Abstract
Although nuclear translocation of NF-κB and subsequent binding to promoters of ICAM-1 and VCAM-1 have been shown to be decisive for their expression, a number of discrepancies in the expression patterns of these adhesion molecules have been reported in both cell culture systems and disease settings, including atherosclerosis, asthma, and autoimmune diseases. Here we show that while p65 NF-κB nuclear translocation in TNF-treated smooth muscle cells (SMCs) was sufficient for the expression of VCAM-1, expression of ICAM-1 showed a critical requirement for PARP-1. I-κBα phosphorylation and subsequent degradation were virtually identical in both TNF-treated wild-type and PARP-1−/− SMCs. VCAM-1 expression in TNF-treated PARP-1−/− SMCs was completely inhibited by the NF-κB inhibitor, pyrrolidine dithiocarbamate, confirming that VCAM-1 expression was indeed NF-κB-dependent. The expression of both VCAM-1 and ICAM-1 was associated with a transient interaction between PARP-1 and p65 NF-κB when examined in the fibroblastic cell line, COS-7, and in the airway epithelial cell line, A549. Such interactions were confirmed using florescence resonance energy transfer analysis. Protein acetylation activity, mediated by p300/CBP, was required for both VCAM-1 and ICAM-1 expression in TNF-treated SMCs; however, the interaction of PARP-1 with p300/CBP was dispensable for VCAM-1 expression. These findings indicate that p65 NF-κB nuclear translocation may be sufficient for certain genes (e.g., VCAM-1) while insufficient for others (e.g., ICAM-1), thus providing a novel insight into the role of NF-κB in driving target gene expression. Furthermore, the data suggest a differential requirement for PARP-1 expression in inflammatory processes.
Introduction
Evidence for the expression of the ICAM-1 and VCAM-1 adhesion molecules on the surfaces of structural and inflammatory cells, as well as endothelial cells, in inflammatory diseases, such as asthma and arthrosclerosis, is mounting [1, 2]. The exact role for the expression of such adhesion molecules on smooth muscle cells (SMCs) is not fully understood, although it has been suggested that such expression facilitates transmigration, accumulation of leukocytes, and cell-cell interactions at inflammatory sites [3–5]. Expression of these adhesion molecules on SMCs was described as prominent in the fibrous caps of advanced atherosclerotic plaques in human specimens and was also associated with disease severity [6]. Similarly, expression of both ICAM-1 and VCAM-1 by SMCs in response to several inflammatory factors within airways has been described in animal models of asthma [7, 8]. In many of these inflammatory situations, the expression of adhesion molecules is induced by cytokines, such as tumor necrosis factor (TNF), which are released by a number of immune and structural cells, including T lymphocytes, macrophages, as well as epithelial and endothelial cells [1]. The expression of these molecules further recruits macrophages and monocytes to sites of lesions, creating conditions that are conducive for chronic inflammation [9]. TNF, as well as the dependent expression of adhesion molecules, can therefore be considered major players not only in the initiation of inflammatory lesions but also in the progression and persistence of inflammation [10, 11].
NF-κB is a pleiotropic transcription factor that plays a critical role in the regulation of the expression of multiple genes involved in inflammatory responses, including ICAM-1 and VCAM-1 [12]. NF-κB binds to the promoter regions of target genes as a dimer of two Rel family proteins, most frequently p50 and p65 [12] [13]. In quiescent cells, NF-κB is sequestered in the cytoplasm as a result of its interaction with a member of the IκB family of proteins, which includes I-κBα and IκBβ. I-κBα is phophorylated, polyubiquitinated, and degraded by the 26S proteasome in response to cell stimulation, resulting in the unmasking of the nuclear localization signal of NF-κB and its consequent translocation to the nucleus [12].
The role of poly(ADP-ribose) polymerase-1 (PARP-1) in the process of inflammation has been intensely investigated in the context of its direct participation in cellular responses to DNA-damaging agents, including oxidative stress, by way of its catalytic activity [14]. In a number of pathological conditions that involve massive DNA damage, the excessive activation of PARP-1 depletes cellular stores of both NAD and its precursor ATP, leading to irreversible cytotoxicity and potentially cell death [15–17]. We have recently shown that PARP-1 plays an important role in allergic asthma and atherosclerosis [18–20]. An emerging role for this protein is its ability to participate, directly or indirectly, in the regulation of a number of inflammatory genes at the promoter level, especially those mediated by NF-κB (reviewed in [21].
In the present study, we examined the role of PARP-1 in the specific expression of the adhesion molecules ICAM-1 and VCAM-1 in response to TNF treatment. We investigated the exact role of PARP-1 in the expression of these two adhesion molecules in SMCs with a special focus on the requirement for NF-κB nuclear translocation. We also studied associated signal transduction pathways, p300/CPB acetylation activity, and the physical interactions between the involved proteins.
Materials and Methods
Animals, isolation of SMCs and treatment protocols
Mice were bred in a specific pathogen-free facility at LSUHSC, New Orleans, LA, and allowed unlimited access to sterilized chow and water. Maintenance, experimental protocols, and procedures were all approved by the LSUHSC Animal Care & Use Committee. C57BL/6 wild-type (Wt) mice were purchased from Jackson Laboratories (Bar Harbor, ME). The generation of C57BL/6 PARP-1−/− mice was previously described [19]. All animals were genotyped by PCR.
SMCs were isolated after removal of whole thoracic aortas from sacrificed C57BL/6 Wt or PARP-1−/− mice. Adventitia and endothelia were removed after digestion of aortic segments with collagenase (175 units/mL) (Sigma-Aldrich, St. Louis MO). The media were further digested with a mixture containing collagenase (175 units/mL) and elastase (1 mg/mL) (Sigma-Aldrich). Cells were grown in DMEM containing 10% fetal bovine serum (FBS, Hyclone, Logan, UT), 100 units/mL penicillin, and 100 μg/mL streptomycin, and incubated at 37 °C in 5% CO2. Purity of SMC preparations was verified by subjecting a fraction of the cells to immunocytochemistry with antibodies to smooth muscle actin (Sigma-Aldrich) (Fig. 1A). Cells were used between passages 4 and 7. Human A549 lung epithelial cells and monkey kidney COS-7 cells (American Type Culture Collection, Manassas, Virginia) were maintained in DMEM supplemented with 10% FBS, penicillin, and streptomycin. Prior to treatment, cells at 50–80% confluence were starved by incubation in DMEM/F12 with 0.5% cell culture-tested BSA (Sigma-Aldrich) for 18 h. Cells were treated with 15 ng/mL TNF (Roche, Nutley, N.J) for the indicated times. For microscopy, cells were incubated in CO2-independent medium.
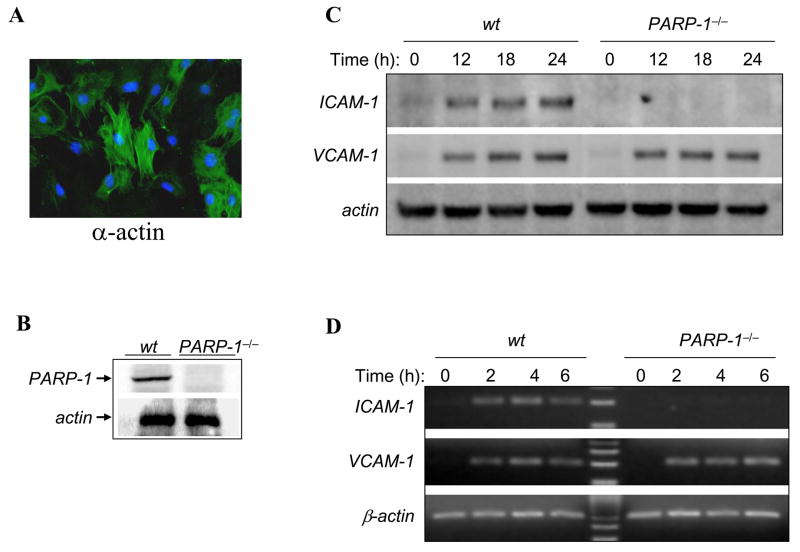
Fig. 1.
Differential expression of ICAM-1 and VCAM-1 in Wt or PARP-1−/− SMCs after TNF exposure. (A) Isolated SMCs were fixed and subjected to immunofluorescence with antibodies to smooth muscle actin (α-actin). (B) SMCs derived from Wt or PARP-1−/− mice were subjected to protein extraction and immunoblot analysis with antibodies to mouse PARP-1 or actin. (C) Wt or PARP-1−/−SMCs were treated with 15 ng/mL TNF for the indicated time intervals after which cell extracts were prepared and subjected to immunoblot analysis with antibodies to mouse ICAM-1, VCAM-1, or actin. (D) SMCs were treated as in (C) but for shorter time intervals after which total RNA was extracted and subjected to RT-PCR using primers specific to mouse ICAM-1, VCAM-1, or β-actin. The amplicons were analyzed by agarose electrophoresis.
Immunoblot analysis
After treatment with TNF, cells were washed with ice-cold PBS by centrifugation, and then incubated for 15 min on ice in lysis buffer supplemented with proteases and phosphatase inhibitors as previously described [22, 23]. A portion (15 μg protein) of each lysate was then fractionated by SDS-PAGE on a 4 to 20% gradient gel, and the separated proteins were transferred to a nitrocellulose filter. The filter was stained with Ponceau S to confirm equal loading and transfer of samples, and was then probed with antibodies to IκBα, VCAM-1, ICAM-1, p300/CBP, c-Myc, Flag, and actin (Santa Cruz Biotechnology, Santa Cruz, CA), as well as antibodies to the phosphorylated form (serine residues 32 and 36) of I-κBα (p-I-κBα) (Cell Signaling Technology, Danvers, MA). Immune complexes were detected with appropriate secondary antibodies and chemiluminescence reagents (PerkinElmer Life Science Inc., Boston, MA).
Conventional and real-time PCR
RNA was extracted from cells using standard methods and cDNA was generated using reverse transcriptase III (Invitrogen, Carlsbad, CA). Oligonucleotide primers to specifically amplify a fragment of ICAM-1, VCAM-1, or β-actin were purchased from Invitrogen. The specific primers were as follows: ICAM-1: forward primer, 5′-TCC TAA AAT GAC CTG CAG ACG -3′; reverse primer, 5′-AGT TTT ATG GCC TCC TCC TGA - 3′; VCAM-1: forward primer, 5′-ACA CTC TTA CCT GTG CGC TGT -3′; reverse primer, 5′-ATT TCC CGG TAT CTT CAA TGG -3′; and β-actin: forward primer, 5′-ACC GTG AAA AGA TGA CCC AGA TC -3′; reverse primer, 5′-TAG TTT CAT GGA TGC CAC AGG -3′. The amplification program was as follows: 3 min at 95 °C, 30 sec at 95 °C, 45 sec at 60 °C, and 2 min. at 72 °C. The cycle numbers were optimized for each primer pair. The PCR products were then incubated for 15 min at 72 °C. The resulting PCR products were subjected to electrophoresis in a 2%-agarose gel and stained with ethidium bromide.
For real-time PCR, the specific primers were as follows: VCAM-1: forward primer, 5′-TGCCGAGCTAAATTACAC ATTG-3′, reverse primer, 5′-CCTTGTGGAGGGATGTACAGA -3′ and β-actin: forward primer, 5′-TACAGCTTCACCACCACAGC-3′, reverse primer, 5′-TCTCCAGGG AGGAAGAGGAT-3′. Quantitative determination of gene expression levels using a 2-step cycling protocol was performed on a MyIQ Cycler (Bio-Rad, Hercules CA). Relative expression levels were calculated using the 2[-Delta Delta C(T)] method [24]. Quantities of all targets from the test samples were normalized to the mouse β-actin housekeeping gene.
Construction of the p65 NF-κB-FLAG (or c-Myc)-YFP and Wt PARP-1-poly-His-YFP plasmids, transfection, and pull-down assay
The p65 NF-κB (GeneBank™ accession number M61909) cDNA was generated by PCR from the pcDNA3.1+-p65 NF-κB plasmid (a kind gift from Dr. C. Giardina, University of Connecticut, Storrs, CT) using PCR. A Kozak sequence (CCACCATGG) was introduced upstream of p65 NF-κB and a Flag-tag was fused at its C-terminus. The fragment was then cloned in-frame into the Xho1 and HindIII site of pEYFP-N1 (CLONTECH Laboratories Inc., Palo Alto, CA). The accuracy of the construct was verified by restriction digestion and sequence analysis. SMCs were transiently transfected with pEYFP-N1-c-myc-p65 NF-κB using the Mirus TransIT-LT1 transfection reagent (Mirus, Madison, WI) according to the manufacturer’s instructions. After starvation, SMCs were cultured in CO2-independent medium (Invitrogen) and treated with TNF as described earlier. Subcellular localization of p65 NF-κB was monitored throughout the treatment using a Carl Zeiss inverted fluorescence microscope. A Wt-PARP-1 fragment was amplified from pcDNA3.1+-wt-PARP-1-6-histidine/flag using PCR and a Kozak sequence was added. The fragment was introduced into the Xho1/Kpn1 site of the pECFP-N1-N1 expression vector. All sequences were confirmed by sequencing. A549 and COS-7 cells were transiently transfected with the indicated expression vectors using Lipofectamine LTX (Invitrogen) or the Mirus TransIT-LT1 transfection reagent, respectively.
Poly-His-PARP-1 was pulled down using Ni-NTA magnetic agarose beads (Qiagen, Valencia, CA) according to the manufacturer’s instructions and as described [25].
Cell Labeling for Fluorescence Resonance Energy Transfer (FRET)
Acceptor photobleaching was performed to test the specificity of FRET. Filters used for observing CFP allowed excitation at 458 nm and emission at 465–495 nm; for YFP excitation at 515 nm and emission at 565–595 nm was used; for FRET of YFP and CFP excitation was at 458 nm and emission at 565–595 nm. ROIs (region of interest) were positioned over only those areas of the nucleus that appeared to possess authentic CFP and YFP fluorescence; several areas were tested within the same nucleus to avoid biased sampling. The increase in donor fluorescence (CFP-PARP-1) following acceptor (YFP-p65 NF-κB) photobleaching was observed in all experiments. The acceptor (YFP-p65 NF-κB) fluorescence was decreased by 90% after photobleaching. To obtain a quantitative measure of FRET efficiency and estimate the distance between donor and acceptor molecules from the same cell, image data was processed using FRET software obtained from Leica microsystems (Leica Confocal Software TCS SP2 156108810 version 2.61.1537, Mannheim, Germany). The FRET efficiency E is given by: FRET eff = Dpost − Dpre/Dpost for all Dpost > Dpre. Quantitative FRET analysis was performed using FRET efficiencies between CFP-PARP-1 and YFP-p65 NF-κB that were higher than 10%. A low resonance energy transfer signal was obtained in untransfected A549 cells (data not shown).
RESULTS
Differential requirements for PARP-1 for ICAM-1 and VCAM-1 proteins and mRNA expression in TNF-induced primary smooth muscle cells
To investigate the role of PARP-1 in the expression of adhesion molecules in SMCs after TNF exposure, we examined the effects of the PARP-1 gene deletion on ICAM-1 and VCAM-1 expression. The purity of SMCs was assessed by immunofluorescence with antibodies to smooth muscle actin (Fig. 1A). Wt and PARP-1−/− SMCs were treated with TNF for the indicated time intervals after which cell extracts were prepared and subjected to western immunoblotting with antibodies to murine ICAM-1, VCAM-1, or actin. Fig. 1B shows a clear induction of ICAM-1 and VCAM-1 expression in Wt SMCs as early as 12 h post-TNF treatment. While ICAM-1 expression was undetectable in PARP-1−/− SMCs, VCAM-1 expression was unaltered in these cells compared with the expression pattern of the protein in Wt SMCs. These results imply a differential requirement for PARP-1 in the respective expression of ICAM-1 and VCAM-1.
We next wished to examine whether the differential expression between these two adhesion molecules was at the level of gene expression. To this end, Wt and PARP-1−/− SMCs were treated with TNF for 2, 4, or 6 h after which total RNA was extracted and subjected to RT-PCR using specific primers to either mRNA. Fig. 1C shows, as expected, that the expression of ICAM-1 and VCAM-1 was relatively similar in Wt SMCs. However, while VCAM-1 expression in PARP-1−/− SMCs was similar to that observed in Wt cells, ICAM-1 expression was undetectable in PARP-1−/− SMCs. These results clearly show that PARP-1 participates in and is required for ICAM-1 expression at the gene level but is dispensable for that of VCAM-1 in SMCs. Our results show a critical difference in the regulation of the expression of these molecules after TNF treatment in SMCs.
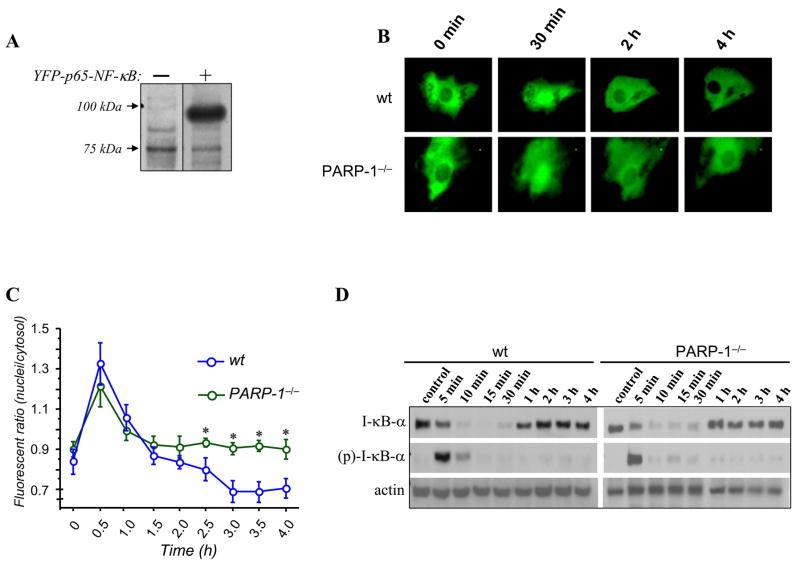
Nuclear translocation of p65 NF-κB is insufficient for the induction of ICAM-1 expression in PARP-1−/−SMCs in response to TNF
NF-κB represents a crucial and important transcription factor necessary for driving ICAM-1 and VCAM-1 gene expression [26–28]. We and others have suggested that PARP-1 may regulate signal transduction events that result in the activation of NF-κB [18, 29–31]. In fact, PARP-1 inhibition by gene deletion prevents NF-κB nuclear translocation in lipopolysaccharide (LPS)-treated peritoneal macrophages and prevents the consequent expression of both inducible nitric oxide synthase (iNOS) and monocyte chemoattractant protein-1 (MCP-1) [32, 33]. Thus, we wished to examine the effect of the PARP-1 gene deletion on NF-κB nuclear translocation in SMCs after TNF treatment. A plasmid encoding fluorescently tagged p65 NF-κB (pEYFP-N1-Flag-p65) was constructed and expression of the protein was verified by subjecting protein extracts of plasmid-transfected COS-7 cells to immunoblot analysis with antibodies to Flag peptide (Fig. 2A). Primary SMCs were transiently transfected with the pEYFP-N1-Flag-p65 NF-κB plasmid and treated with TNF (15 ng/mL) after which the subcellular localization of p65 NF-κB was monitored throughout the treatment using fluorescence microscopy. Fig. 2B shows that p65 NF-κB was strictly cytoplasmic prior to stimulation in Wt cells and that the subcellular localization quickly changed to nuclear when cells were stimulated with TNF (30 min). After 2 h of treatment, p65 NF-κB became mostly cytoplasmic, which is consistent with the transient nature of this transcription factor in the nucleus [34]. The pattern of NF-κB translocation from the cytoplasm to the nucleus in SMCs derived from PARP-1−/− mice was nearly identical to that observed in Wt cells. Interestingly, however, the exit of p65 from the nucleus to the cytoplasm appeared to be delayed in TNF-treated PARP-1−/− cells. Fig. 2C provides a quantitative assessment of NF-κB subcellular localization with additional time points. Such lack of a difference in NF-κB translocation was confirmed in fibroblasts derived from the two mouse strains by electrophoretic mobility shift assay (data not shown). These results clearly show that p65 NF-κB nuclear translocation was sufficient to drive only the expression of VCAM-1 but was completely insufficient to drive the expression of ICAM. Furthermore, these results show that unlike H2O2 or LPS treatment [18, 31–33], the PARP-1 gene deletion does not affect NF-κB translocation. Accordingly, these findings provide new insight into the role of NF-κB in driving target gene expression and the differential requirement for PARP-1 in such a process.
Fig. 2.
Effects of PARP-1 gene deletion on p65 NF-κB nuclear translocation and associated phosphorylation and degradation of I-κBα. (A) COS 7 cells were transiently transfected with an expression vector encoding p65 NF-κB tagged with the yellow fluorescent protein (YFP). Ectopic expression of p65 NF-κB was assessed by immunoblot analysis with antibodies to YFP. (B) Wt and PARP-1−/− SMCs were transiently transfected with p65 NF-κB-YFP plasmid. Cells were then incubated in CO2-independent medium and specific SMCs were then identified using a temperature-controlled inverted fluorescence microscope. TNF was added to the culture plates and images were then taken at the times indicated. (C) p65 NF-κB nuclear translocation was quantitated and assessed as the ratios between nuclear and cytosolic fluorescence in at least 10 cells and expressed as mean ± SEM; *, difference from Wt SMCs treated with TNF, p < 0.05. (D) Wt and PARP-1−/− SMCs were treated with TNF for the indicated time intervals after which protein extracts were prepared and subjected to immunoblot analysis with antibodies to mouse IκBα, phophorylated I-κBα (at serine residues 32 and 36), or actin.
The PARP-1 gene deletion does not alter I-κBα phosphorylation or its subsequent degradation in TNF-treated SMCs
As mentioned above, signal transduction events, such as the phosphorylation of I-κBα by I-κB kinase at serine residues 32 and 36, as well as its subsequent degradation, are critical for the activation and nuclear translocation of NF-κB in response to TNF and a variety of other inflammatory factors [34, 35]. Accordingly, we assessed the status of I-κBα phosphorylation and its fate during TNF treatment in SMCs derived from Wt and PARP-1−/− mice. To this end, cells were treated with TNF for different time intervals after which total proteins were extracted and subjected to immunoblot analysis with antibodies specific for mouse I-κBα, phosphorylated I-κBα at serine residues 32 and 36, or actin. Fig. 2D shows that I-κBα was phosphorylated and degraded in a matter of minutes (5–10 minutes) and that the PARP-1 gene deletion exerted no apparent effects on such events. Furthermore, no differences in the fate of I-κBβ were observed between Wt and PARP-1−/− SMCs (data not shown). These results strongly suggest that the signal transduction cascade of NF-κB, such as the activities of I-κBα kinase and the 26S proteasome, were unaffected by the PARP-1 gene deletion. These results also suggest that differences in the regulation of ICAM and VCAM-1 gene expression reside in the nucleus, likely at the promoter level post-NF-κB nuclear translocation.
PARP-1 interaction with NF-κB is transient after TNF treatment
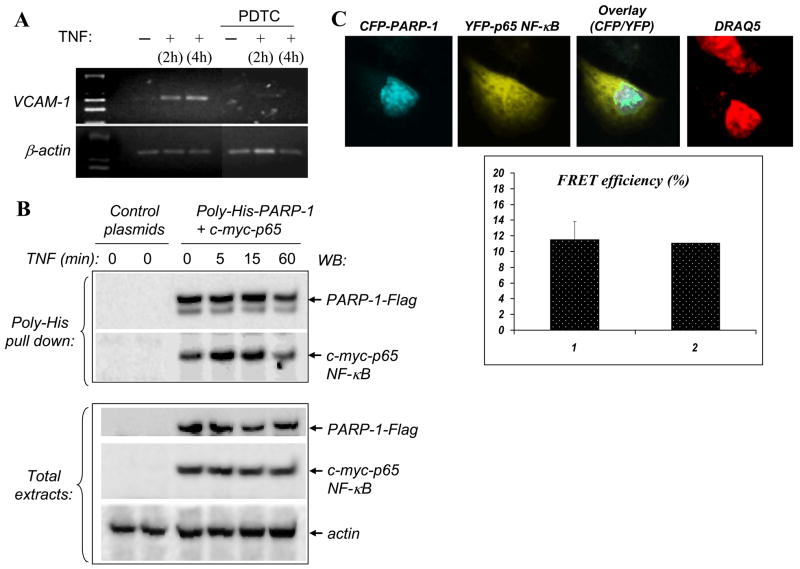
A rather important question is whether VCAM-1 expression is indeed regulated by NF-κB in PARP-1−/− SMCs. This became important in light of the finding by Lavi et al. [36] who showed that VCAM-1 can be NF-κB-independent in differentiated SMCs; in this study, VCAM-1 expression was shown to be sensitive to PDTC, a known inhibitor of NF-κB, only in undifferentiated SMCs. To this end, we stimulated PARP-1−/− SMCs with TNF in the presence of PDTC after which expression of VCAM-1 was assessed by RT-PCR. PDTC almost completely abrogated VCAM-1 expression in response to TNF treatment (Fig. 3A), suggesting that VCAM-1 expression is certainly regulated by NF-κB in PARP-1−/− SMCs in a manner similar to that observed in Wt cells (data not shown).
Fig. 3.
Dependence of VCAM-1 expression on NF-κB in PARP-1−/− SMCs and interaction between PARP-1 and p65 NF-κB upon TNF exposure. (A) PARP-1−/− SMCs were stimulated with TNF in the presence of 100 μM PDTC after which expression of VCAM-1 was assessed by RT-PCR; β-actin was used as an internal control. (B) COS-7 cells were co-transfected with expression vectors encoding PARP-1 tagged with poly-His and Flag and p65 NF-κB tagged with c-Myc or with empty vectors. Cells were treated with TNF for the indicated time intervals after which total protein extracts were prepared and subjected to a nickel bead-based pull-down assay. Precipitates were then subjected to immunoblot analysis (WB) with antibodies to either Flag or c-Myc. A portion of the total proteins was subjected to immunoblot analysis to confirm equal expression of PARP-1 and p65 NF-κB in the different conditions; antibodies to actin were used to confirm equal loading. (C) A549 cells were transiently co-transfected with plasmids encoding CFP-PARP-1 and YFP-p65 NF-κB. Cells were then treated with TNF for 15 min. Protein-protein interactions were assessed by FRET analysis using a Leica confocal microscope. The panels represent CFP, YFP, an overlay of both, or DRAQ5 (nuclear marker), respectively; the graph shows an average of FRET efficiency [1] detected in 15 cells in addition to the efficiency of the particular cell shown in the images [2].
PARP-1 interactions with the NF-κB subunits, including p65, were shown to be required for specific NF-κB transcriptional activation in cultured cells and that both NF-κB and PARP-1 formed a stable immunoprecipitable nuclear complex [29]. Given our results showing that VCAM-1 expression was NF-κB-dependent and did not require PARP-1 expression, it became important to determine whether, in our experimental system, p65 NF-κB interacted with PARP-1 after TNF stimulation. For this purpose, COS7 cells were co-transfected with expression vectors encoding poly-His-PARP-1 and c-Myc-tagged p65 NF-κB. Cells were then treated for different time intervals with TNF after which whole cell extracts were prepared and subjected to pull-down assays using magnetic nickel beads. Fig. 3B (top panel) shows that PARP-1 interacted with p65 NF-κB soon after TNF stimulation and that such an interaction was rather transient in nature. A maximal interaction was detected 5 to 15 min after stimulation and immediately began to subside during the course of treatment. A similar interaction pattern was observed in the airway epithelial cell line A549 (Fig. 4A). The transient nature of this interaction is in sharp contrast to that reported by Hassa el al. [29] who described a persistent and stable interaction between these two proteins.
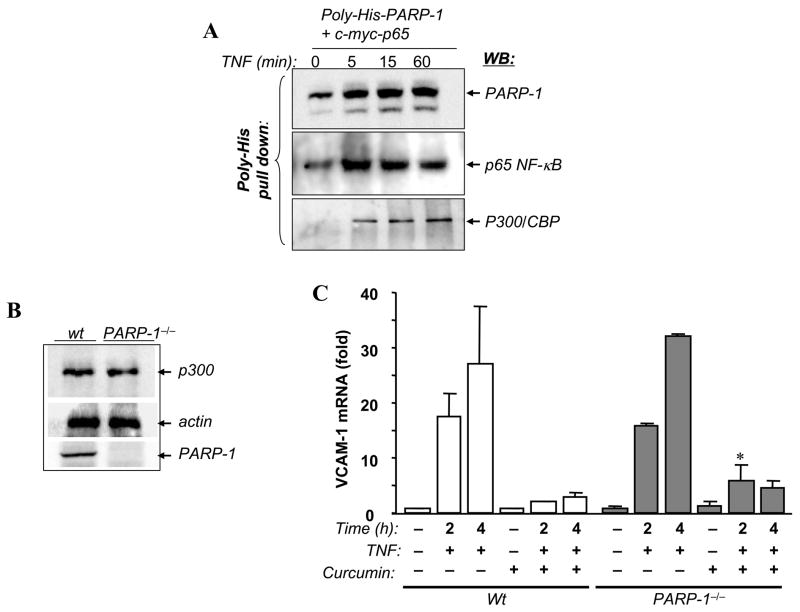
Fig. 4.
Transient interactions between PARP-1 and p300/CBP and the requirement of p300/CBP acetylation activity for VCAM-1 expression after TNF treatment. (A) A549 cells were co-transfected with expression vectors encoding PARP-1 tagged with poly-His and p65 NF-κB or with empty vectors. Cells were treated with TNF for the indicated time intervals after which total protein extracts were prepared and subjected to a nickel bead-based pull-down assay. Precipitates were then subjected to immunoblot analysis (WB) with antibodies to PARP-1, p65 NF-κB, or p300/CBP. (B) Protein extracts from Wt or PARP-1−/− SMCs were subjected to immunoblot analysis with antibodies to p300/CBP PARP-1 or actin. (C) Wt or PARP-1−/− SMCs were treated with TNF for the indicated time intervals in the presence or absence of 10 μM curcumin, a specific inhibitor of p300/CBP, after which total RNA was extracted and subjected to cDNA generation followed by real-time PCR analysis using primers specific to mouse VCAM-1 or β-actin. *, difference from the Wt counterpart, p < 0.05.
To fully confirm the interaction between PARP-1 and p65 NF-κB in cells without the interference of additional pull-down assay-associated manipulations, we examined such interactions in whole cells using FRET. A549 cells were transfected with CFP-PARP-1 and YFP-p65 NF-κB, incubated for 36 h, and cells were then treated with TNF for 15 min and fixed with methanol. The interaction between PARP-1 and NF-κB was monitored using a Leica confocal microscope. The nuclear distribution of CFP-PARP-1 did not differ from that of endogenous Wt PARP-1 during the course of treatment with TNF as assessed by immunofluorescence (data not shown). Fig. 3C shows that PARP-1 physically interacted with p65 NF-κB after its translocation to the nuclei of TNF-treated A549 cells with a rather high FRET efficiency (10–19%). A low resonance energy transfer signal was obtained in untransfected A549 cells or in cells with cytosolic CFP-PARP-1 (data not shown). These results strongly demonstrate a robust interaction between PARP-1 and p65-NF-κB after TNF stimulation and validate our findings using the pull-down assay.
Relationship between PARP-1 and p300/CBP is dispensable for VCAM-1 expression in response to TNF treatment in SMCs
Recently, Hassa et al. [37] reported that PARP-1 directly interacts with p300/CBP and that such an interaction results in the acetylation of PARP-1 at specific lysine residues. Furthermore, they suggested that such a relationship is required for NF-κB-dependent transcriptional activity. Therefore, it became important to determine whether an interaction between PARP-1, p65 NF-κB, and p300 takes place after TNF treatment in our experimental system. To this end, A549 cells were transiently co-transfected with expression vectors encoding poly-His-PARP-1 or p65 NF-κB. Cells were then treated for different time intervals after which whole cell extracts were prepared and subjected to a pull-down assay using magnetic nickel beads followed by immunoblot analysis with antibodies to PARP-1, p65 NF-κB, or p300/CBP. Similar to results obtained using COS7 cells, NF-κB transiently interacted with PARP-1 (Fig. 4A). Such an interaction was accompanied by an interaction with p300/CBP (Fig. 4A, bottom panel). The interaction between PARP-1 and p300/CBP increased with time of treatment. Maximal interaction between PARP-1 and p300/CBP appeared to take place after that with p65 NF-κB. These results confirm the reported interaction [37] between the two proteins but show that such interaction is dynamic in nature.
Given the suggested crucial role of PARP-1 acetylation by p300/CBP in the regulation of NF-κB-dependent gene expression, we next wished to determine whether p300/CBP-acetylation activity was important for VCAM-1 expression in PARP-1−/− SMCs. Before conducting these experiments, we wished to ascertain that the PARP-1 gene deletion did not have a direct effect on the expression of p300/CBP in SMCs. Fig. 4B shows that the PARP-1 gene deletion indeed exerted no effect on p300/CBP expression as assessed by immunoblot analysis with antibodies to p300/CBP, which is consistent with those reported for fibroblasts [37]. We next examined the effect of the inhibition of p300/CBP-acetylation activity by curcumin, a specific inhibitor of the enzyme, on VCAM-1 expression in VCAM-1 Wt and PARP-1−/− SMCs. Cells were treated with TNF for 2 or 4 h in the presence or absence of curcumin after which total RNA was extracted and subjected to cDNA generation followed by an assessment of mRNA by real-time PCR. Fig. 4C shows that the inhibition of p300/CBP enzymatic activity completely abrogated the ability of TNF to induce VCAM-1 expression in Wt SMCs, consistent with a recent report using human tracheal SMCs [38]. The PARP-1 gene deletion, however, exerted little to no effect on the ability of curcumin to modulate VCAM-1 expression after TNF treatment. These results clearly show that the interaction between PARP-1 and p300/CBP and the subsequent acetylation of PARP-1 are not necessary for VCAM-1 expression.
DISCUSSION
Evidence for the expression of ICAM-1 and VCAM-1 in SMCs in inflammatory diseases, such as arthrosclerosis and asthma, is mounting [1, 2]. The exact roles for the expression of such adhesion molecules are not fully understood, although it has been suggested that such expression facilitates transmigration and accumulation of leukocytes at inflammatory sites [39, 40]. The expression of these adhesion molecules in SMCs was described as prominent in the fibrous caps of advanced atherosclerotic plaques in human specimens and was also associated with disease severity [6]. Similarly, expression of both ICAM-1 and VCAM-1 by SMCs in response to several inflammatory factors within airways has been described in animal models of asthma [7, 39]. In many of these inflammatory situations, the expression of adhesion molecules is induced by cytokines, such as TNF, which are released by a number of immune and structural cells, including T lymphocytes, macrophages, as well as epithelial and endothelial cells [1].
For many years, VCAM-1 and ICAM-1 have been described as closely related in their mechanisms of regulation given the number of similarities in the regulatory sequences of their respective promoters, with NF-κB exerting a dominant role [41–44]. NF-κB has been shown to bind to the promoter regions of both ICAM-1 and VCAM-1 to drive their expression [12, 35, 43, 45]. Inhibition of NF-κB activation, pharmacologically, by gene knockout, or through expression of dominant-negative mutants of I-κBα or I-κB kinase has been shown to hinder the expression of these adhesion molecules [43, 46, 47]. Initially, it was considered that PARP-1 may indiscriminately regulate all NF-κB-dependent genes and that its inhibition by gene knockout compromises the expression of these genes [21, 31]. Also, it was also considered that via mere translocation of NF-κB to the nuclei of cytokine-stimulated cells, NF-κB-regulated genes would be expressed [45, 48]. The results of the present study challenge these two concepts. PARP-1 may be required for certain NF-κB-regulated gene (e.g., ICAM-1) but not for all (e.g., VCAM-1). Preliminary results show that the PARP-1 gene deletion prevented the expression of ICAM-1, but not of VCAM-1, in airway epithelial cells after intranasal administration of TNF in mice (data not shown), lending additional support to our findings.
Our results show that NF-κB can discriminate between ICAM-1 and VCAM-1 and that localization of transcription to the nucleus does not necessarily culminate in the activation of all genes that harbor its consensus sequence within their promoters. Indeed, the kinetics of p65 NF-κB translocation to nuclei of TNF-treated Wt or PARP-1−/− SMCs was very similar. Interestingly, however, PARP-1 gene deletion appears to affect p65 NF-κB exit from the nucleus at later time points after TNF treatment (Fig. 2B–C). The mechanism behind such delay is unclear and is currently under investigation. Kanai et al. very recently reported that PARP-1 plays an important role in p53 exit from the nucleus as PARP-1-mediated poly(ADP-ribosyl) ation blocks the interaction between p53 and Crm1 nuclear export receptor, resulting in nuclear accumulation of p53 [49]. The role of PARP-1 in exit of p53 and p65 NF-κB from the nucleus appears to involve contrasting mechanisms. Clearly, much experimentation is needed to clarify the role of PARP-1 in p65 NF-κB exit from the nucleus. It is important to note, through work by us and others [18, 29, 32, 33], that in response to inducers such as LPS or H2O2, NF-κB nuclear translocation appears to require PARP-1 expression.
The transient nature of the interaction between PARP-1 and p65 NF-κB is rather interesting. What regulates this transient interaction is not clear and may involve the enzymatic activity of PARP-1. Chang and Alvarez-Gonzalez [30] showed that the auto-poly(ADP-ribosyl)ation reaction catalyzed by PARP-1 facilitates the binding of p50 NF-κB to its consensus DNA sequence via the inhibition of specific protein-protein interactions between p50 NF-κB and PARP-1. It is tempting to speculate that similar events take place with p65 NF-κB in TNF-treated cells. However, a clear assessment of such a relationship remains to be determined. Hassa et al. reported that such interactions are more stable and independent of DNA [37]. It is important to note that gene expression is a dynamic event that involves a number of transient processes which allows regulatory systems to function properly, particularly for inflammatory genes. An aberration of these processes can lead to uncontrolled expression of these genes, rendering the environment conducive to chronic inflammation. Recently, Nelson et al. [50] showed, using single-cell time-lapse imaging and computational modeling of p65 NF-κB localization, asynchronous oscillations in conditions similar to ours (i.e., TNF treatment). Such oscillations decreased in frequency with increased I-κBα transcription.
The role of p300/CBP-mediated acetylation in regulating the expression of NF-κB-driven genes is well established [51]. Our results confirm the critical role of the enzymatic activity of this protein in VCAM-1 expression. ICAM-1 expression was completely inhibited by curcumin after TNF treatment in wild-type SMCs (data not shown), which clearly suggests that expression of these two adhesion molecules is highly dependent on p300/CBP enzymatic activity. The critical difference between ICAM-1 and VCAM-1 in this context is that ICAM-1 requires PARP-1 expression and VCAM-1 does not. Whether the interaction between p300/CBP and PARP-1 is required for ICAM-1 is not clear. We do know, however, that the interaction between these proteins does take place [37]. Such an interaction was shown to be critical for the expression of a number of inflammatory genes in lung fibroblasts and macrophages [37]. Whether the differential requirement for ICAM-1 and VCAM-1 expression in SMCs is general and operative in all cell types remains to be determined.
The results described above may have great implications in understanding the mechanism(s) by which adhesion molecules are expressed during the onset of inflammatory diseases and may provide insight into the observed discrepancies in the expression of their proteins in a number of conditions, including atherosclerosis [4, 5, 52, 53], asthma [54, 55], and autoimmune diseases [56]. These results may also help in the design of therapeutic strategies that target adhesion molecules for the treatment of a number of inflammatory diseases. Indeed, initial studies using monoclonal antibodies or antisense strategies targeted to ICAM-1 in patients with rheumatoid arthritis or mild to moderate descending ulcerative colitis, respectively, have provided promising results [57, 58]. These results advance our efforts toward presenting PARP-1 inhibition as a viable therapeutic strategy for the treatment of airway and cardiovascular inflammatory diseases.
Supplementary Material
Acknowledgments
We thank Drs. Alberto Martinez and Cooper Woods, and the Ochsner Clinic Foundation for providing us with laboratory space to pursue our research in the aftermath of Hurricane Katrina. We also thank Dr. Emel Songu-Mize and the Cell Culture Core for continuously providing us with primary SMCs from the different mouse strains. This work was supported in part by grants HL072889 and 1P20RR18766 (overall PI: D. Kapusta) from the NIH to H. Boulares.
The abbreviations used are
- PARP-1
poly(ADP-ribose) polymerase-1
- SMCs
smooth muscle cells
- FRET
fluorescence resonance energy transfer
- YFP
yellow fluorescent protein
- CFP
cyan fluorescent protein
- NF-κB
nuclear factor-kappa B
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Braun M, Pietsch P, Schror K, Baumann G, Felix SB. Cardiovasc Res. 1999;41(2):395–401. doi: 10.1016/s0008-6363(98)00302-2. [DOI] [PubMed] [Google Scholar]
- 2.Hughes JM, Arthur CA, Baracho S, Carlin SM, Hawker KM, Johnson PR, Armour CL. Mediators Inflamm. 2000;9(2):93–99. doi: 10.1080/096293500411550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rojas AI, Ahmed AR. Crit Rev Oral Biol Med. 1999;10(3):337–358. doi: 10.1177/10454411990100030601. [DOI] [PubMed] [Google Scholar]
- 4.Huo Y, Ley K. Acta Physiol Scand. 2001;173(1):35–43. doi: 10.1046/j.1365-201X.2001.00882.x. [DOI] [PubMed] [Google Scholar]
- 5.Cybulsky MI, Iiyama K, Li H, Zhu S, Chen M, Iiyama M, Davis V, Gutierrez-Ramos JC, Connelly PW, Milstone DS. J Clin Invest. 2001;107(10):1255–1262. doi: 10.1172/JCI11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasper HU, Schmidt A, Roessner A. Gen Diagn Pathol. 1996;141(5–6):289–294. [PubMed] [Google Scholar]
- 7.Lazaar AL, Albelda SM, Pilewski JM, Brennan B, Pure E, Panettieri RA., Jr J Exp Med. 1994;180(3):807–816. doi: 10.1084/jem.180.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazaar AL, Panettieri RA., Jr J Allergy Clin Immunol. 2005;116(3):488–495. doi: 10.1016/j.jaci.2005.06.030. quiz 496. [DOI] [PubMed] [Google Scholar]
- 9.Shin WS, Szuba A, Rockson SG. Atherosclerosis. 2002;160(1):91–102. doi: 10.1016/s0021-9150(01)00571-8. [DOI] [PubMed] [Google Scholar]
- 10.Lucas AD, Greaves DR. Expert Rev Mol Med. 2001;2001:1–18. doi: 10.1017/S1462399401003696. [DOI] [PubMed] [Google Scholar]
- 11.Ngoc PL, Gold DR, Tzianabos AO, Weiss ST, Celedon JC. Curr Opin Allergy Clin Immunol. 2005;5(2):161–166. doi: 10.1097/01.all.0000162309.97480.45. [DOI] [PubMed] [Google Scholar]
- 12.Karin M, Cao Y, Greten FR, Li ZW. Nat Rev Cancer. 2002;2(4):301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto Y, Gaynor RB. Curr Mol Med. 2001;1(3):287–296. doi: 10.2174/1566524013363816. [DOI] [PubMed] [Google Scholar]
- 14.Burkle A. Bioessays. 2001;23(9):795–806. doi: 10.1002/bies.1115. [DOI] [PubMed] [Google Scholar]
- 15.Ha HC, Snyder SH. Proc Natl Acad Sci U S A. 1999;96(24):13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiarugi A, Meli E, Calvani M, Picca R, Baronti R, Camaioni E, Costantino G, Marinozzi M, Pellegrini-Giampietro DE, Pellicciari R, Moroni F. J Pharmacol Exp Ther. 2003;305(3):943–949. doi: 10.1124/jpet.103.048934. [DOI] [PubMed] [Google Scholar]
- 17.Nicoletti VG, Stella AM. Neurochem Res. 2003;28(2):187–194. doi: 10.1023/a:1022316914492. [DOI] [PubMed] [Google Scholar]
- 18.Boulares AH, Zoltoski AJ, Sherif ZA, Jolly P, Massaro D, Smulson ME. Am J Respir Cell Mol Biol. 2003;28(3):322–329. doi: 10.1165/rcmb.2001-0015OC. [DOI] [PubMed] [Google Scholar]
- 19.Oumouna M, Datta R, Oumouna-Benachour K, Suzuki Y, Hans C, Matthews K, Fallon K, Boulares AH. J Immunol. 2006;177(9):6489–6496. doi: 10.4049/jimmunol.177.9.6489. [DOI] [PubMed] [Google Scholar]
- 20.Oumouna-Benachour K, Oumouna M, Zerfaoui M, Hans C, Fallon K, Boulares AH. Mol Carcinog. 2007 doi: 10.1002/mc.20351. In press. [DOI] [PubMed] [Google Scholar]
- 21.Hassa PO, Hottiger MO. Cell Mol Life Sci. 2002;59(9):1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulares HA, Giardina C, Navarro CL, Khairallah EA, Cohen SD. Toxicol Sci. 1999;48(2):264–274. doi: 10.1093/toxsci/48.2.264. [DOI] [PubMed] [Google Scholar]
- 23.Boulares AH, Yakovlev AG, Ivanova V, Stoica BA, Wang G, Iyer S, Smulson M. J Biol Chem. 1999;274:22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Boulares AH, Zoltoski AJ, Contreras FJ, Yakovlev AG, Yoshihara K, Smulson ME. J Biol Chem. 2002;277(1):372–378. doi: 10.1074/jbc.M107738200. [DOI] [PubMed] [Google Scholar]
- 26.Pande V, Ramos MJ. Curr Med Chem. 2005;12(3):357–374. doi: 10.2174/0929867053363180. [DOI] [PubMed] [Google Scholar]
- 27.Jones WK, Brown M, Wilhide M, He S, Ren X. Cardiovasc Toxicol. 2005;5(2):183–202. doi: 10.1385/ct:5:2:183. [DOI] [PubMed] [Google Scholar]
- 28.Aggarwal BB, Takada Y, Shishodia S, Gutierrez AM, Oommen OV, Ichikawa H, Baba Y, Kumar A. Indian J Exp Biol. 2004;42(4):341–353. [PubMed] [Google Scholar]
- 29.Hassa PO, Hottiger MO. Biol Chem. 1999;380(7–8):953–959. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- 30.Chang WJ, Alvarez-Gonzalez R. J Biol Chem. 2001;276(50):47664–47670. doi: 10.1074/jbc.M104666200. [DOI] [PubMed] [Google Scholar]
- 31.Ha HC. Proc Natl Acad Sci U S A. 2004;101(14):5087–5092. doi: 10.1073/pnas.0306895101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver FJ, Menissier-de Murcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet JC, de Murcia G. Embo J. 1999;18(16):4446–4454. doi: 10.1093/emboj/18.16.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oumouna-Benachour K, Hans CP, Suzuki Y, Naura A, Datta R, Belmadani S, Fallon K, Woods C, Boulares AH. Circulation. 2007;115(18):2442–2450. doi: 10.1161/CIRCULATIONAHA.106.668756. [DOI] [PubMed] [Google Scholar]
- 34.Silverman N, Maniatis T. Genes Dev. 2001;15(18):2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 35.Wright JG, Christman JW. Am J Respir Med. 2003;2(3):211–219. doi: 10.1007/BF03256650. [DOI] [PubMed] [Google Scholar]
- 36.Lavie J, Dandre F, Louis H, Lamaziere JM, Bonnet J. J Biol Chem. 1999;274(4):2308–2314. doi: 10.1074/jbc.274.4.2308. [DOI] [PubMed] [Google Scholar]
- 37.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. J Biol Chem. 2005;280(49):40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 38.Lee CW, Lin WN, Lin CC, Luo SF, Wang JS, Pouyssegur J, Yang CM. J Cell Physiol. 2006;207(1):174–186. doi: 10.1002/jcp.20549. [DOI] [PubMed] [Google Scholar]
- 39.Lazaar AL. Expert Opin Ther Targets. 2002;6(4):447–459. doi: 10.1517/14728222.6.4.447. [DOI] [PubMed] [Google Scholar]
- 40.Schober A, Weber C. Antioxid Redox Signal. 2005;7(9–10):1249–1257. doi: 10.1089/ars.2005.7.1249. [DOI] [PubMed] [Google Scholar]
- 41.Collins T, Read MA, Neish AS, Whitley MZ, Thanos D, Maniatis T. Faseb J. 1995;9(10):899–909. [PubMed] [Google Scholar]
- 42.Chen CC, Manning AM. Agents Actions Suppl. 1995;47:135–141. doi: 10.1007/978-3-0348-7343-7_12. [DOI] [PubMed] [Google Scholar]
- 43.MacKenzie CJ, Ritchie E, Paul A, Plevin R. Cell Signal. 2007;19(1):75–80. doi: 10.1016/j.cellsig.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Gloire G, Horion J, El Mjiyad N, Bex F, Chariot A, Dejardin E, Piette J. J Biol Chem. 2007 doi: 10.1074/jbc.M610728200. [DOI] [PubMed] [Google Scholar]
- 45.Wang T, Zhang X, Li JJ. Int Immunopharmacol. 2002;2(11):1509–1520. doi: 10.1016/s1567-5769(02)00058-9. [DOI] [PubMed] [Google Scholar]
- 46.Ogawara K, Kuldo JM, Oosterhuis K, Kroesen BJ, Rots MG, Trautwein C, Kimura T, Haisma HJ, Molema G. Arthritis Res Ther. 2006;8(1):R32. doi: 10.1186/ar1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mo SJ, Son EW, Lee SR, Lee SM, Shin DH, Pyo S. J Ethnopharmacol. 2007;109(1):78–86. doi: 10.1016/j.jep.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 48.Denk A, Goebeler M, Schmid S, Berberich I, Ritz O, Lindemann D, Ludwig S, Wirth T. J Biol Chem. 2001;276(30):28451–28458. doi: 10.1074/jbc.M102698200. [DOI] [PubMed] [Google Scholar]
- 49.Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, Fukasawa K. Nat Cell Biol. 2007 doi: 10.1038/ncb1638. In press. [DOI] [PubMed] [Google Scholar]
- 50.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MR. Science. 2004;306(5696):704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 51.Perkins ND. Oncogene. 2006;25(51):6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 52.Iiyama K, Hajra L, Iiyama M, Li H, DiChiara M, Medoff BD, Cybulsky MI. Circ Res. 1999;85(2):199–207. doi: 10.1161/01.res.85.2.199. [DOI] [PubMed] [Google Scholar]
- 53.Ley K, Huo Y. J Clin Invest. 2001;107(10):1209–1210. doi: 10.1172/JCI13005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamamoto H, Sedgwick JB, Busse WW. J Immunol. 1998;161(2):971–977. [PubMed] [Google Scholar]
- 55.Wong CK, Wang CB, Li ML, Ip WK, Tian YP, Lam CW. Int Immunopharmacol. 2006;6(12):1859–1871. doi: 10.1016/j.intimp.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Sallum AM, Kiss MH, Silva CA, Wakamatsu A, Vianna MA, Sachetti S, Marie SK. Autoimmun Rev. 2006;5(2):93–100. doi: 10.1016/j.autrev.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 57.Kavanaugh AF, Davis LS, Nichols LA, Norris SH, Rothlein R, Scharschmidt LA, Lipsky PE. Arthritis Rheum. 1994;37(7):992–999. doi: 10.1002/art.1780370703. [DOI] [PubMed] [Google Scholar]
- 58.van Deventer SJ, Wedel MK, Baker BF, Xia S, Chuang E, Miner PB., Jr Aliment Pharmacol Ther. 2006;23(10):1415–1425. doi: 10.1111/j.1365-2036.2006.02910.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.