle C, Forbush B., III Am J Physiol. 1996;270:C437–C448. doi: 10.1152/ajpcell.1996.270.2.C437. [DOI] [PubMed] [Google Scholar]
- 16.Isenring P, Jacoby S C, Payne J A, Forbush B., III J Biol Chem. 1998;273:11295–11301. doi: 10.1074/jbc.273.18.11295. [DOI] [PubMed] [Google Scholar]
- 17.Isenring P, Forbush B., III J Biol Chem. 1997;272:24556–24562. doi: 10.1074/jbc.272.39.24556. [DOI] [PubMed] [Google Scholar]
- 18.Isenring P, Behnke R D, Forbush B. J Am Soc Nephrol. 1995;6:341. (abstr.). [Google Scholar]
- 19.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–381. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 20.Haas M, McManus T J. Am J Physiol. 1983;245:C235–C240. doi: 10.1152/ajpcell.1983.245.3.C235. [DOI] [PubMed] [Google Scholar]
- 21.Turner R J, George J N. J Membr Biol. 1988;102:71–77. doi: 10.1007/BF01875354. [DOI] [PubMed] [Google Scholar]
- 22.Hegde R S, Palfrey H C. J Membr Biol. 1992;126:27–37. doi: 10.1007/BF00233458. [DOI] [PubMed] [Google Scholar]
- 23.Lytle C, McManus T J. J Gen Physiol. 1986;88:36a. [Google Scholar]
- 24.Rose B D. Clinical Physiology of Acid-Base and Electrolyte Disorders. New York: McGraw–Hill; 1994. [Google Scholar]