Abstract
The interferon inducible transmembrane (IFITM) proteins mediate several cellular processes such as homotypic cell adhesion functions of interferons (IFNs) and cellular anti-proliferative activities. We show that the BAF complex mediated induction of IFITM3 is dependent on binding of the transcriptional enhancer factor 1 (TEF-1/TEAD1) to the M-CAT like elements of its promoter. TEF-1 knock-down reduced the BAF complex mediated activation of IFITM3 promoter. In the absence of the BAF complex, TEF-1 is repressive to IFITM3 expression. The regulation of IFITM3 by TEF-1 demonstrates that TEF-1 dependent regulation is more widespread than its previously established role in the expression of muscle specific genes.
1. INTRODUCTION
Interferons (IFNs) play critical roles in tumor surveillance by controlling apoptosis and through cellular anti-proliferative and differentiating activities. They also play major roles in cellular defense against viral and parasitic infection [1,2]. The expression of several interferon inducible genes require the chromatin remodeling SWI/SNF-like BAF complexes for their basal as well as the IFN inducible expression [3-6]. Among these are the interferon inducible transmembrane protein (IFITM) family genes, which comprise of IFITM1 (9−27), IFITM2 (1−8U) and IFITM3 (1−8D) [7,8]. These genes have been implicated in several cellular processes such as homotypic cell adhesion functions of IFN and cellular anti-proliferative activities [7,9,10]. The expression level of IFITM genes have also been found to be up-regulated in a number of cancer cells [1,10].
We previously found that the BAF complex is constitutively associated with the promoters of the IFITM genes and maintains an open chromatin structure at the promoter for rapid induction by IFNs or viral infection [3]. We had also shown that the BAF complex-mediated induction of the IFITM3 is dependent on two critical DNA elements in its promoter [5]. The first element located between the positions −152 and −138, contains an Sp1 binding motif. Sp1 recruited BAF complex to the promoter and a mutation in this site reduced BAF complex-mediated activation significantly. The second element was identified upstream of the Sp1 binding motif, between the positions −173 and −152. Deletion of this region resulted in a significant reduction in the BAF complex-mediated activation as well. Though the sequence analysis of this region did not reveal any consensus protein binding motifs [5], a sequence, 5’-AGGAATTTGT-3’, resembling an M-CAT motif was identified. In this study, we present evidence that the second critical element in the IFITM3 promoter is bound by the transcriptional enhancer factor 1 (TEF-1/TEAD1). TEF-1 regulates the BRG1-dependent activation and IFN-induction of the IFITM3 gene.
2. MATERIALS AND METHODS
2.1. Constructs and antibodies
pREP4-Luc, pREP7-BRG1 and pREP4-puro were described earlier [3,6]. pREP4-TM3-Luc was constructed as reported earlier, by PCR amplifying the IFITM3 promoter from position −238 to −25 [5]. Point mutations of the IFITM3 promoter were performed using the QuickChange kit (Stratagene). TEF-1 antibodies were obtained from BD transduction laboratories (610923).
2.2. Cell culture, transfection and luciferase assay
SW-13 and Hela cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1 mM glutamine. For reporter assays, the reporter constructs were transfected into SW-13 or HeLa cells and luciferase activity was measured 72 hrs after transfection using the dual luciferase assay kit (Promega). Transfections were carried out using superfect transfection reagent (Qiagen).
2.3. Electrophoretic mobility shift assay (EMSA)
For EMSA, the nuclear extracts from SW-13 and HeLa cells or the in vitro translated TEF-1 were used. Detailed nuclear extracts isolation and EMSA procedures are provided in the supplemental information.
2.4. DNA affinity protein purification and mass spectrometry
For DNA affinity purification, biotinylated oligonucleotides containing the 7-bp protein-binding site were concatamerized using the self primed PCR technique [11,12]. Complete details of the DNA affinity protein purification are provided in the supplemental information. MALDI-TOF mass spectrometry was carried out at the Lerner Research Institute, Mass Spectrometry Laboratory for Protein Sequencing, the Cleveland clinic foundation, Cleveland, Ohio.
2.5. RNA interference, RNA isolation, cDNA synthesis and quantitative PCR
TEF-1 siRNAs were obtained from Qiagen. Total RNAs were isolated from HeLa or SW-13 cells as described earlier [6]. cDNA was synthesized using SuperScript III RNase H- reverse transcriptase (Invitrogen). TaqMan probes and universal RT-PCR master mix were obtained from Applied Biosystems Inc.
2.6. In vitro translation
HeLa cell mRNAs were used to synthesize the cDNA encoding TEF-1. The cDNA was cloned into the vector, pBluescript KS (Stratagene). TnT Quick Coupled Transcription/Translation system was used for in vitro translation (Promega).
3. RESULTS
3.1. Identification of the sites essential for IFITM3 promoter activation
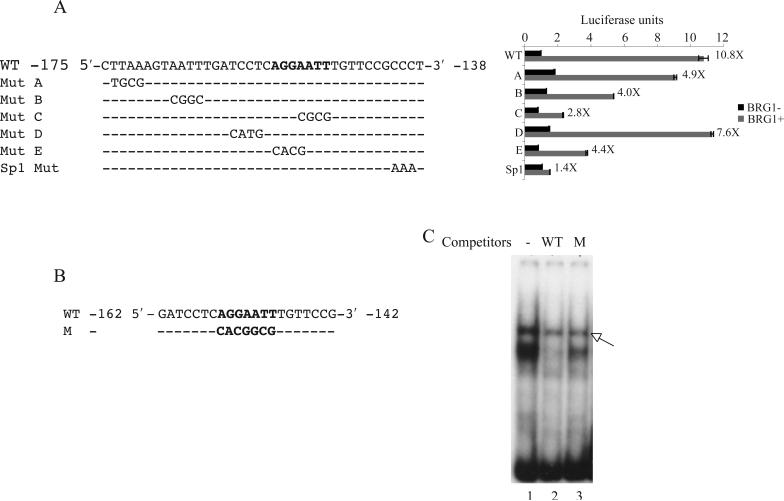
SW-13 cells do not express detectible levels of either BRG1 or hBRM, the essential ATPase subunits of the BAF complexes. Reconstitution of active BAF complex by the transient expression of BRG1 activates a number of genes including the IFN-inducible IFITM3 [6]. We previously found that a 213 bp promoter fragment of IFITM3 contains sequences responsive to BRG1 activation in SW-13 cells [5]. Though the Sp1 binding site between −144 and −139 plays a critical role, the region between −173 and −152 also contribute significantly to the BRG1-mediated activation of the IFITM3 promoter [5], suggesting that more elements may mediate the BRG1 activation. To identify the BRG1 response sequences between the positions −173 and −152 of the IFITM3 promoter, we generated luciferase constructs containing the sequences from −238 to −25 in pREP4 episomal vector, which forms a regular chromatin structure. Mutants were constructed with mutations spanning the promoter region between −175 and −138. Activities of the constructs were analyzed by transient transfection assays (Fig. 1A). The wild-type promoter construct showed an activation of over 10-fold when BRG1 was transiently transfected into the cells. Mutation C resulted in the most significant decrease in promoter activity, whereas the mutations A and B modestly reduced the promoter activity (Fig. 1A). Mutation E, which overlaps the mutation C also showed a modest reduction in the promoter activity. Thus the mutations C and E together are a 7-bp region that is important in mediating the BRG1 activity.
Fig.1.
Identification of the sites critical for the BAF complex dependent regulation of IFITM3 promoter. (A) Left panel: Sequence between the positions −175 and −138 of the IFITM3 promoter in the luciferase reporter constructs (pREP4-TM3-Luc). WT:wild type promoter. The 7-bp site important for the IFITM3 promoter activation is shown in bold. Mut A to Mut E : promoter mutants, mutated sites are indicated; Sp1 Mut: Sp1 binding site mutant. Right panel: luciferase activities of the corresponding constructs. (B) Sequence of the oligonucleotide probes used for EMSA in Figures 1C and 3A, B and C. WT: wild-type probe; M: mutant probe, mutated site shown in bold. (C) EMSA with SW-13 nuclear extracts. A 100-fold excess of the wild-type or mutant competitors were used. WT: wild-type competitor (Lane 2), M: mutant competitor (Lane 3), Lane 1: no competitor. Arrow - non-specific band
To analyze the protein-DNA interactions in this region we carried out electrophoretic mobility shift assays (EMSA) using a 21-bp [γ−32P]ATP labeled oligonucleotide probe spanning the positions from −162 to −142 (Fig. 1B). When incubated with the nuclear extracts of SW-13 cells, retardation in the migration of the wild-type probe due to the formation of a protein/DNA complex was observed (Fig. 1C, lane 1). Competition with a 100-fold excess of the unlabelled probe diminished the signal intensity of the complex (Fig. 1C, lane 2). Furthermore, an excess of the unlabelled mutant probe with mutations in the 7-bp sequence (Fig. 1B, highlighted in bold), did not compete with the complex (Fig. 1C, lane 3), showing that the protein-DNA interaction was specific.
3.2. Transcriptional enhancer factor 1 binds to IFITM3 promoter
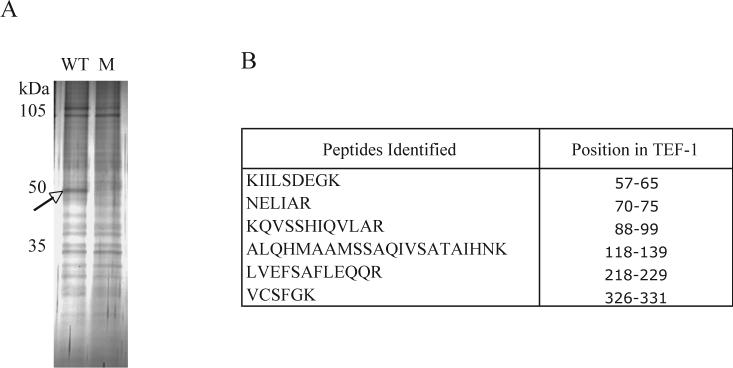
To identify the proteins that bind to the probe, we used DNA affinity chromatography. In this procedure, biotinylated oligonucleotides containing the 7-bp protein-binding site (Fig. 1B, WT) were concatamerized by PCR amplification [11,12]. The PCR products were bound to streptavidin coated magnetic beads and were incubated with SW-13 nuclear extracts. The bound factors were resolved on an SDS-PAGE gel and stained with silver nitrate (Fig. 2A, WT). In parallel, control purification was carried out with mutant oligonucleotides (Fig. 1B, M). A comparison of the protein bands from the two samples revealed a 50-kDa band in the wild-type eluate, which was absent in the mutant eluate (Fig. 2A, compare lanes WT and M). The protein band was excised from the gel and analyzed by matrix-assisted laser desorption time-of-flight (MALDI-TOF) mass spectrometry. Six peptides were identified from this sample, which were identical to the sequence of the protein, transcriptional enhancer factor 1 (TEF-1) (Fig. 2B). TEF-1 is a member of the eukaryotic TEA/ATTS family of transcription factors, which contains the highly conserved DNA binding domain that recognize the canonical M-CAT sequence motif, 5’-AGGAATG-3’ [13,14].
Fig.2.
Purification and identification of the protein binding to the 7-bp region of the IFITM3 promoter. (A) Proteins purified from the SW-13 nuclear extracts by DNA affinity chromatography using the wild-type (Fig. 1B, WT) and mutant probes (Fig. 1B, M) were resolved by SDS-PAGE and stained with silver nitrate. The 50-kDa band (arrow) was excised from the gel and analyzed by MALDI-TOF mass spectrometry. (B) The six peptides that were identified by MALDI-TOF MS and their positions in the TEF-1 protein.
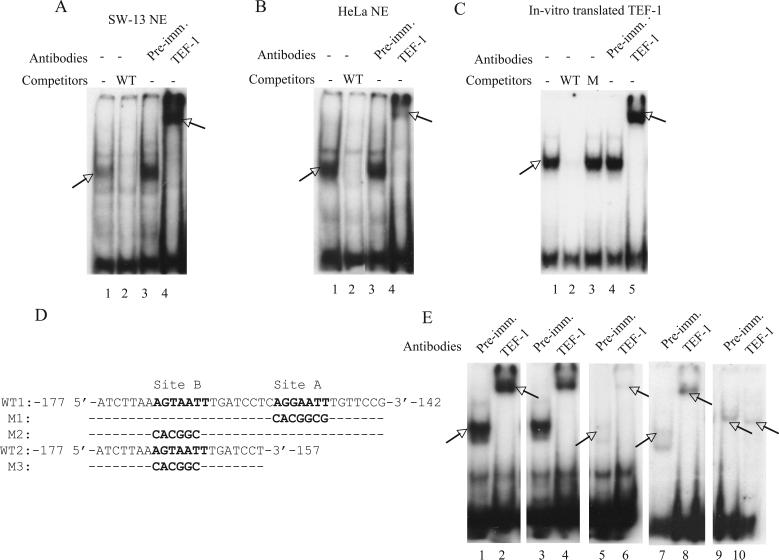
To investigate whether the probe shift in the EMSA assays (Fig. 1C) were caused by the binding of TEF-1, we carried out EMSA supershift assays using anti TEF-1 antibodies. TEF-1 antibodies supershifted the protein/DNA complex formed with both SW-13 (Fig. 3A, lane 4) and HeLa nuclear extracts (Fig. 3B, lane 4). Addition of pre-immune serum did not result in any supershifted complexes (Fig. 3A and B, lanes 3). We then used the in vitro translated TEF-1 to confirm its binding to the probe. As shown in figure 3C, the in vitro translated TEF-1 formed a complex with the probe, with migration similar to that observed with the endogenous TEF-1 in SW-13 (Fig. 1C and 3A) and HeLa nuclear extracts (Fig. 3B). While 100-fold excess of wild-type unlabelled probe competed with the complex formation, the mutant probe did not (Fig. 3C, compare lanes 2 and 3). In addition, the in vitro translated TEF-1/probe complex was supershifted when incubated with TEF-1 antibodies (Fig. 3C, lane 5). Thus the properties of the in vitro translated TEF-1 were similar to that of the endogenous protein, confirming that the protein bound to the probe was indeed TEF-1.
Fig.3.
TEF-1 binds to the IFITM3 promoter. (A) EMSA of the wild-type probe (Fig. 1B) with SW-13 nuclear extracts. A protein/DNA complex was formed (lane 1) which was competed by a 100-fold excess of the unlabeled wild-type probe (WT). TEF-1 antibody super-shifted the complex (lane 4), while the pre-immune serum did not (lane 3). The shifted and supershifted bands are indicated by arrows. (B) EMSA was performed as above, with HeLa nuclear extracts. (C) EMSA was performed as above, with in vitro translated TEF-1. M: 100-fold excess of the mutant probe (Fig. 1B, M). (D) WT1, M1 and M2: probes used in the EMSA shown in figure 3E, lanes 1−6 (WT1: lanes 1 and 2; M1: lanes 5 and 6; M2: lanes 3 and 4). Sites A and B: primary and secondary TEF-1 binding sites, respectively, in the wild-type probe (WT). M1 and M2: site A and site B mutant probes, respectively. Mutated sites are in bold. WT2 and M3: probes used in the EMSA shown in Fig. 3E, lanes 7−10 (WT2:lanes 7 and 8; M3: lanes 9 and 10). WT2 :wild-type probe, M3: mutant probe. Mutated sites are in bold. (E) EMSA with the SW-13 nuclear extracts and in vitro translated TEF-1.The protein/DNA complex formed between the SW-13 nuclear extracts and the wild-type probe (WT1, lane 1) is supershifted by TEF-1 antibodies (lane 2). A protein/DNA complex is formed with the site B mutant probe (M2, lane 3) and is supershifted by the TEF-1 antibodies (lane 4). The complex formation is significantly reduced when the site A mutant (M1) probe was used. A very faint complex formation is observed (lane 5, arrow) that is supershifted by TEF-1 antibodies (lane 6, arrow). The protein/DNA complex formed between the in vitro translated TEF-1 and the wild-type probe (WT2, lane 7) is supershifted by TEF-1 antibodies (lane 8). The mutant probe (M3, lanes 9 and 10) did not form a specific complex. The non-specific bands in lane 9 and 10 are marked with arrows.
The IFITM3 promoter contains another sequence, between the positions −170 and −164 (5′-AGTAATT-3′), that resembles the M-CAT sequence. Mutation of this sequence also caused a modest decrease in BRG1-induced activation (Fig. 1A, Mut B). We used EMSA to examine whether TEF-1 can bind to this sequence. For this, we used a 36-bp probe (Fig 3D, probe WT1), encompassing both the −170 5′-AGTAATT-3′ −164 region (Fig. 3D, site B, in bold) and the identified TEF-1 binding site (Fig. 3D, site A, in bold) in the IFITM3 promoter. When incubated with SW-13 nuclear extracts, the probe formed a protein/DNA complex, which was supershifted by TEF-1 antibodies (Fig. 3E, lanes 1 and 2). Mutation in site B (Fig 3D, probe M2) did not alter the gel mobility shift or the supershift with TEF-1 antibodies (Fig 3E, lanes 3 and 4). There was no significant variation in the signal intensity of the shifted complexes formed with the wild-type and mutant probe, M2 (Fig 3E, compare lanes 1,2 with 3,4) showing that the TEF-1 binding is largely unaffected. When site A in the probe was mutated (Fig 3D, probe M1) the protein/DNA complex signal intensity was greatly reduced (Fig. 3E, lanes 5 and 6), showing that TEF-1 bound the probe much less efficiently. Interestingly, the migration pattern in the EMSA and EMSA supershift assays was similar to that of the wild-type probe showing that indeed TEF-1 has a secondary binding site between the positions −170 and −164 of the IFITM3 promoter. Thus the reduction in the BRG1 mediated activation in the IFITM3 promoter mutant B (Fig. 1A, Mut B) could be attributed to the loss of a TEF-1 binding site. To confirm that the bands observed in figure 3E, lanes 5 and 6, were due to the binding of TEF-1 to site B and not due to some residual binding to the mutated site A, we carried out EMSA with a shorter probe which lacked site A (Fig. 3D, probe WT2). The migration of the wild-type probe was retarded by the in vitro translated TEF-1 and the complex was supershifted by TEF-1 antibodies (Fig. 3E, lanes 7 and 8). When site B in the probe was mutated (Fig. 3D, M3), no retardation of the probe migration was seen (Fig. 3E, lanes 9 and 10), showing clearly that the TEF-1 bound to a secondary site in the IFITM3 promoter, though much less efficiently.
Next, we tried to demonstrate binding of TEF-1 to the IFITM3 promoter in vivo, using chromatin immunoprecipitation (ChIP) assays. Unfortunately, we could not detect enrichment with the TEF-1 antibodies compared to pre-immune controls (data not shown), which can be explained by inaccessible epitopes or antibodies not being suitable for ChIP analysis. Therefore, we used the following methods to confirm the involvement of TEF-1 in IFITM3 regulation.
3.3. IFITM3 promoter activity is dependent on TEF-1
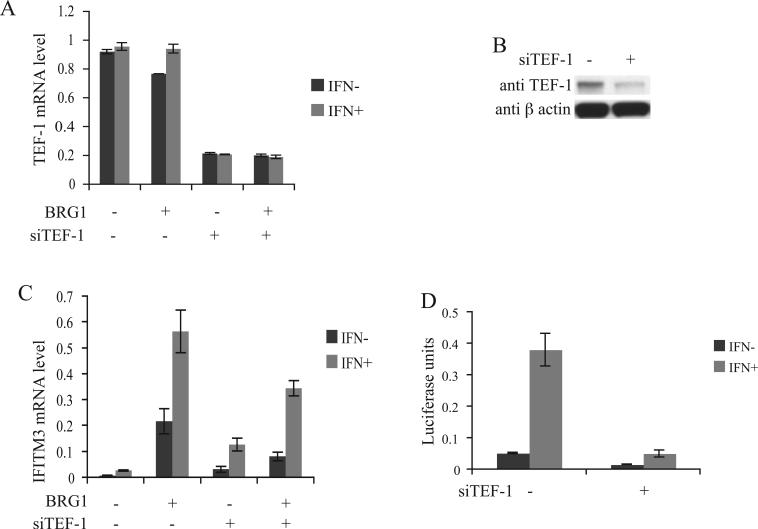
In order to determine whether TEF-1 is involved in the regulation of IFITM3 we knocked-down TEF-1 in cultured cells. As shown in figure 4A, knock-down of TEF-1 in SW-13 cells by transfection with siRNAs targeting TEF-1 mRNA resulted in its reduction by over 70% and a similar reduction in the protein levels was also observed (Fig. 4B). Neither BRG1 expression nor IFN-α treatment affected the TEF-1 expression level (Fig. 4A). The basal expression of IFITM3 was low in SW-13 cells (Fig. 4C). Induction with IFN-α resulted in a slight increase in the expression. Transient transfection of BRG1 and thereby reconstitution of the active BAF complex resulted in a significant increase in the basal and the IFN-α induced IFITM3 expression levels (Fig. 4C). When TEF-1 levels in the cells were reduced by siTEF-1, a two-fold decrease of the BRG1-mediated induction of the basal IFITM3 expression was observed (Fig. 4C). A corresponding decrease in the IFN-α induced expression level also was also observed. Interestingly, the basal and the IFN-α induced levels of IFITM3 expression were higher in TEF-1 knock-down SW-13 cells in the absence of the active BAF complex, when compared to the cells without TEF-1 knock-down (Fig. 4C). This suggests a repression activity caused or mediated by TEF-1 in the absence of the BAF complex on the IFITM3 promoter.
Fig.4.
TEF-1 knock-down results in the reduction of IFITM3 promoter activity and expression levels. (A) SW-13 cells were transfected with siTEF-1 RNA oligonucleotides or the control RNA oligonucleotides and co-transfected with either pREP7-BRG1-Hygromycin or pREP7-Hygromycin control vector. Total RNAs were isolated from the cells selected with hygromycin for 48 h and TEF-1 mRNA levels were determined using quantitative real-time PCR. The cells were induced with 1000 u/ml α-interferon (IFN-α) for 12 h. (B) Western blot showing the TEF-1 levels in SW-13 cells transfected with siTEF-1 RNA oligonucleotides or the control RNA oligonucleotides and co-transfected with pREP7-Hygromycin. Protein was isolated from the cells selected with hygromycin for 48 h. β actin was used as control (C) IFITM3 mRNA levels in SW-13 cells. Conditions same as in figure 4A. (D) IFITM3 promoter activity in the wild-type and TEF-1 knock-down HeLa cells. pREP4-TM3-Luc was co-transfected into HeLa cells with siTEF-1 RNA oligonucleotides or the control RNA oligonucleotides. The cells were induced with 1000 u/ml IFN-α for 12 h. Luciferase activity was measured 48 h after transfection.
TEF-1 regulation of IFITM3 promoter was also tested in HeLa cells, which has an active BAF complex. As shown in figure 4D, IFN-α treatment induced the promoter activity significantly. Transfection of the siTEF-1 into HeLa cells, which caused 70% reduction in the TEF-1 expression (data not shown), resulted in a significant decrease in both the basal and IFN-α induced promoter activity (Fig. 4D).
4. DISCUSSION
TEF-1 belongs to a family of four transcription factors that also include TEF-3, TEF-4 and TEF-5. They contain the highly conserved N-terminal TEA-DNA binding domain (TEAD) [13-16] recognizing the canonical M-CAT motif in several muscle-specific gene promoters [17-19]. While TEF-1 is known to regulate several M-CAT dependent promoters that are muscle specific, TEF-1 mRNA has been detected in several other tissues, suggesting that the M-CAT binding activity could be involved in the regulation of non-muscle specific genes as well [20]. In this study, we have found that TEF-1 regulates the interferon-inducible IFITM3 gene in non-muscle cells.
EMSA assays showed that the TEF-1 protein binds specifically to the 5′-AGGAATT-3′ sequences between the positions −155 and −149 in the IFITM3 promoter. This is a one base pair variation from the canonical M-CAT motif, 5′-AGGAATG-3′. Mutation in the TEF-1 binding site resulted in significant reduction in the promoter activity (Fig. 1A). Knock-down of the TEF-1 by siRNAs resulted in decreased IFITM3 mRNA levels (Fig. 4C) and promoter activity in reporter assays (Fig. 4D), thereby clearly showing the involvement of TEF-1 in IFITM3 gene regulation.
In addition to the sequences between −155 and −149 (Fig. 3D, site A) in the IFITM3 promoter, TEF-1 also exhibits a weak binding to a secondary site, 5′-AGTAATT-3′ (Fig 3D, site B) between positions −170 and −164, which varies from the canonical M-CAT motif in the first and fifth positions. A systematic study to identify nucleotides in the M-CAT motif, most important for the activation of the c-TNT promoter in the chick embryo primary muscle cells has shown that a shift in the fifth position of the TEF-1 binding site 5′-CATTCCT-3′ from C to A resulted in a significant decrease in the promoter activity [21]. This could explain the weaker binding of TEF-1 to site B and stronger binding to site A (Fig. 3E). However, even though the majority of the activation potential seems to arise from site A, site B also contributes to the activity of the IFITM3 promoter as its mutation resulted in a modest decrease in BRG1-mediated activation (Fig 1A, mutants A and B). Both the TEF-1 binding sites A and B are completely conserved between IFITM3, IFITM2 and IFITM1 [7], suggesting a similar mechanism in the regulation of all the IFITM genes.
Several cofactors that are required for TEF-1 activity have been identified [13,22,23]. TONDU, the mammalian homolog of Drosophila vestigial gene, binds to all the four TEAD proteins [24]. YAP65, that interacts with all the four TEAD proteins has been shown to be a general transcriptional co-activator in mammalian cells [15]. TAZ also has been shown to be a co-activator of TEF-1 [13]. This study showed that TEF-1 and the BAF complex cooperatively activate the IFITM3 gene, which suggests that the BAF complex could be a co-activator of TEF-1 during the regulation of IFITM3.
In this study, we found that TEF-1 knock-down resulted in an increase in the basal expression levels of the IFITM3 gene in SW-13 cells, suggesting that TEF-1 could play a repressive role in the absence of the co-activating BAF complex. TEF-1 has been suggested to act as a repressor of the α-tropomyosin transgene in the early embryo. The lack of the co-repressors or the presence of co-activators has been speculated as the reason for the loss of repressive activity of TEF-1 during the later stages of development [25]. Over-expression of TEF-1 resulting in the repression activity of TEF-1 is also well known. It has been suggested that this phenomenon could be due the squelching, where the co-activator could be titrated out by the binding of the over-expressed TEF-1 [15,26-28]. Thus the repressive effect of TEF-1 could be due to a co-repressor activity, which may be recruited by TEF-1 in the absence of the co-activator, the BAF complex being such a co-activator in our studies.
We previously found the involvement of Sp1 in the BRG1 mediated activation of the IFITM3 gene. In this study, we have identified a novel player, TEF-1 in its regulation. Our data indicate that the expression of the IFITM genes is regulated by multiple transcription factors through interaction with the chromatin remodeling complexes and suggests a role for TEF-1 in the IFN signaling pathway
ACKNOWLEDGEMENTS
This work was supported by the Intramural Research Program of the National Heart, Lung and Blood Institute, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
REFERENCES
- 1.Yang Y, et al. The interferon-inducible 9−27 gene modulates the susceptibility to natural killer cells and the invasiveness of gastric cancer cells. Cancer Lett. 2005;221:191–200. doi: 10.1016/j.canlet.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 2.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–64. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Cui K, Tailor P, Liu H, Chen X, Ozato K, Zhao K. The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming. Mol Cell Biol. 2004;24:4476–86. doi: 10.1128/MCB.24.10.4476-4486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang M, Qian F, Hu Y, Ang C, Li Z, Wen Z. Chromatin-remodelling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nat Cell Biol. 2002;4:774–81. doi: 10.1038/ncb855. [DOI] [PubMed] [Google Scholar]
- 5.Liu H, Kang H, Liu R, Chen X, Zhao K. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol Cell Biol. 2002;22:6471–9. doi: 10.1128/MCB.22.18.6471-6479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu R, Liu H, Chen X, Kirby M, Brown PO, Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–18. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 7.Lewin AR, Reid LE, McMahon M, Stark GR, Kerr IM. Molecular analysis of a human interferon-inducible gene family. Eur J Biochem. 1991;199:417–23. doi: 10.1111/j.1432-1033.1991.tb16139.x. [DOI] [PubMed] [Google Scholar]
- 8.Lange UC, Saitou M, Western PS, Barton SC, Surani MA. The fragilis interferon-inducible gene family of transmembrane proteins is associated with germ cell specification in mice. BMC Dev Biol. 2003;3:1. doi: 10.1186/1471-213X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans SS, Collea RP, Appenheimer MM, Gollnick SO. Interferon-alpha induces the expression of the L-selectin homing receptor in human B lymphoid cells. J Cell Biol. 1993;123:1889–98. doi: 10.1083/jcb.123.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreu P, Colnot S, Godard C, Laurent-Puig P, Lamarque D, Kahn A, Perret C, Romagnolo B. Identification of the IFITM family as a new molecular marker in human colorectal tumors. Cancer Res. 2006;66:1949–55. doi: 10.1158/0008-5472.CAN-05-2731. [DOI] [PubMed] [Google Scholar]
- 11.Hemat F, McEntee K. A rapid and efficient PCR-based method for synthesizing high-molecular-weight multimers of oligonucleotides. Biochem Biophys Res Commun. 1994;205:475–81. doi: 10.1006/bbrc.1994.2690. [DOI] [PubMed] [Google Scholar]
- 12.Xue HH, et al. GA binding protein regulates interleukin 7 receptor alpha-chain gene expression in T cells. Nat Immunol. 2004;5:1036–44. doi: 10.1038/ni1117. [DOI] [PubMed] [Google Scholar]
- 13.Mahoney WM, Jr., Hong JH, Yaffe MB, Farrance IK. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J. 2005;388:217–25. doi: 10.1042/BJ20041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anbanandam A, Albarado DC, Nguyen CT, Halder G, Gao X, Veeraraghavan S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci U S A. 2006;103:17225–30. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–41. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang SW, Dong M, Trujillo MA, Miller LJ, Eberhardt NL. DNA binding of TEA/ATTS domain factors is regulated by protein kinase C phosphorylation in human choriocarcinoma cells. J Biol Chem. 2001;276:23464–70. doi: 10.1074/jbc.M010934200. [DOI] [PubMed] [Google Scholar]
- 17.Nikovits W,, Jr., Kuncio G, Ordahl CP. The chicken fast skeletal troponin I gene: exon organization and sequence. Nucleic Acids Res. 1986;14:3377–90. doi: 10.1093/nar/14.8.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin SB, Farrance IK, Ordahl CP. Flanking sequences modulate the cell specificity of M-CAT elements. Mol Cell Biol. 1996;16:3742–55. doi: 10.1128/mcb.16.7.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang SW, Desai D, Khan S, Eberhardt NL. Cooperative binding of TEF-1 to repeated GGAATG-related consensus elements with restricted spatial separation and orientation. DNA Cell Biol. 2000;19:507–14. doi: 10.1089/10445490050128430. [DOI] [PubMed] [Google Scholar]
- 20.Farrance IK, Ordahl CP. The role of transcription enhancer factor-1 (TEF-1) related proteins in the formation of M-CAT binding complexes in muscle and non-muscle tissues. J Biol Chem. 1996;271:8266–74. doi: 10.1074/jbc.271.14.8266. [DOI] [PubMed] [Google Scholar]
- 21.Farrance IK, Mar JH, Ordahl CP. M-CAT binding factor is related to the SV40 enhancer binding factor, TEF-1. J Biol Chem. 1992;267:17234–40. [PubMed] [Google Scholar]
- 22.Yoshida T. MCAT Elements and the TEF-1 Family of Transcription Factors in Muscle Development and Disease. Arterioscler Thromb Vasc Biol. 2007 doi: 10.1161/ATVBAHA.107.155788. In Press. [DOI] [PubMed] [Google Scholar]
- 23.Hucl T, Brody JR, Gallmeier E, Iacobuzio-Donahue CA, Farrance IK, Kern SE. High cancer-specific expression of mesothelin (MSLN) is attributable to an upstream enhancer containing a transcription enhancer factor dependent MCAT motif. Cancer Res. 2007;67:9055–65. doi: 10.1158/0008-5472.CAN-07-0474. [DOI] [PubMed] [Google Scholar]
- 24.Vaudin P, Delanoue R, Davidson I, Silber J, Zider A. TONDU (TDU), a novel human protein related to the product of vestigial (vg) gene of Drosophila melanogaster interacts with vertebrate TEF factors and substitutes for Vg function in wing formation. Development. 1999;126:4807–16. doi: 10.1242/dev.126.21.4807. [DOI] [PubMed] [Google Scholar]
- 25.Pasquet S, Naye F, Faucheux C, Bronchain O, Chesneau A, Thiebaud P, Theze N. Transcription enhancer factor-1-dependent expression of the alpha-tropomyosin gene in the three muscle cell types. J Biol Chem. 2006;281:34406–20. doi: 10.1074/jbc.M602282200. [DOI] [PubMed] [Google Scholar]
- 26.Chaudhary S, Brou C, Valentin ME, Burton N, Tora L, Chambon P, Davidson I. A cell-specific factor represses stimulation of transcription in vitro by transcriptional enhancer factor 1. Mol Cell Biol. 1994;14:5290–9. doi: 10.1128/mcb.14.8.5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao JH, Davidson I, Matthes H, Garnier JM, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–68. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 28.Jiang SW, Eberhardt NL. TEF-1 transrepression in BeWo cells is mediated through interactions with the TATA-binding protein, TBP. J Biol Chem. 1996;271:9510–8. doi: 10.1074/jbc.271.16.9510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.