Abstract
LINE-1 elements represent a significant proportion of mammalian genomes. The impact of their activity on the structure and function of the host genomes has been recognized from the time of their discovery as an endogenous source of insertional mutagenesis. L1 elements contain numerous functional internal polyadenylation signals and splice sites that generate a variety of processed L1 transcripts. These sites are also reported to contribute to the generation of hybrid transcripts between L1 elements and host genes. Using northern blot analysis we demonstrate that L1 splicing, but not L1 polyadenylation, is delayed during the course of L1 expression. L1 splicing can also be negatively regulated by EBV SM protein known to alter mRNA splicing. These results suggest a potential for L1 mRNA processing to be regulated in a tissue- and/or development-specific manner. The delay in L1 splicing may also serve to protect host genes from the excessive burden of L1 interference with their normal expression via aberrant splicing.
Keywords: LINE-1, alternative splicing, retroelement, gene regulation, mobile elements, RNA processing
1. Introduction
Long Interspersed Element-1, LINE-1 or L1, is a long-term resident of mammalian genomes that occupies up to 17% of the total DNA (Lander et al., 2001; Waterston et al., 2002). Insertional mutagenesis of L1 and its parasites, Small Interspersed Elements (SINEs) and SVA elements, has long been recognized to contribute to human disease and cancer origins and progression (Deininger et al., 2003; Kazazian and Goodier, 2002; Ostertag et al., 2003).
The majority of the L1 inserts in mammalian genomes are 5’ truncated (Grimaldi et al., 1984; Lander et al., 2001; Waterston et al., 2002). Full-length L1 (FL1) elements contain an internal polymerase II promoter that drives expression of a bicistronic mRNA terminating at the polyadenylation (polyA) site located at the 3’ end of the L1 sequence (Belancio et al., 2007; Kazazian et al., 1988; Swergold, 1990). The L1 mRNA encodes two open reading frames ORF1 and 2 that are essential for successful integration (Moran et al., 1996). Insertional mutagenesis associated with L1 activity relies on the production of the FL1 mRNA, ORF1 and ORF2 proteins for L1 integration (Moran et al., 1996), and only functional L1 ORF2 protein for mobilization of SINEs (Dewannieux et al., 2003; Moran et al., 1996).
Multiple mechanisms exist to control retrotransposition and expression of L1 elements. Cellular factors involved in retroviral defense, such as the APOBEC family of proteins are known to modulate L1 integration (Bogerd et al., 2006; Muckenfuss et al., 2006; Stenglein and Harris, 2006). L1 promoter strength and methylation serve as a first limiting factor in the production of the functional full-length L1 mRNA (Hata and Sakaki, 1997; Matlik et al., 2006; Swergold, 1990). However, efficient transcription from the L1 promoter does not manufacture high levels of the FL1 due to the complex and extensive processing of L1 RNA via premature polyadenylation at the internal polyA sites and splicing (Belancio et al., 2006; Perepelitsa-Belancio and Deininger, 2003). These processes restrain L1 expression even in cancer cells where L1 promoter methylation is lessened (Belancio et al., 2006; Ehrlich, 2002; Perepelitsa-Belancio and Deininger, 2003).
RNA processing by splicing and polyadenylation is often regulated in a developmental and tissue-specific manner, and altered upon malignant transformation (Kalnina et al., 2005; Wallace et al., 1999; Yeo et al., 2004). There is also a complex interplay between the two processes (Tian et al., 2007). Most of the splice and polyadenylation sites subjected to such regulation are weak (Batt et al., 1994; Garg and Green, 2007). The majority of the sites (functional and predicted) present within the L1 sequence fall into this category (Belancio et al., 2006; Perepelitsa-Belancio and Deininger, 2003). We have previously reported variation in the L1 processing in cancer cell lines of different origin (Belancio et al., 2006). This variation suggests a possibility that L1 mRNA splicing and polyadenylation could be regulated not only in normal cells but also in malignances.
Cis- signals for splicing and premature polyadenylation present in the L1 RNA sequence can also contribute to pre-mRNA processing of host genes that contain L1 elements. Multiple examples of the hybrid transcripts generated by splicing between L1 and human genes have been reported (Belancio et al., 2006; Matlik et al., 2006; Wheelan et al., 2005). We demonstrate that L1 RNA splicing is delayed, while premature polyadenylation of the L1-related transcripts occurs rapidly. This is likely to be due to the inefficient recognition/processing of the L1 splice sites. We propose a model in which this delay minimizes the negative impacts of L1 sequences residing within the introns of mammalian genes.
2. Materials and methods
2.1. Cell culture
NIH 3T3 (ATCC CRL-1658) and HeLa (ATCC CCL2) cells were maintained as described (Belancio et al., 2006; Perepelitsa-Belancio and Deininger, 2003).
2.2. Transient transfections
5×106 NIH 3T3 or HeLa cells were plated per T75 tissue culture flask (CORNING) and transiently transfected with 6 μg of L1.3 or L1Neo expression vectors 17−20 hours after plating using Lipofectamine Plus Reagent (InVitrogen) as described before (Perepelitsa-Belancio and Deininger, 2003). For co-transfection experiments with EBV SM protein the same number of cells was transfected with 5 μg of the L1.3 expression cassette and 3μg of vector expressing EBV SM protein in either reverse or forward orientations using Lipofectamine Plus Reagent (InVitrogen). For the experiments that test the effect of the L1-related products on the onset of L1 splicing, 2.5×106 cells were transfected with 2 μg of L1 ORF2, L1spa, or L15’UTRluc expression vectors using Lipofectamine Plus Reagent (InVitrogen). 24 hours later cells were transfected again with 6μg of the L1 expression cassette and 1 μg of the expression vectors mentioned above using Lipofectamine Plus Reagent (InVitrogen). RNAs were harvested 9 and 24 hours post transfection. Note that the post-transfection time is indicated as the time after the transfection cocktail was added to the cells. The pBudCE4.1 basic expression vector was used as a negative control for transfections with L1 ORF2, and L1spa expression vectors. Firefly luciferase driven by SV40 promoter (pGL3-Promoter, Promega) was used as a negative control for the pGL3 vector expressing Firefly luciferase driven by the L1.3 5’ UTR.
2.3. Northern blot analysis
RNA extractions and northern blot analysis was performed as previously described (Belancio et al., 2006; Perepelitsa-Belancio and Deininger, 2003).
2.4. Plasmids
JM101/L1.3 (L1.3) (Wei et al., 2001) and JM101/L1.3 (L1.3Neo) vectors are a gift from Dr. J. Moran (Sassaman et al., 1997). L1spa expression vector is a gift from Dr. Kazazian (Naas et al., 1998). EBV aSM and SM expression vectors are a gift from Dr. S. Swaminathan (Ruvolo et al., 1998). pBudORF2opt (Gasior et al., 2006) was created using the codon optimized L1RP as a source for the ORF2 coding sequences. The open reading frames were cloned into the expression vector pBudCE4.1 (Invitrogen), under control of the CMV promoter. The 5’-UTR expression vector was constructed by subcloning L1.3 5’ UTR sequence into pGL3-basic vector (Promega) to drive the expression of the Firefly luciferase gene (El Sawy et al., 2005).
3. Results
3.1. LINE-1 polyadenylation and splicing are differentially regulated
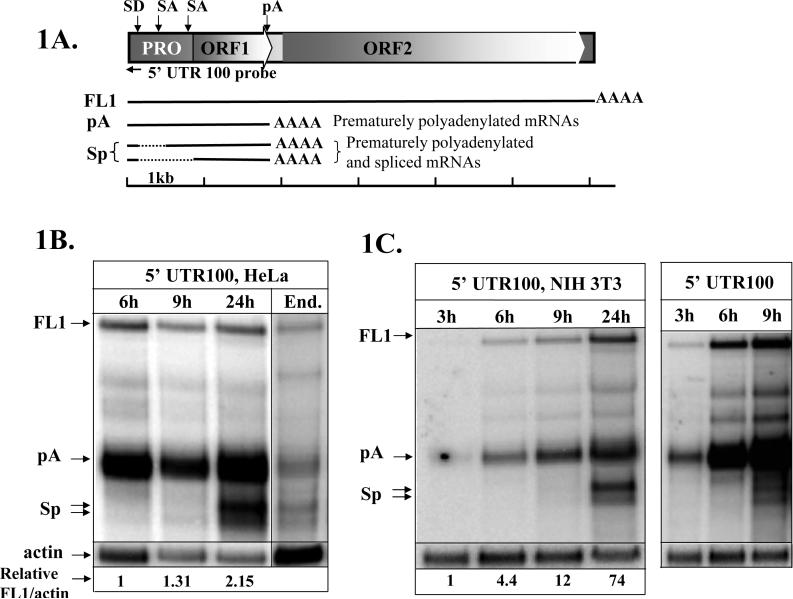
Multiple L1 loci likely undergo de-repression by the loss of promoter methylation during embryogenesis and carcinogenesis (Ehrlich, 2002; Lees-Murdock et al., 2003; Mays-Hoopes et al., 1986) or activation due to altered transcription factor availability (Tchenio et al., 2000; Yang et al., 2003). To analyze RNA species produced during progression of L1 expression, we performed a northern blot analysis of the time course of the transiently transfected wild-type L1.3 expression vector in HeLa cells (Figure 1A) with a strand-specific RNA probe. This probe is complementary to the first 100 bp of the L1 5’ UTR (5’ UTR 100 probe) and therefore detects both spliced and prematurely polyadenylated L1 transcripts (Figure 1A) (Belancio et al., 2006). This northern blot analysis demonstrated a steady accumulation of the full-length L1 mRNA (1, 1.31, and 2.15 relative units at 6, 9, and 24 hours post transfection, respectively) and the previously characterized prematurely polyadenylated L1-related products (1, 1.26, and 2.3 relative units at 6, 9, and 24 hours post transfection, respectively) between 6 and 24 hours after the addition of the transfection cocktail post. Intriguingly, spliced L1-related mRNAs were almost undetectable in the early time points (under 10% of the polyadenylated L1 mRNAs) but accumulated to amounts similar to those of the prematurely polyadenylated mRNAs (86%) by 24 hours (Figure 1B). The L1 RNA profiles produced by the transiently transfected L1 expression cassette at 24 hours are very similar to those observed for the endogenously expressed L1 elements in HeLa cells (Figure 1B, End). The similarity of the RNA processing between the transiently transfected and endogenous L1 elements in HeLa cells indicates that the delay in L1 splicing is likely to be due to the kinetics of splice site recognition and/or processing rather than due to any potential imbalance of the cellular factors associated with transfection. A similar delay in L1 mRNA splicing was also observed in mouse cells when the human L1.3 expression cassette was transiently transfected in NIH 3T3 cells (Figure 1C) indicating conservation of the process between the species.
Figure 1. L1 mRNA splicing is delayed during the course of L1 expression.
A. A schematic representation of the human L1 element and RNA species produced during L1 transcription. PRO, ORF1, and ORF2 designate internal polymerase II promoter in the 5’ UTR, and Open Reading Frames 1 and 2, respectively. FL1 corresponds to the full-length L1 mRNA, pA stands for prematurely polyadenylated L1 products that utilize internal polyA sites, and Sp refers to the spliced L1 transcripts. Solid black lines represent the different L1 transcripts, where the dashed lines represent the L1 sequences removed by splicing. A black horizontal arrow indicates relative position of the strand-specific RNA probe (5’ UTR 100 probe) in the L1 sequence. Black vertical arrows above the L1 schematic mark the positions of the splice donor (SD), splice acceptor (SA), and polyadenylation (pA) sites that generate the described L1 products. A scale bar indicates relative sizes of the depicted L1 products. B. Northern blot analysis of the transiently transfected wild-type L1.3 expression vector in HeLa cells with the strand-specific RNA probe complementary to the first 100 bp of the L1.3 sequence (5’ UTR 100) at 6, 9, and 24 hours post transfection. Note that the post-transfection time is indicated as the time after the transfection cocktail was added to the cells. mRNA profiles of the transiently transfected L1 element at 24h post transfection match well those of the endogenous (End.) L1 expressed in HeLa cells. Actin marks endogenously expressed actin mRNA. Numbers at the bottom of the northern blot represent relative ratios of the full-length L1 (FL1) mRNA to actin normalized to the FL1/actin ratio detected at 6h. Note that the levels of the endogenous L1 expression cannot be directly compared to the levels of the transiently transfected L1 mRNA because the blots were not carried out in parallel. C. Northern blot analysis of the transiently transfected wild-type L1.3 expression vector in NIH 3T3 cells with the 5’ UTR 100 probe at 3, 6, 9, and 24 hours after addition of the transfection cocktail. Right panel in C is a longer exposure of the same blot shown in the left panel. Full-length L1 mRNA (FL1), prematurely polyadenylated L1 mRNAs (pA), and spliced and prematurely polyadenylated L1 products (Sp) are indicated.
3.2. Strong splice signals constitutively used in human genes are not subject to regulation observed for the L1 splice sites
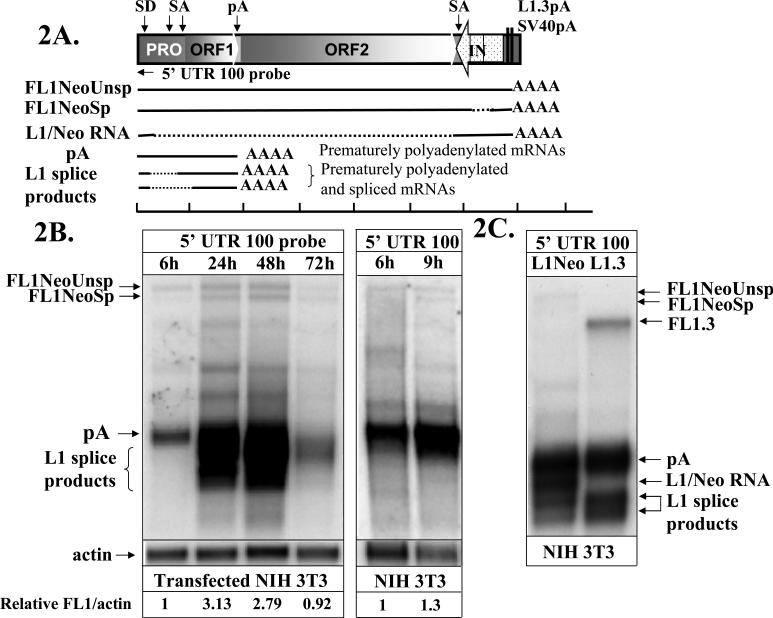
To determine whether the observation of delayed splicing is specific to L1 processing, we transiently transfected NIH 3T3 cells with an L1.3 expression cassette tagged with the neomycin resistance gene that is interrupted by the γ-globin intron (L1Neo) (Sassaman et al., 1997). Northern blot analysis of the mRNA profiles produced by the L1Neo construct (Figure 2A) demonstrated that splicing of the γ-globin intron is observed by 9 hours after the addition of the transfection cocktail (Figure 2B), while the splicing of the L1-specific products is detected only at later time points much like it is in the case of the L1 without the Neo tag (Figure 1). This observation indicates that the delay in recognition and/or processing of L1 splice sites is specific to L1 sequences. We have previously reported that the L1.3Neo expression vector produces a hybrid splice product that is the result of the usage of the L1 splice donor site at position 97 and a splice acceptor site located in the beginning of the Neo cassette (Figure 2A and C, L1/Neo splice) (Belancio et al., 2006). Figure 2 shows that the production of this product is also delayed indicating that L1 sequences dictate the onset of L1 splice site utilization even when they are used in conjunction with the non-L1 splice sites.
Figure 2. L1 but not neomycin gene splicing is delayed during the course of L1 expression.
A. A schematic representation of the neomycin tagged human L1.3 element expression cassette (L1Neo) and some of the relevant mRNA products that it makes. PRO, ORF1, ORF2, and 5’ UTR 100 probe are designated as in Figure 1. L1.3pA and SV40pA indicate the position of the respective polyadenylation signals. Stippled arrow indicates the position and orientation of the neomycin (NeoR) gene, which is interrupted by an intron (IN) in the same orientation as the L1 ORFs. Solid black lines correspond to the portions of the L1Neo expression cassette that are included into the mature mRNA product. Dashed black lines indicate sequences that are removed by splicing. Black horizontal arrow indicates relative position of the strand-specific RNA probe (5’ UTR100) used to detect both spliced and polyadenylated products. A scale bar and splice donor, acceptor, and polyadenylation sites are indicated as in 1A.B. Northern blot analysis of the transiently transfected L1Neo expression vector in NIH 3T3 cells with the 5’ UTR 100 probe at 6, 9, 24, 48 and 72 hours after the addition of the transfection cocktail. FL1NeoUnsp and FL1NeoSp indicate unspliced and spliced (intron positioned in the Neo cassette) full-length L1Neo mRNAs, respectively. pA labels prematurely polyadenylated mRNA. L1 splice products and L1/NeoRNA indicate spliced and prematurely polyadenylated mRNAs and a hybrid splice between the L1 sequence and the Neo gene as depicted in 2A (Belancio et al., 2006). C. A northern blot analysis of the transiently transfected tagged L1.3 (L1Neo) and wild-type L1.3 (L1.3) expression vectors in NIH 3T3 cells probed with the strand-specific RNA probe (5’ UTR 100). L1/Neo splice product is a hybrid mRNA formed by splicing of L1 and Neo sequences (Belancio et al., 2006). FL1.3 marks the full-length L1 mRNA produced by the wild-type L1.3 expression vector.
3.3.Regulation of L1 RNA processing is independent of L1 proteins or 5’ UTR RNA expression
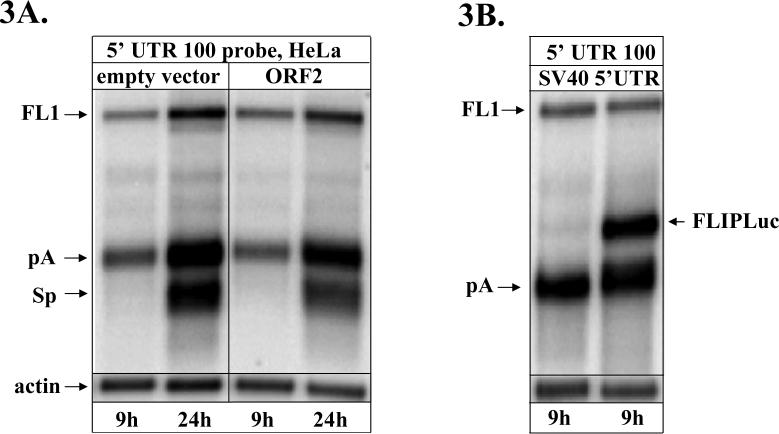
L1 elements are distant relatives of retroviruses (Nakamura and Cech, 1998) that share some aspects of their biology (reverse transcription, integration etc). Retroviruses are known to have regulatory mechanisms that control specific steps of their life cycle, such as regulation of RNA splicing by viral and cellular proteins (Kammler et al., 2006; Ropers et al., 2004; Stoltzfus and Madsen, 2006). In light of recent discoveries indicating that L1 proteins most likely interact with a broad spectrum of cellular proteins (Bogerd et al., 2006; Gasior et al., 2006; Grimaldi et al., 1984; Muckenfuss et al., 2006; Stenglein and Harris, 2006) we wished to investigate any potential involvement of the L1 proteins in the regulation of L1 splicing. To investigate this possibility HeLa or NIH 3T3 cells were transfected with expression constructs that produce large amounts of ORF2 proteins 24 hours prior to transfection with the human L1 expression cassette. Expression of the L1 ORF1 protein with the L1 expression cassette had no effect on the kinetics of the L1 splice products (data not shown). Northern blot analysis of the L1 RNA profiles in both human and mouse cell lines demonstrated that expression of either of the L1 ORFs preceding the expression of the full-length L1 elements had no influence the onset of L1 splicing (Figure 3A and D).
Figure 3. Delay in L1 splicing is not affected by L1 proteins, 5’ UTR, or L1spa.
A. Northern blot analysis of the wild-type L1.3 expression vector expressed in HeLa cells with the 5’ UTR 100 probe at 9 and 24 hours post transfection. HeLa cells were first transfected with the empty vector or a vector expressing codon optimized human L1 ORF2 protein then 24h later transfected with the L1.3 expression vector and the empty vector or the ORF2 expression vector. B. Northern blot analysis of the HeLa cells transiently transfected with the L1.3 expression vector and a Firefly luciferase expression vector driven by either the SV40 promoter (SV40, left lane) or by the L1 5’ UTR (5’ UTR, right lane) 24h after they were transfected with the Firefly luciferase expression vectors. FLIPLuc denotes mRNA produced by the Firefly luciferase expression vector driven by the L1 5’ UTR. C. Northern blot analysis of HeLa cells transiently transfected L1.3 expression vector and a vector expressing mouse L1spa element 24h after the cells were transiently transfected with the L1spa expression vector. D. Northern blot analysis of the RNA from the NIH 3T3 cells transiently transfected with L1.3 expression vector and either an empty vector or vectors expressing codon optimized human L1 ORF2 protein or mouse L1spa element. 24h before the transfection the same cells were transfected with the empty vector or vectors expressing codon optimized human L1 ORF2 protein or mouse L1spa element. 5’ UTR 100 probe was used to detect RNA isolated at 6 and 24 hours post transfection.
The most frequently used L1 splice sites are located in the L1 5’ UTR (Belancio et al., 2006). To rule out the involvement of any potential signals within the L1.3 5’ UTR that might influence L1 processing in trans, HeLa cells were transiently transfected with the expression vector containing Firefly luciferase reporter gene driven by either the SV40 promoter or the L1 5’ UTR 24 hours prior to transient transfection with the L1.3 expression vector. Northern blot analysis of the mRNA species demonstrated the same pattern and onset of L1 processing in the presence or absence of the L1 5’ UTR (Figure 3B). This result indicates that L1 proteins or 5’ UTR sequence do not alter L1 processing in trans suggesting that accumulation of L1 proteins or L1 RNA does not influence L1 processing.
To determine whether overexpression of a transcript that requires extensive pre-mRNA processing may perturb the balance of cellular proteins and lead to aberrant splicing events, human and mouse cells were pre-transfected with the mouse L1spa expression cassette that also undergoes extensive processing (Perepelitsa-Belancio and Deininger, 2003). 24 hours later the same cells were transfected with L1spa and L1.3 expression vectors. Northern blot analysis of the L1.3 mRNAs with the 5’ UTR 100 strand-specific RNA probe demonstrated no alteration in the timing of the L1 splicing or profiles of the L1-related products (Figure 3C and D).
These data indicate that the observed splice-timing phenomenon is not a mere result of saturation of the cellular splicing machinery but rather a process intrinsic to the sequence environment of the L1 splice sites that dictates their recognition and/or processing.
3.4. LINE-1 splicing is regulated by the Epstein-Barr virus (EBV) SM protein
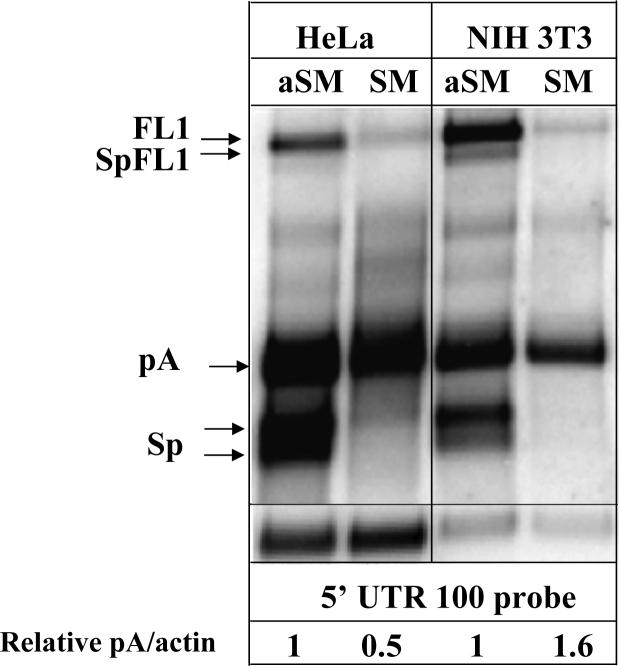
EBV SM protein has multiple functions (Ruvolo et al., 1998; Swaminathan, 2005)and it is reported to influence splicing of various cellular transcripts by preferentially decreasing splicing efficiency of the weak splice sites (Buisson et al., 1999). To determine whether L1 splicing can be regulated by the EBV SM protein we used northern blot analysis of the L1 mRNAs in HeLa and NIH 3T3 cells transiently transfected with the human L1.3 expression vector and a cassette expressing EBV SM protein in the reverse (aSM) or forward (SM) orientation (Ruvolo et al., 1998). This analysis demonstrated a dramatic effect of the SM expression on the production of the full-length L1 mRNA and almost completely abolished L1 splicing (undetectable levels) but did not alter L1 polyadenylation (relative units 1 and 0.5 and 1 and 1.6 in HeLa and NIH 3T3 cells, respectively) (Figure 4). In addition to the regulation of cellular mRNA splicing, the SM protein is also known to alter some mRNA stabilities and influence subcellular localization of unspliced cellular transcripts. We speculate that the reduced levels of the full-length L1 mRNA in the presence of the SM protein could be due to one or both of these effects. These data indicate that expression of a single protein can drastically change the processing of L1 RNAs suggesting the possibility that cellular proteins involved in regulation of RNA splicing may play a role in regulation of L1 expression and retrotransposition. Expression of cellular proteins influencing RNA splicing often exhibit some degree of tissue specificity suggesting that L1 processing can vary among different tissues.
Figure 4. The effect of EBV SM protein on L1 splicing.
Northern blot analysis of the transiently transfected wild-type L1.3 expression vector co-transfected with the vectors expressing EBV SM protein in either antisense (aSM) or sense (SM) orientations in HeLa and NIH 3T3 cells with the 5’ UTR 100 probe at 24 hours after the addition of the transfection cocktail. L1-related mRNAs are labeled as above. SpFL1 is a spliced L1 that contains both ORFs (Belancio et al., 2006). The numbers at the bottom of the figure indicate relative pA to actin ratios in each cell line normalized to the pA/actin ratio produced by the L1.3 vector co-transfected with the control (aSM) vector.
4. Discussion
Our data indicate that, even though both premature polyadenylation and splicing of L1 elements contribute almost equally to the limitation of the full-length L1 mRNA production (Belancio et al., 2006), the onset of the production of the spliced species varies greatly from the accumulation kinetics of the prematurely polyadenylated mRNAs. Prematurely polyadenylated L1 transcripts (Perepelitsa-Belancio and Deininger, 2003) are detected rather early post-transfection and accumulate steadily in the following hours. In contrast, splicing of the L1 mRNA is significantly delayed. This phenomenon of the delayed splicing is specific to the L1-encoded splice sites because the processing of the constitutive splice sites defining γ-globin intron placed in the L1 3’UTR is observed very early.
L1 sequence contains numerous splice sites, the majority of which are predicted to be weak (Belancio et al., 2006). Weak splice sites are more likely to be used for regulated splicing (Batt et al., 1994; Garg and Green, 2007). In addition to the strength of a particular splice site, its usage is often controlled by the surrounding sequences that may contain auxiliary cis-regulatory elements (Soret et al., 2006; Watakabe et al., 1993). A-richness of the L1 sequence provides ample amount of predicted recognition sites for the variety of splice proteins (Belancio et al., 2006). These ‘weak’ splice sites may also be manifested in terms of RNA molecules that take longer to assemble an effective splicing complex.
Splicing of cellular RNAs has been well established to regulate gene expression during development and in a tissue-specific manner (Elliott and Grellscheid, 2006; Yeo et al., 2004). Previous data indicate that there is some degree of variation in the processing of the endogenously expressed L1 mRNA in cancer cell lines (Belancio et al., 2006). The data presented here provide experimental evidence that non-L1 proteins known to alter splicing of cellular mRNAs can negatively influence L1 splice site usage. Expression of EBV SM protein results in the complete abolishment of the L1 splice products leaving prematurely polyadenylated mRNAs at relatively unchanged levels. EBV SM protein can regulate RNA splicing through a direct interaction with RNA thus masking cis-acting signals from the splicing machinery (Ruvolo et al., 2004) or through down-regulation of splicing factors such as SC35 (Chen et al., 2001). Either one of these possibilities suggests that the extent of L1 RNA processing may fluctuate significantly in various tissues depending on the composition and/or availability of the regulatory proteins.
Delayed L1 splicing may play a role in protecting the host from post-insertional L1 damage. Multiple reports indicate that splicing of cellular mRNAs is intimately linked to RNA transport to the cytoplasm so that unprocessed mRNAs are recognized for retention in the nucleus. L1 splicing, therefore, may serve to lower the load of L1 insertional damage in two complementary ways. First, the nuclear transport machinery may recognize the full-length L1 mRNA as an intron containing unprocessed mRNA making L1 export to the cytoplasm rather inefficient and make L1 mRNA a potential target for degradation. Trapping of the full-length L1 mRNA in the nucleus would translate into lower amount of RNP complexes in the cytoplasm destined for retrotransposition. Second, the delay in L1 splicing could be due to either inefficient recognition of the splice sites during the early stages of L1 expression followed by their proficient identification and processing later or due to the very slow processing of the splice sites throughout the time course of L1 expression. The former model would imply some sort of saturation or alteration in the composition of the splice factors during the course of L1 expression. Because pre-transfection of the extensively processed heterologous (L1spa) or L1 5’ UTR RNAs that could have saturated splicing machinery did not influence the onset of L1 splicing we believe that the latter model is a more likely one. If true, very inefficient splicing of the L1 mRNA would be halting production of the full-length L1 mRNA ultimately leading to less efficient retrotransposition.
Additionally, delayed L1 splicing may decrease the negative impact of L1 sequences on the expression of some genes. L1 interference with the normal gene expression by causing alternative, L1-hybrid splicing has been reported (Belancio et al., 2006; Matlik et al., 2006; Wheelan et al., 2005). Clearly the negative impact of the intronic full-length L1 insertions in the forward orientation on gene expression can be dramatic (Chen et al., 2006) and, therefore, it is expected that those inserts would be eliminated in utero or in a relatively short period of time during evolution. As a result there is a significant bias against the L1 inserts in the forward orientation within and near genes (Chen et al., 2006; Medstrand et al., 2002). Even then, it is intriguing that L1 interference with the normal gene expression via splicing is not as dominant as expected given the fact that there are copious amounts of intronic L1 inserts in mammalian genes (Lander et al., 2001; Waterston et al., 2002). We hypothesize that slow recognition and/or processing of the weak L1 splice sites during the transcription of cellular genes allows proper recognition of the intron/exon boundaries of cellular genes promoting normal splicing. This affect is likely to be variable with different genes, depending on the strength of their splice signals and regulatory elements. Under this scenario efficient processing of the strong splice sites identifying gene introns would leave no time for recognition of the L1 splice sites in the context of pre-mRNA producing predominantly properly spliced cellular transcripts despite the presence of the intronic L1 insert (Figure 5, bottom). Only in the instances where gene introns are defined by the weak splice sites, would there be sufficient time for the L1 splice sites to compete and interfere with the proper splicing leading to the production of a significant proportion of aberrantly spliced mRNAs containing L1 sequences (Figure 5, top). This may translate into a tissue-specific effect of L1 sequences on gene expression. For example, brain and testes are reported to support the highest amount of alternative splicing compared to other tissues (Yeo et al., 2004). The same applies to the L1 mRNA processing during the developmental process, tissue differentiation, or upon malignant transformation.
Figure 5. A model of the intronic L1 inserts interference with the normal gene expression.
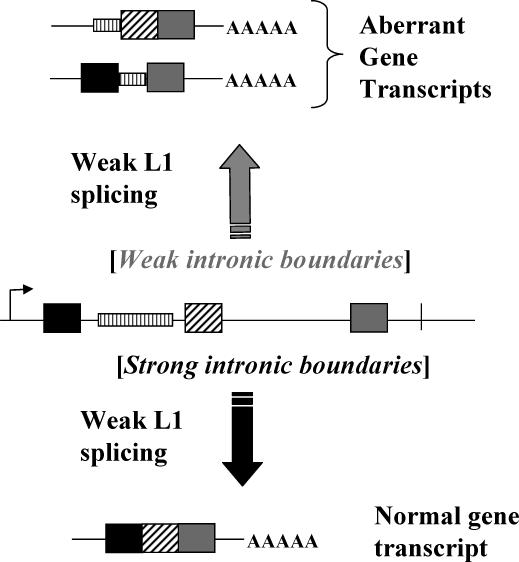
The central part of the figure depicts a gene containing exons (black, diagonally striped, and gray boxes) and introns (solid black lines separating the exons). Vertically striped rectangle represents a L1 insertion into the first intron of the gene. A scenario represented at the bottom indicates production of predominantly correctly processed mRNA transcripts despite the presence of the L1 insert. This is possible when the splice sites used by the gene are strong and the L1-specific splice sites are weak. Efficient utilization of the strong genomic splice sites would not leave enough time for the processing of the weak L1 sites in the context of pre-mRNA resulting in the production of the normal gene transcript. A scenario represented at the top of the figure reflects a situation when genomic splice sites are weak. Under this scenario recognition and processing of genomic splice sites requires a longer period of time, which may allow for the L1 splice sites to interfere with the normal RNA processing resulting in predominantly aberrant transcripts.
Acknowledgments
This work was supported by grants from the USPHS grant R01GM45668, NIH P20 RR020152, National Science Foundation EPS-0346411 and the State of Louisiana Board of Regents Support Fund.
Glossary
- APOBEC
cytidine deaminases family of proteins
- EBV SM protein
Epstein-Barr virus SM protein
- FL1
Full-length L1
- LINE-1, L1
Long interspersed element-1
- L1.3
one of the active human L1 elements
- L1spa
one of the active mouse L1 elements
- ORF1
open reading frame 1
- ORF2
open reading frame 2
- PolyA
polyadenylation site
- SINE
Short interspersed element
- SC35
splicing factor
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Batt DB, Luo Y, Carmichael GG. Polyadenylation and transcription termination in gene constructs containing multiple tandem polyadenylation signals. Nucleic Acids Res. 1994;22:2811–2816. doi: 10.1093/nar/22.14.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belancio VP, Whelton M, Deininger P. Requirements for polyadenylation at the 3' end of LINE-1 elements. Gene. 2007;390:98–107. doi: 10.1016/j.gene.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, O'Shea KS, Moran JV, Cullen BR. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson M, Hans F, Kusters I, Duran N, Sergeant A. The C-terminal region but not the Arg-X-Pro repeat of Epstein-Barr virus protein EB2 is required for its effect on RNA splicing and transport. J. Virol. 1999;73:4090–4100. doi: 10.1128/jvi.73.5.4090-4100.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rattner A, Nathans J. Effects of L1 retrotransposon insertion on transcript processing, localization and accumulation: lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum. Mol. Genet. 2006;15:2146–2156. doi: 10.1093/hmg/ddl138. [DOI] [PubMed] [Google Scholar]
- Chen L, Liao G, Fujimuro M, Semmes OJ, Hayward SD. Properties of two EBV Mta nuclear export signal sequences. Virology. 2001;288:119–128. doi: 10.1006/viro.2001.1057. [DOI] [PubMed] [Google Scholar]
- Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr. Mobile elements and mammalian genome evolution. Curr. Opin. Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- El Sawy M, Kale SP, Dugan C, Nguyen TQ, Belancio V, Bruch H, Roy-Engel AM, Deininger PL. Nickel stimulates L1 retrotransposition by a post-transcriptional mechanism. J. Mol. Biol. 2005;354:246–257. doi: 10.1016/j.jmb.2005.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DJ, Grellscheid SN. Alternative RNA splicing regulation in the testis. Reproduction. 2006;132:811–819. doi: 10.1530/REP-06-0147. [DOI] [PubMed] [Google Scholar]
- Garg K, Green P. Differing patterns of selection in alternative and constitutive splice sites. Genome Res. 2007;17:1015–1022. doi: 10.1101/gr.6347907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi G, Skowronski J, Singer MF. Defining the beginning and end of KpnI family segments. EMBO J. 1984;3:1753–1759. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189:227–234. doi: 10.1016/s0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes. Cancer. 2005;42:342–357. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- Kammler S, Otte M, Hauber I, Kjems J, Hauber J, Schaal H. The strength of the HIV-1 3' splice sites affects Rev function. Retrovirology. 2006;3:89–109. doi: 10.1186/1742-4690-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH, Goodier JL. LINE drive: Retrotransposition and genome instability. Cell. 2002;110:277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- Kazazian HH, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Hemophilia-A Resulting from Denovo Insertion of L1 Sequences Represents A Novel Mechanism for Mutation in Man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lees-Murdock DJ, De Felici M, Walsh CP. Methylation dynamics of repetitive DNA elements in the mouse germ cell lineage. Genomics. 2003;82:230–237. doi: 10.1016/s0888-7543(03)00105-8. [DOI] [PubMed] [Google Scholar]
- Matlik K, Redik K, Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J. Biomed. Biotechnol. 2006;2006:1–16. doi: 10.1155/JBB/2006/71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mays-Hoopes L, Chao W, Butcher HC, Huang RC. Decreased methylation of the major mouse long interspersed repeated DNA during aging and in myeloma cells. Dev. Genet. 1986;7:65–73. doi: 10.1002/dvg.1020070202. [DOI] [PubMed] [Google Scholar]
- Medstrand P, van de Lagemaat LN, Mager DL. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 2002;12:1483–1495. doi: 10.1101/gr.388902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, Flory E, Schumann GG, Munk C. APOBEC3 Proteins Inhibit Human LINE-1 Retrotransposition. J. Biol. Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 1998;17:590–597. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Cech TR. Reversing time: origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Goodier JL, Zhang Y, Kazazian HH., Jr. SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 2003;73:1444–1451. doi: 10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 2003;35:363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- Ropers D, Ayadi L, Gattoni R, Jacquenet S, Damier L, Branlant C, Stevenin J. Differential effects of the SR proteins 9G8, SC35, ASF/SF2, and SRp40 on the utilization of the A1 to A5 splicing sites of HIV-1 RNA. J. Biol. Chem. 2004;279:29963–29973. doi: 10.1074/jbc.M404452200. [DOI] [PubMed] [Google Scholar]
- Ruvolo V, Sun L, Howard K, Sung S, Delecluse HJ, Hammerschmidt W, Swaminathan S. Functional analysis of Epstein-Barr virus SM protein: identification of amino acids essential for structure, transactivation, splicing inhibition, and virion production. J. Virol. 2004;78:340–352. doi: 10.1128/JVI.78.1.340-352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo V, Wang E, Boyle S, Swaminathan S. The Epstein-Barr virus nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8852–8857. doi: 10.1073/pnas.95.15.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr. Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- Soret J, Gabut M, Tazi J. SR proteins as potential targets for therapy. Prog. Mol. Subcell. Biol. 2006;44:65–87. doi: 10.1007/978-3-540-34449-0_4. [DOI] [PubMed] [Google Scholar]
- Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- Stoltzfus CM, Madsen JM. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr. HIV. Res. 2006;4:43–55. doi: 10.2174/157016206775197655. [DOI] [PubMed] [Google Scholar]
- Swaminathan S. Post-transcriptional gene regulation by gamma herpesviruses. J. Cell Biochem. 2005;95:698–711. doi: 10.1002/jcb.20465. [DOI] [PubMed] [Google Scholar]
- Swergold GD. Identification, characterization, and cell specificity of a human LINE- 1 promoter. Mol. Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchenio T, Casella JF, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–415. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Pan Z, Lee JY. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007;17:156–165. doi: 10.1101/gr.5532707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AM, Dass B, Ravnik SE, Tonk V, Jenkins NA, Gilbert DJ, Copeland NG, MacDonald CC. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6763–6768. doi: 10.1073/pnas.96.12.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheelan SJ, Aizawa Y, Han JS, Boeke JD. Gene-breaking: a new paradigm for human retrotransposon-mediated gene evolution. Genome Res. 2005;15:1073–1078. doi: 10.1101/gr.3688905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Zhang L, Zhang Y, Kazazian HH. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Research. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74.1–R74.15. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]