Abstract
Current techniques for identifying mutations that convey a small increased cancer risk or those that modify cancer risk in carriers of highly penetrant mutations are limited by the statistical power of epidemiologic studies, which require screening of large populations and candidate genes. To identify dosage-sensitive genes that mediate genomic stability, we performed a genomewide screen in Saccharomyces cerevisiae for heterozygous mutations that increase chromosome instability in a checkpoint-deficient diploid strain. We used two genome stability assays sensitive enough to detect the impact of heterozygous mutations and identified 172 heterozygous gene disruptions that affected chromosome fragment (CF) loss, 45% of which also conferred modest but statistically significant instability of endogenous chromosomes. Analysis of heterozygous deletion of 65 of these genes demonstrated that the majority increased genomic instability in both checkpoint-deficient and wild-type backgrounds. Strains heterozygous for COMA kinetochore complex genes were particularly unstable. Over 50% of the genes identified in this screen have putative human homologs, including CHEK2, ERCC4, and TOPBP1, which are already associated with inherited cancer susceptibility. These findings encourage the incorporation of this orthologous gene list into cancer epidemiology studies and suggest further analysis of heterozygous phenotypes in yeast as models of human disease resulting from haplo-insufficiency.
CANCERS are commonly characterized as having an abnormal number of chromosomes, termed aneuploidy, which arises due to genomic instability. Aneuploidy has been proposed as a mutagenic mechanism and a driving force of tumor progression and not just a phenotype of the disease (Weaver and Cleveland 2006). Many of the genes whose disruption results in aneuploidy were first identified in budding yeast on the basis of analysis of haploid strains containing null mutations. Aneuploidy can be caused by inherited or somatic mutations in genes that function in the maintenance of genomic stability, such as those involved in centrosome formation, chromosome metabolism, and cell cycle checkpoints. Among these, cell cycle checkpoint pathways play an especially important role in maintaining genomic integrity when cells are challenged with genotoxic agents or other stressors (Zhou and Elledge 2000).
Mutations in checkpoint genes including, BRCA1, PTEN, ATM, and CHEK2, lead to increases in breast cancer susceptibility (Li et al. 1997; Nathanson and Weber 2001; Meijers-Heijboer et al. 2002; King et al. 2003). Women who inherit BRCA1 mutations have an increased lifetime breast cancer risk of 50–80% (12% risk in the general population) and an ovarian cancer risk of 15–65% (1.5% risk in the general population). However, the age of cancer onset and type and number of cancers can vary, even among women carrying the same mutation. One mechanism for this variation is the presence of unlinked genetic loci, which modify this risk (Rebbeck 2002). For instance, ovarian cancer risk in BRCA1 carriers is influenced by specific alleles of a variable number of tandem repeats polymorphism in HRAS1 (Phelan et al. 1996).
Current methods of identifying genetic modifiers and low-penetrant alleles for cancer susceptibility require large patient cohorts and are limited by the statistical power of epidemiologic studies (Chenevix-Trench et al. 2007). New approaches that are unbiased and comprehensive in their search for genes modulating genomic stability, and putatively cancer development, are needed.
Conservation of the spindle, DNA replication, and DNA damage pathways between humans and Saccharomyces cerevisiae has been well documented, allowing for use of this model organism in studies focused on control of genomic stability (Elledge 1996). However, the majority of yeast genetic screens have focused on null mutations in haploid strains, restricting analysis to nonessential genes. In contrast, cancer susceptibility is often seen in heterozygous mutation carriers; however, screens for the impact of haplo-insufficiency in a diploid setting have not been extensively explored. Here we perform a genomewide heterozygous screen in S. cerevisiae to identify haplo-insufficient mutations that result in modest increases in genomic instability, as a novel strategy for the identification of candidate cancer susceptibility and modifier loci in humans.
MATERIALS AND METHODS
Strains and assay markers:
Parental strains were made by mating DVL142-35B to HKY965-12B, kind gifts from Vicki Lundblad and Hannah Klein, respectively, and followed by dissection, genotyping, and re-creation of desired strains (Klein 2001). The chromosome fragment (CF) used in this screen was originally created and provided by Phil Hieter (personal communication): ES75, rad9Δ/rad9Δ, leu2-3/leu2-3, his3-Δ200/his3-Δ200, trp1-Δ1/trp1-Δ1, lys2-801/LYS2, ura3-52/ura3-52, can1-100/CAN1, ade2-101/ade2-101, 2× [CF:(ura3∷TRP1, SUP11, CEN4, D8B)]; ES76, leu2-3/leu2-3, his3-Δ200/his3-Δ200, trp1-Δ1/trp1-Δ1, lys2-801/LYS2, ura3-52/ura3-52, can1-100/CAN1, ade2-101/ade2-101, 2× [CF:(ura3∷TRP1, SUP11, CEN4, D8B)].
Insertional mutagenesis:
Heterozygous mutations were created using the mTn-lacZ/LEU2-mutagenized library (Kumar et al. 2002). Briefly, the mTn-lacZ/LEU2 library was received as DNA pools, which were amplified using Escherichia coli and then NotI excised from the bacterial vector and transformed into ES75 yeast cultures via the lithium acetate method (Gietz and Woods 2006). Leu+ transformants were then struck out onto 1/8 of a low-adenine concentration plate (6 μg/ml), which allowed visualization of ∼185 single colonies/insertionally mutagenized strain. After growth for 3 days at 30° and 1 night at 4° (for color development), colonies were examined for the appearance of pink/red sectors by one observer. Sectors arise due to loss of a CF, which harbors a SUP11 ochre-suppressing tRNA responsible for suppression of the homozygous ade2-101 ochre mutation that causes yeast to be red due to build-up of adenine precursors. S. cerevisiae containing two copies of the CF are white, one copy is pink, and zero copies are red. The use of two CFs in the parental strain enabled us to more easily identify pink and red sectors on a white colony background. A positive sectoring phenotype was set at four or more sectors among the ∼185 colonies inspected compared with, on average, 0.5 sectors in a similar number of the rad9Δ/rad9Δ parental strain (P-value of <0.001, compared with parental rate). Fully pink or red colonies were not counted as sectored colonies.
Fluctuation analysis: estimation of chromosome V instability rate via method of the median:
Chromosome V instability rate was measured as the conversion of a heterozygous can1-100/CAN1 strain to canavanine resistance using a modification of the method described in Klein (2001). Strains were struck out so that colonies arose from single cells and were allowed to grow for 3 days at 30°. Twenty-four separate colonies per strain were then chosen and dispersed in 200 μl of water each in 96-well plates. Absorbance was measured using the Tecan Spectroflour Plus at 620 nm, and the 15 colonies with the closest optical densities were used for analysis. The canavanine-resistant (k) and viable numbers of cells (N) in each colony were determined by plating the appropriate dilution on SC–Arg− plus canavanine at 60 μg/ml and nonselective (YPD) plates, respectively. Plates were grown for 3 days at 30°, and then colonies were counted using the aCOLyte SuperCount colony counter. Chromosome V instability rates were calculated using fluctuation analysis and a modification of the Lea and Couslon method of the median (Lea and Coulson 1949). To accommodate variations in N, chromosome V instability rate estimates were separately computed for the 15 colonies, setting the initial cell count at N0 = 1 and utilizing individual N and k values for each colony. A median of these estimates was accepted as a final estimate. Independently, estimates were computed using the maximum-likelihood method (Zheng 2005) modified to accommodate variation in N; estimates from both methods showed <10% difference.
Statistical analysis of chromosome V instability rates:
To determine if estimated chromosome V instability rates of mutant strains were different from the parental strain rate, we performed one-sided unequal Behrens–Fisher (Lix et al. 2005) variance t-tests for the estimated chromosome V instability rate of each strain (using the estimated mutation rate data from all parallel cultures) against the estimated parental chromosome V instability rate to test if this rate was smaller.
Transposon insertion site identification—vectorette PCR and inverse PCR:
Identification of the site of insertion of the transposon in 300 strains was performed using two different methods: Vectorette PCR and inverse PCR. The remaining 98 insertional mutants were unidentifiable through extensive rounds of Vectorette and inverse PCR with different individual restriction enzymes and combinations and thus were not pursued further. Vectorette PCR methods utilizing RsaI, AluI, and DraI in different experiments were carried out. Briefly, genomic DNA was isolated from strains of interest and digested. Anchor bubbles were ligated onto the blunt ends created by use of frequent blunt-end cutter restriction enzymes, and PCR was performed utilizing a primer to the anchor bubble and a primer to the mTn-lacZ/LEU2 transposon. A total of 80 μl of the PCR reaction was electrophoresed on a 0.7% TAE gel, and unique bands were gel purified and sequenced using a primer from the transposon. Inverse PCR techniques were then carried out on the remaining strains utilizing RsaI, AluI, DraI, TaqI, HpaII, AciI, and HaeIII restriction enzymes. Genomic DNA was isolated from strains of interest and digested with a single restriction enzyme, ligated at low density, precipitated, and used as template DNA for PCR with opposing primers from within the mTn-lacZ/LEU2 transposon. Unique bands were again gel purified, sequenced, and then analyzed against the S. cerevisiae genomic sequence to identify the transposon insertion site.
Gene ontology analysis and interactome mapping:
Gene ontology annotations were analyzed using the Generic Gene Ontology (GO) Term Mapper (http://go.princeton.edu/cgi-bin/GOTermmapper) from the Lewis-Sigler Institute for Integrated Genomics at Princeton University and the S. cerevisiae GO slim gene association files. The P-values were calculated using standard χ2 tests.
An interactome map of the network of genetic and physical interactions among the 172 unique genes identified was created utilizing the Osprey visualization system (Breitkreutz et al. 2003). Node colors depict biological process gene ontologies, while edge colors denote the genetic or physical interaction identified.
Creation of precise heterozygous mutations utilizing the KAN-MX cassette:
Precise heterozygous deletions were created for a subset of the genes utilizing the KAN-MX cassette homologous recombination switch-out method (Wach et al. 1994). KAN-MX module deletions for each gene were amplified using PCR on DNA isolated from individual strains from the haploid deletion collection or by long flanking homology techniques. Primers were designed 500–800 bp upstream and downstream of the KAN-MX cassette to create large areas of homology to facilitate recombination between the deletion cassette and endogenous gene. After amplification, the PCR-generated products were transformed into ES75 and ES76 strains, and positive transformants were selected on 200 μg/ml G418 plates. Positive transformants were then checked for proper integration via PCR with a forward primer upstream of the gene of interest (GOI) and a reverse primer within the KAN-MX cassette.
Genotoxic stress assays:
Precise heterozygous deletion strains were tested for sensitivity to phleomycin, methyl methanesulfonate (MMS), hydroxyurea (HU), ultraviolet light (UV), and benomyl. Cultures for each strain were grown overnight at 30° to saturation in 96-well plates and then diluted down to 0.2 OD and allowed to double twice. Each assay plate contained two parental controls and six tester strains and was spotted in duplicate. Five fivefold serial dilutions were performed, and equal aliquots of each dilution were transferred (frogged) onto three plates per genotoxic stress agent: phleomycin—0.1, 0.5, and 1 μg/ml; benomyl—10, 20, and 30 μg/ml; MMS—0.0075, 0.015, and 0.030%; HU—25, 50, and 75 mm; and onto YPD plates for exposure to UV at 20, 30, and 40 J/m2/sec. After several hours (to allow drying), the plates were inverted and placed at 30° for 3 days. Each plate was photographed and visually analyzed for changes in sensitivity.
RESULTS
A genomewide yeast genetic screen for heterozygous mutations enhancing genome instability:
The S. cerevisiae RAD9 gene encodes a putative functional ortholog of mammalian BRCA1, NFBD1 (MDC1), and 53BP1 proteins. All of these proteins contain two BRCT functional domains and function as mediators of the DNA damage checkpoint (Weinert and Hartwell 1990; Bork et al. 1997; Rappold et al. 2001; Peng and Chen 2003, 2005). We developed this screen in a diploid rad9Δ/rad9Δ-deficient background to model mammalian checkpoint-deficient cells, which could potentially lead to identification of specific genetic modifiers of cancer predisposition. In addition, use of the rad9Δ/rad9Δ strain background increases the baseline instability of S. cerevisiae strains ∼5- to 10-fold (Weinert and Hartwell 1990; Klein 2001). For our specific sectoring assay, use of the rad9Δ/rad9Δ background raised the absolute instability of the starting strain, allowing identification of insertional mutants with only a 3- to 10-fold increase in sectors. This would not be feasible in a wild-type background where the starting strain demonstrates only one sector/1500 colonies on average.
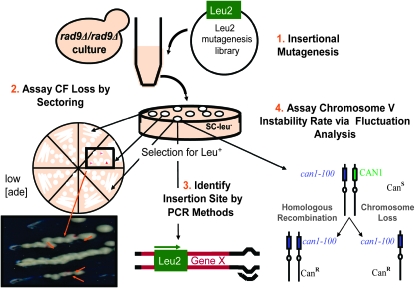
To focus on heterozygous mutations that increase chromosome loss, we developed a diploid parental strain, ES75, which includes the features required for a primary qualitative assay based on the spontaneous loss of a nonessential linear chromosome, the CF, and a secondary quantitative assay, which determines the rate of loss or mitotic recombination of a natural endogenous chromosome. The overall screen strategy is depicted in Figure 1. The initial assay measured genome instability through visual assessment of the loss of one or two copies of the CF, resulting in pink or red sectors on white colonies due to an inability to synthesize adenine (Hieter et al. 1985).
Figure 1.—
Screen strategy. (1) The rad9Δ/rad9Δ strain ES75 was mutagenized by transformation with the NotI-digested mTn-lacz/Leu2 insertional library and positive transformants were selected on leucine-deficient plates. (2) Approximately 185 colonies from each mutant strain were then struck out on low-adenine media and monitored for sectoring colonies as a measure of CF loss. Strains that met the sectoring threshold were restruck three times and strains that consistently showed increased CF loss were carried forward. (3) The transposon insertion site was then identified in mutagenized strains that showed increased CF loss. (4) The rate of chromosome V loss or recombination as quantitatively measured by conversion to canavanine resistance was estimated using fluctuation analysis for those strains that demonstrated increased CF loss.
At the initiation of this study, use of the diploid heterozygous deletion collection was not feasible since this collection did not carry the reporters for chromosome instability or the homozygous rad9 deletion necessary for our screen. Therefore, we decided to use an insertional mutagenesis technique based on the Snyder mTn-lacZ/LEU2-mutagenized library (Kumar et al. 2002) to create heterozygous insertions potentially disrupting nonessential, essential, and novel/uncharacterized genes. As recommended by Snyder, to cover ∼95% of yeast genes with insertions, we visually inspected 30,000 Leu+ mutant strains for a sectoring phenotype.
In the initial screen, 1225 of 30,000 mutant strains met our threshold of having four or more sectors in ∼185 colonies. To test for reproducibility of the sectoring phenotype, these 1225 mutant strains were then restruck three additional times. The mutant strains were then categorized into three groups:
Insertional mutants (398), which had four or more sectors in at least three of the four streak-outs. This increased CF loss group represents 1.33% of the 30,000 mutant strains and was carried forward to the next step of the screen.
Mutant strains (761), which consistently sectored but below the threshold level.
Strains (66) that only had sectored colonies on the initial screen.
The increased CF loss group typically demonstrated 4–10 sectors/∼185 colonies, compared with 0–1 in the parental ES75 strain (see Figure 1). This small but significant increase in the number of sectored colonies (P-value of <0.001 over parental strain) supports the hypothesis that heterozygous mutations may convey modest changes in genomic stability.
Mutant strains with increased chromosome fragment loss also demonstrate instability of an endogenous chromosome:
The second assay for genomic instability monitored the endogenous chromosome V's, one containing the wild-type CAN1 gene conferring sensitivity to canavanine and the second containing the can1-100 recessive allele conferring resistance. Quantitative assessment of conversion to canavanine resistance assessed the combined effects of loss of the endogenous chromosome V containing the wild-type CAN1 gene, mitotic recombination of the CAN1 locus to can1-100, or point mutation in the CAN1 gene. However, spontaneous point mutations account for only 1–2% of the canavanine-resistant mutants (Ohnishi et al. 2004) and thus do not significantly contribute to this assay.
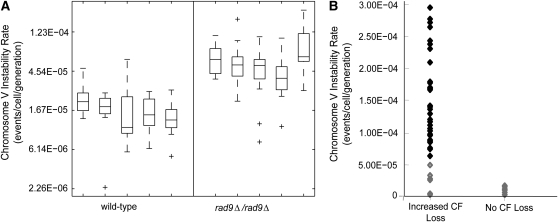
Chromosome V instability was measured by fluctuation analysis using a modification of the Lea and Coulson method of the median (see materials and methods) Five independent estimations of chromosome V instability for the rad9Δ/rad9Δ and wild-type strains were performed to show reproducibility of the assay with estimated mutation rates of 3.37E-05 and 7.86E-06, respectively (see Figure 2A). To confirm that the mutant strains exhibiting increased CF loss also had increased instability of chromosome V, we performed fluctuation analysis on 40 of the 398 increased CF loss strains and 10 mutagenized strains that did not show sectored colonies. We performed one-sided unequal variance t-tests (Welch 1938) on the estimated chromosome V instability rate of each of the strains compared to the estimated rate of the rad9Δ/rad9Δ parental strain (see Figure 2B). We found that 29/40 (72.5%) of the strains from the increased CF loss group showed statistically increased chromosome V instability rates, while none of the 10 strains from the nonsectoring group were statistically different from the parental strain. These data demonstrate that the heterozygous mutants identified in the screen on the basis of increased CF loss are highly enriched for strains that exhibit increased instability of endogenous chromosomes.
Figure 2.—
Validation of chromosome V instability rate estimates and enrichment of increased CF loss strains for increases in chromosome V instability. (A) Fluctuation analysis experiments were performed on five wild-type and five rad9-deficient yeast strains to test reproducibility of the chromosome V instability rate estimations. These data are shown in a combined box and whisker plot where each box represents summary statistics of instability rate estimates in the 15 parallel cultures of each strain. The box has lines at the lower quartile, median, and upper quartile of the data. The whiskers are lines extending from each end of the box to show the extent of the rest of the data. The whiskers indicate the minimum and maximum data values, unless outliers (marked by “+”) are present in which case the whiskers extend to a maximum of 1.5 times the interquartile range. (B) We measured the chromsome V instability rates of 40 mutant strains from the increased CF loss category and 10 mutant strains from the nonsectoring category. One-sided unequal variance t-tests were performed on the estimated rate of chromosome V instability of each of the 50 strains compared to the rad9Δ/rad9Δ parental strain (3.37E-05 by summation of five separate rad9Δ/rad9Δ estimations). Strains that were statistically increased as determined by the t-test are shown as solid black diamonds; strains not significantly increased are shown as open gray diamonds. Seventy percent of the strains from the high CF loss group showed statistically increased chromosome V instability rates, while none of the strains from the no CF loss group were statistically different.
Identification of the site of transposon insertion in 300 high CF loss strains:
The transposon insertion sites in 300 of the 398 increased CF loss strains were identified using Vectorette and inverse PCR techniques (see materials and methods). Analysis of the 300 insertion sites revealed 172 insertions within previously defined gene coding sequences, which likely disrupt or modify activity of the encoded proteins; 53 insertions in regions that do not contain a previously annotated ORF; and 75 sites that are replicas of those in the previous two groups. See supplemental Table 1 at http://www.genetics.org/supplemental/ for all identified transposon insertion sites.
We performed analysis of the chromosomal coordinates of the 106 genes with the potential to have promoter regions disrupted by transposon insertions in nonannotated regions. We find that 64 of these genes are in an orientation with their 5′ region adjacent to the insertion site. We searched the limited S. cerevisiae promoter database (Zhu and Zhang 1999) for insertions within the promoter region. However, only 5 of the 64 genes in the correct orientation have documented promoter regions, and in only two cases do the insertion sites fall between the promoter region and the transcription start site (TSS): STE2 and FAS1. However, by mapping the distance to the TSS (base pair 0) for the insertions not in documented promoters, we found that in 28.6% of these cases the insertion site of the transposon was within 250 bp of the TSS and may be expected to disrupt transcription (see supplemental Table 2 at http://www.genetics.org/supplemental/).
Notably, of the 172 unique gene disruptions, 20% were in essential genes or genes identified only as hypothetical ORFs. Since these two groups are not included in the haploid deletion collection (Winzeler et al. 1999), a substantial portion of genes identified in this screen have not been phenotypically characterized by other investigators employing this collection.
GO annotations and interactome mapping to characterize the 172 unique gene disruptions:
We focused on the 172 strains containing unique gene-disrupting insertions, given the possible varying impact of the other insertions on gene expression. To characterize this large set of insertions, we used GO, a vocabulary of annotated gene assignments into ontologies on the basis of their known biological process, compartment location, or molecular function (Ashburner et al. 2000).
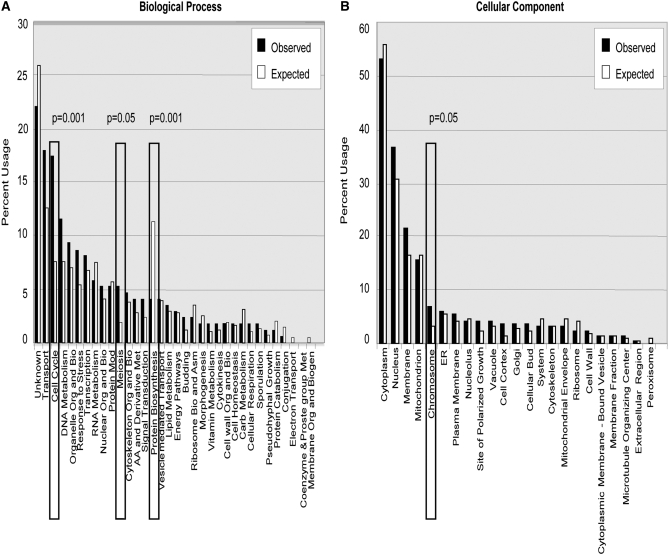
We characterized the 172 genes by comparing their ontology representation against the ontology of the entire budding yeast genome. Using GO processes at the third level of complexity, the groups that were statistically significantly overrepresented in the biological process division included cell cycle and meiosis; the only underrepresented group was protein biosynthesis (Figure 3A). Of note, all of the genes identified in the meiosis group are also contained in the cell cycle group. The analysis of gene ontology by cellular compartment revealed that chromosome location was overrepresented (Figure 3B). Thus, the nonrandom nature of this list as well as the overrepresentation in genes known to be involved in genomic stability further supports the validity of this screen.
Figure 3.—
Gene ontology analysis of biological processes and cellular component categories for S. cerevisiae. This representation shows the percentage of usage in the gene ontology at the third level of both biological process (A) and cellular component (B). Solid bars show the observed percentage of representation in list of genes isolated in the screen; open bars show the representation observed in the yeast genome and demonstrate the expected rate in the list of genes if the list is random. The boxed ontologies of cell cycle, meiosis process, and chromosome (location) are statistically significantly overrepresented while the boxed ontology of protein biosynthesis is underrepresented.
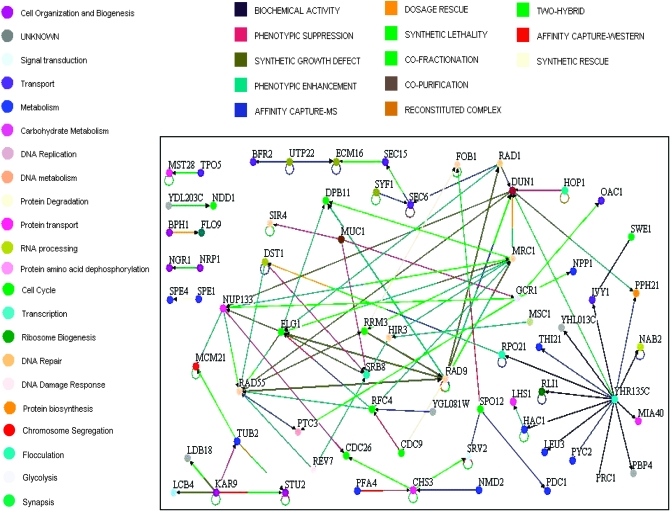
For further analysis of the gene list, we created an interactome map of the known genetic and physical interactions (Figure 4). Notably, 69 of 172 genes had identified interactions with at least one other gene from our list, with the majority of these being genes that mapped to cell cycle, metabolism, and DNA repair ontologies. We included RAD9 on the map and 8 of the genes identified in the screen had previously established genetic or biochemical interactions with RAD9. The YHR135C node shows interactions with 14 other genes/proteins within our list, 13 of which are biochemical activity interactions; however, further inquiry shows that YHR135C has biochemical interactions with a total of 285 proteins (Saccharomyces Genome Database, http://www.yeastgenome.org/), suggesting that it may represent just a “sticky” protein.
Figure 4.—
Network interactome of 69 genes identified by coding region disruptions. This representation shows the 69 of 172 genes, identified by transposon insertion into their coding region, which had been previously identified to physically or genetically interact with another member of this group or with RAD9. The node color indicates the primary biological process gene ontology into which the gene is classified. The edge color indicates the identified physical or genetic interaction.
Quantifying the genomic instability of increased CF loss mutant strains:
Chromosome V instability assays (as previously described) were performed for all 172 unique gene disruptions found in our set of increased CF loss strains (see supplemental Table 3 at http://www.genetics.org/supplemental/). We again performed one-sided unequal variance t-tests for the estimated rate of each unique locus compared to the estimated chromosome V instability rate of the rad9Δ/rad9Δ parental strain. In this larger sample, we found that 45% of the tested strains have a significantly increased chromosome V instability rate compared to the rad9Δ/rad9Δ parental rate of 3.37E-05 and these increases ranged from 3- to 25-fold.
Assays of chromosome instability and rad9 dependence in strains with precise heterozygous deletions of genes identified in the screen:
A subset of 65 of the 172 genes with insertions in a coding region were chosen for further characterization on the basis of one of the following criteria: (1) member of overrepresented groups from the GO analysis, (2) the seven insertional mutant strains with the highest chromosome V instability rate, (3) novel or newly annotated genes, and (4) essential genes. We used directed gene targeting to make precise heterozygous gene deletions of the coding region in nonmutagenized backgrounds to (1) test reproducibility of the phenotype in a nonmutagenized background, (2) eliminate potential hypomorphic effects of the insertion, and (3) test the impact of heterozygous precise deletions in both wild-type and rad9-deficient backgrounds.
Chromosome V instability was determined by fluctuation analysis on these 130 strains (65 precise deletions in both wild-type and rad9Δ/rad9Δ backgrounds). As shown in Table 1, the chromosome V instability phenotype was reproducible, with 80% of the reconstructed deletions in a rad9Δ/rad9Δ background having an equal (within twofold) or greater rate than the original insertional mutant strain isolated in the screen. Surprisingly, 85% of the heterozygous deletions resulted in a greater than twofold increase in chromosome V instability in a wild-type ES76 background. Thus, these deletions identify dosage-sensitive genes that mediate genomic stability independently of RAD9 status. We further characterized the type of effect with regard to rad9 specificity in Table 1. Heterozygous deletion of only three genes, sam1Δ/SAM1, tma17Δ/TMA17, and dml1Δ/DML1, demonstrated increases in chromosome V instability rates that were completely rad9 dependent. There were 8 heterozygous gene deletions where the impact of combining rad9Δ/rad9Δ and the heterozygous deletion resulted in increased instability above a multiplicative effect, and 12 where the level met what would be expected for a simple multiplicative effect. The former group may represent genes that have a synergistic effect on genome instability when combined with a checkpoint deficiency, and the latter group of genes likely causes instability independent of checkpoint deficiencies. The remaining 45 of 65 tested heterozygous gene deletions demonstrated a larger impact in a wild-type vs. a rad9Δ/rad9Δ parental background. Unexpectedly, a small group of these genes had significantly more instability in wild-type compared with rad9Δ/rad9Δ backgrounds. Notably, a mcm21Δ/MCM21 strain has a 500-fold increase in chromosome V instability rate (4.03E-03 chromosome V instability rate, standard deviation 3.49E-03) over the wild-type parental strain. This rate was derived from performing replicate fluctuation analyses of four mcm21Δ/MCM21 strains created from independent transformations with the deletion cassette. In contrast, the rad9Δ/rad9Δ mcm21Δ/MCM21 strain has a rate close to that of a wild-type level (4.21E-06 chromosome V instability rate, standard deviation 3.48E-06). CF loss results were similar where the sectoring of the mcm21Δ/MCM21 strain was much higher than in the rad9Δ/rad9Δ mcm21Δ/MCM21 strain, although some increase in sectoring still occurred compared to the rad9-deficient starting strain, as expected on the basis of the original identification of the insertional mutant in this screen. Thus, although the initial screen was carried out in a rad9Δ/rad9Δ strain background, 62 of 65 heterozygous deletions also increased chromosome instability in checkpoint-proficient strains.
TABLE 1.
Chromosome V instability rate of mutant strains for 65 genes identified in the screen
| GOI | Mutation rate (μ) of insertional mutant | μ of precise deletion (rad9Δ/rad9Δ, goiΔ/GOI) | Ratio of μ (rad9Δ/rad9Δ goiΔ/GOI) to μ (ISM) | A: ratio of μ (rad9Δ/rad9Δ goiΔ/GOI) to μ (rad9Δ/rad9Δ) | μ (goiΔ/GOI) | B: ratio of μ (goiΔ/GOI) to μ (wild type) | A:B ratio |
|---|---|---|---|---|---|---|---|
| Wild type | 7.86E-06 | ||||||
| rad9Δ/rad9Δ | 3.37E-05 | ||||||
| SAM1 (A) | 4.36E-04 | 2.12E-04 | 0.49 | 6.29 | 8.67E-06 | 1.1 | 5.72 |
| TMA17 (B) | 1.15E-04 | 9.87E-05 | 0.86 | 2.93 | 6.90E-06 | 0.88 | 3.33 |
| ARO4 (A) | 9.56E-05 | 5.14E-05 | 0.54 | 1.53 | 3.99E-06 | 0.51 | 3 |
| DML1 (A) | 7.59E-05 | 1.35E-04 | 1.78 | 4.01 | 1.08E-05 | 1.37 | 2.93 |
| MSH5 (A) | 4.01E-05 | 1.79E-04 | 4.46 | 5.32 | 2.48E-05 | 3.16 | 1.68 |
| YBL104C (B) | 1.91E-04 | 8.17E-05 | 0.43 | 2.42 | 1.13E-05 | 1.44 | 1.68 |
| DPB11 (A) | 2.67E-05 | 1.69E-04 | 6.33 | 5 | 2.54E-05 | 3.23 | 1.55 |
| SPP2 (C) | 6.33E-04 | 2.07E-04 | 0.33 | 6.13 | 3.33E-05 | 4.24 | 1.45 |
| CHS3 (A) | 6.06E-05 | 8.19E-05 | 1.35 | 2.43 | 1.51E-05 | 1.92 | 1.27 |
| TUB2 (A) | 5.73E-05 | 2.85E-04 | 4.97 | 8.44 | 6.48E-05 | 8.24 | 1.02 |
| TOM1 (A) | 1.99E-04 | 9.79E-05 | 0.49 | 2.91 | 2.27E-05 | 2.89 | 1.01 |
| RRN3 (A) | 2.49E-05 | 6.00E-05 | 2.41 | 1.78 | 1.44E-05 | 1.83 | 0.97 |
| YLR307C-A (B) | 1.77E-04 | 9.16E-05 | 0.52 | 2.72 | 2.21E-05 | 2.81 | 0.97 |
| MNN4 (A) | 1.76E-04 | 6.86E-05 | 0.39 | 2.03 | 1.72E-05 | 2.19 | 0.93 |
| NDD1 (A) | 2.42E-06 | 7.97E-05 | 32.93 | 2.36 | 2.11E-05 | 2.68 | 0.88 |
| HOP1 (A) | 1.60E-04 | 1.21E-04 | 0.76 | 3.6 | 3.25E-05 | 4.13 | 0.87 |
| REV7 (A) | 1.58E-04 | 8.95E-05 | 0.57 | 2.66 | 2.52E-05 | 3.21 | 0.83 |
| RLI1 (C) | 1.90E-04 | 1.36E-04 | 0.72 | 4.02 | 3.83E-05 | 4.87 | 0.83 |
| HAC1 (A) | 5.12E-05 | 9.18E-05 | 1.79 | 2.72 | 2.65E-05 | 3.37 | 0.81 |
| CDC26 (A) | 3.38E-05 | 9.59E-05 | 2.84 | 2.84 | 2.97E-05 | 3.78 | 0.75 |
| DUN1 (A) | 5.06E-05 | 1.51E-04 | 2.98 | 4.48 | 5.14E-05 | 6.54 | 0.69 |
| TSC11 (C) | 9.44E-05 | 1.05E-04 | 1.11 | 3.11 | 3.72E-05 | 4.73 | 0.66 |
| SDC25 (D) | 8.31E-05 | 1.32E-04 | 1.59 | 3.9 | 4.72E-05 | 6.01 | 0.65 |
| ATG13 (D) | 9.77E-05 | 6.60E-05 | 0.68 | 1.96 | 2.44E-05 | 3.1 | 0.63 |
| CDC9 (A) | 4.08E-06 | 7.00E-05 | 17.16 | 2.08 | 2.72E-05 | 3.46 | 0.6 |
| SWE1 (A) | 9.25E-05 | 8.45E-05 | 0.91 | 2.51 | 3.44E-05 | 4.38 | 0.57 |
| LEU3 (A) | 8.54E-05 | 5.60E-05 | 0.66 | 1.66 | 2.31E-05 | 2.94 | 0.56 |
| YIR035C (B) | 6.75E-05 | 6.43E-05 | 0.95 | 1.91 | 2.76E-05 | 3.51 | 0.54 |
| ACK1 (B) | 1.33E-04 | 1.23E-04 | 0.92 | 3.66 | 5.39E-05 | 6.86 | 0.53 |
| SSK1 (D) | 1.18E-04 | 9.87E-05 | 0.84 | 2.93 | 4.50E-05 | 5.73 | 0.51 |
| DST1 (A) | 1.89E-04 | 8.61E-05 | 0.46 | 2.55 | 3.92E-05 | 4.99 | 0.51 |
| YKL044W (B) | 7.20E-05 | 1.08E-04 | 1.5 | 3.19 | 5.01E-05 | 6.37 | 0.5 |
| IME2 (A) | 3.12E-05 | 5.22E-05 | 1.67 | 1.55 | 2.52E-05 | 3.21 | 0.48 |
| SEC6 (C) | 1.33E-04 | 1.21E-04 | 0.91 | 3.59 | 5.89E-05 | 7.49 | 0.48 |
| GDA1 (D) | 1.54E-04 | 1.49E-04 | 0.97 | 4.41 | 7.73E-05 | 9.83 | 0.45 |
| FOB1 (A) | 5.99E-05 | 6.47E-05 | 1.08 | 1.92 | 3.48E-05 | 4.43 | 0.43 |
| ECM16 (C) | 5.65E-05 | 1.16E-04 | 2.05 | 3.46 | 6.49E-05 | 8.26 | 0.42 |
| SYF1 (A) | 3.31E-06 | 5.63E-05 | 17.01 | 1.67 | 3.16E-05 | 4.02 | 0.42 |
| SPO12 (A) | 7.20E-05 | 5.68E-05 | 0.79 | 1.69 | 3.59E-05 | 4.57 | 0.37 |
| BFR2 (C) | 9.86E-04 | 6.78E-05 | 0.07 | 2.01 | 4.48E-05 | 5.7 | 0.35 |
| MSC1 (A) | 4.22E-05 | 5.68E-05 | 1.35 | 1.68 | 3.87E-05 | 4.92 | 0.34 |
| SPE4 (A) | 2.98E-06 | 4.99E-05 | 16.74 | 1.48 | 3.48E-05 | 4.43 | 0.33 |
| DBR1 (A) | 5.76E-05 | 1.10E-04 | 1.91 | 3.26 | 7.79E-05 | 9.91 | 0.33 |
| SIR4 (A) | 1.76E-04 | 1.22E-04 | 0.69 | 3.62 | 8.66E-05 | 11.02 | 0.33 |
| DIP5 (A) | 1.06E-04 | 9.95E-05 | 0.94 | 2.95 | 7.31E-05 | 9.3 | 0.32 |
| TEA1 (A) | 1.36E-04 | 3.93E-05 | 0.29 | 1.17 | 2.91E-05 | 3.7 | 0.32 |
| STU2 (A) | 1.60E-04 | 5.34E-05 | 0.33 | 1.58 | 3.98E-05 | 5.06 | 0.31 |
| ELG1 (A) | 5.10E-05 | 6.85E-05 | 1.34 | 2.03 | 5.12E-05 | 6.51 | 0.31 |
| LHS1 (A) | 2.34E-04 | 6.27E-05 | 0.27 | 1.86 | 4.98E-05 | 6.34 | 0.29 |
| RAD55 (A) | 8.48E-05 | 8.52E-05 | 1 | 2.53 | 7.84E-05 | 9.97 | 0.25 |
| NGR1 (A) | 1.37E-04 | 2.29E-06 | 0.02 | 0.07 | 2.29E-06 | 0.29 | 0.24 |
| RAD1 (A) | 2.51E-04 | 7.43E-05 | 0.3 | 2.2 | 7.17E-05 | 9.12 | 0.24 |
| NMD2 (D) | 1.47E-04 | 1.03E-05 | 0.07 | 0.31 | 1.03E-05 | 1.31 | 0.24 |
| PPH21 (A) | 2.01E-05 | 2.30E-05 | 1.14 | 0.68 | 2.30E-05 | 2.93 | 0.23 |
| PTC3 (A) | 9.81E-05 | 4.76E-05 | 0.49 | 1.41 | 4.89E-05 | 6.22 | 0.23 |
| UTP22 (C) | 9.52E-05 | 4.69E-05 | 0.49 | 1.39 | 5.00E-05 | 6.37 | 0.22 |
| RFC4 (A) | 1.78E-04 | 6.64E-05 | 0.37 | 1.97 | 7.33E-05 | 9.33 | 0.21 |
| HIR3 (A) | 1.72E-04 | 4.45E-05 | 0.26 | 1.32 | 6.96E-05 | 8.85 | 0.15 |
| MRC1 (A) | 4.42E-05 | 2.89E-05 | 0.65 | 0.86 | 7.37E-05 | 9.38 | 0.09 |
| YDR066C (B) | 6.18E-05 | 3.13E-05 | 0.51 | 0.93 | 9.13E-05 | 11.62 | 0.08 |
| ATG11 (D) | 1.61E-04 | 2.22E-05 | 0.14 | 0.66 | 7.70E-05 | 9.8 | 0.07 |
| TRX1 (A) | 1.22E-04 | 3.78E-05 | 0.31 | 1.12 | 1.44E-04 | 18.32 | 0.06 |
| SPE1 (D) | 1.69E-04 | 1.18E-04 | 0.7 | 3.5 | 6.87E-04 | 87.4 | 0.04 |
| YAL066W (B) | 6.74E-05 | 1.33E-04 | 1.97 | 3.95 | 7.83E-04 | 99.62 | 0.04 |
| MCM21 (A) | 8.63E-05 | 4.21E-06 | 0.04 | 0.12 | 4.03E-03 | 512.72 | 0 |
Column 1 identifies the GOI isolated in the mutagenesis screen by transposon insertion and selected for construction of a precise heterozygous deletion (goiΔ/GOI) in both rad9-deficient and wild-type backgrounds. The basis for inclusion of this gene in this subset is denoted by the letter in parentheses (A, B, C, and D) after the gene name. A, gene ontology statistically significantly overrepresented group; B, hypothetical open reading frame; C, essential; D, high chromosome V instability rate (μ). Column 2 shows μ (events/cell/generation) as measured by conversion to canavanine resistance of the insertionally mutagenized strains (ISM) in a rad9Δ/rad9Δ background. Column 3 shows the μ of the precise heterozygous deletion in the rad9-deficient background. Column 4 shows the ratio between the two to determine the reproducibility of the instability phenotype. Column 5 shows the ratio (A) of the μ of the heterozygous gene deletion in the rad9Δ/rad9Δ background to the μ of the rad9Δ/rad9Δ parental strain. Column 6 shows the μ of the heterozygous deletion in a wild-type background. Column 7 shows the ratio (B) of the μ of the heterozygous gene deletion in the wild-type background to the wild-type parental strain. Column 8, the calculated A/B ratio, shows the fold change between the increases in μ due to the heterozygous gene deletion in the rad9Δ/rad9Δ vs. wild-type backgrounds. Column 8 was then used to categorize these chromosome V instability rates on the basis of whether the fold change due to the heterozygous deletion was larger or smaller in the rad9Δ/rad9Δ or wild-type backgrounds. The first group (A > B) represents 8 strains where the heterozygous deletion of the GOI appears to have a larger effect in a rad9Δ/rad9Δ background than in a wild-type background. The second group (A ≈ B) represents 12 strains where heterozygous deletion of the GOI has a similar effect in both backgrounds. The third group (A < B) represents 45 strains where the heterozygous gene deletion has a larger fold effect on chromosome V instability in the wild-type background compared to the rad9Δ/rad9Δ background.
The majority of the heterozygous gene deletion strains do not show changes in the response to exogenous genotoxic stress:
The original sectoring and canavanine resistance assays measured spontaneous events. We wanted to determine if these heterozygous mutations also had an impact on the response to exogenous damage. Sensitivity of the wild-type and rad9Δ/rad9Δ strains containing the 65 reconstructed heterozygous deletions to a variety of different genotoxic stresses was tested by exposure to agents including MMS, HU, UV, phleomycin, and benomyl, a microtubule-depolymerizing agent. The strains were exposed to three doses of each drug or UV radiation, and viability was measured in a semiquantitative assay.
The majority (65%) of heterozygous mutant strains did not show a change in sensitivity to any of the agents tested. The minority of strains that had altered growth in response to agents are shown in Table 2. This group includes strains with deletions of several genes in which null mutations in haploid or homozygous strains have been previously shown to be sensitive. For example, rad52Δ/RAD52 and rad52Δ strains show sensitivity to MMS and UV and both dun1Δ/DUN1 and dun1Δ strains are sensitive to MMS (Table 2 and Begley et al. 2004). Our results demonstrate dosage sensitivity of heterozygous mutations for these genes. However, the finding that the majority of heterozygous mutant strains do not demonstrate increased sensitivity to genotoxic stress reinforces that our screen predominantly identified heterozygous mutations that affect spontaneous genomic events distinct from pathways that mediate damage sensitivity.
TABLE 2.
Genotoxic stress phenotypes for precise heterozygous gene deletions
| Heterozygous deletion | Strain background | UV | MMS | Benomyl | Phleomycin | HU |
|---|---|---|---|---|---|---|
| ATG11 | Wild type | All | ||||
| ATG11 | rad9Δ/rad9Δ | Resistant | Resistant | |||
| ARO4 | Wild type | All | ||||
| ARO4 | rad9Δ/rad9Δ | All | Mild | Resistant | Mild | |
| ACK1 | rad9Δ/rad9Δ | Mild | ||||
| CHS3 | Wild type | All | All | All | All | All |
| DUN1 | Wild type | All-mild | Mild | All | All | |
| GDA1 | Wild type | All | ||||
| GDA1 | rad9Δ/rad9Δ | All | Mild | Resistant | All | |
| HIR3 | rad9Δ/rad9Δ | Resistant | Resistant | Mild | ||
| HOP1 | Wild type | All | ||||
| HOP1 | rad9Δ/rad9Δ | All | Mild | Resistant | ||
| MRC1 | Wild type | All | ||||
| MRC1 | rad9Δ/rad9Δ | Resistant | ||||
| MCM21 | Wild type | All | Mild | Resistant | Mild | |
| NDD1 | Wild type | All | Mild | Mild | ||
| NGR1 | Wild type | Mild | Mild | Resistant | ||
| NGR1 | rad9Δ/rad9Δ | All | Mild | Resistant | ||
| PPH21 | Wild type | Mild | ||||
| RAD1 | Wild type | All | ||||
| RAD55 | Wild type | All | All | All | All | |
| RRN3 | Wild type | Mild | ||||
| SAM1 | Wild type | Resistant | Resistant | Resistant | ||
| SAM1 | rad9Δ/rad9Δ | Resistant | ||||
| SIR4 | Wild type | All | ||||
| SPE1 | Wild type | Resistant | Resistant | Resistant | ||
| TMA17 | Wild type | Resistant | Resistant | |||
| TRX1 | Wild type | All | ||||
| TRX1 | rad9Δ/rad9Δ | Resistant | ||||
| YAL066W | Wild type | Resistant | Resistant | |||
| YAL066W | rad9Δ/rad9Δ | Resistant | ||||
| YBL104C | Wild type | All | All | |||
| YDR066C | Wild type | All | ||||
| YCR066C | rad9Δ/rad9Δ | Resistant | Resistant |
Sixty-five precise heterozygous deletions in rad9Δ/rad9Δ and wild-type backgrounds, as described in Table 1, were tested for response to genotoxic stress. Forty-one of the 65 tested heterozygous gene deletions were not sensitive to any of the agents tested in either background. Strains that showed an altered response to any agent are shown. Exposures to UV doses of 20, 30, and 40 J/m2/sec; methyl methanesulfonate doses of 0.0075, 0.015, and 0.03%; benomyl doses of 10, 20, and 30 μg/ml; phleomycin doses of 0.1, 0.5, and 1 μg/ml; and HU doses of 25, 50, and 75 mm were tested. All, sensitive to all doses; mild, sensitive only at high doses; All-mild, slight decreases in growth seen at every dose.
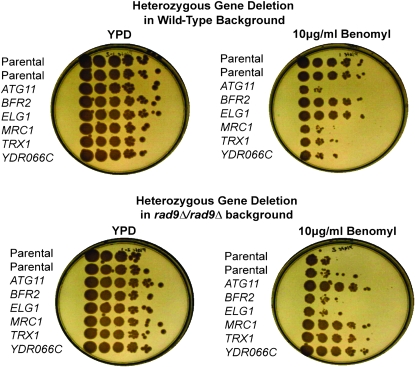
Most of the agents that we tested result in DNA damage; however, we also included benomyl in this study due to its impact on the mitotic spindle and chromosome segregation. Although a few heterozygous strains showed benomyl sensitivity (Table 2), we also identified a group of seven heterozygous deletions, e.g., atg11Δ/ATG11, which demonstrated relative resistance to benomyl in a rad9-deficient state and sensitivity in a wild-type background (see Figure 5 and discussion).
Figure 5.—
Altered benomyl sensitivity and resistance of a subset of mutant strains. Wild-type and rad9Δ/rad9Δ strains with heterozygous deletions in atg11, bfr2, elg1, mrc1, trx1, and ydr066c were grown on rich media (YPD) without or with 10 μg/ml benomyl. Although atg11Δ/ATG11, mrc1Δ/MRC1, trx1Δ/TRX1, and ydr066cΔ/YDR066C strains are sensitive to benomyl compared to wild-type strains, the heterozygous mutation confers relative resistance to benomyl in a rad9Δ/rad9Δ background. The rad9Δ/rad9Δ strain itself shows an increased sensitivity to benomyl vs. the wild-type counterpart.
Heterozygous mutations of MCM21 and the genes encoding other members of the COMA kinetochore complex are very unstable in a wild-type background:
The mcm21Δ/MCM21 heterozygous strains showed the greatest instability with a 500-fold increase in a wild-type background. mcm21 was originally identified as minichromosome maintenance mutant number 21 in a haploid EMS mutagenesis screen for instability of a monocentric circular minichromosome (Maine et al. 1984). This gene was later identified to be part of the COMA complex involved in binding the kinetochore to centromeric regions (Ortiz et al. 1999). We further determined the dosage sensitivity of mcm21 mutants as well as the other COMA complex members (Table 3). The mcm21Δ/mcm21Δ homozygous null strain had an even further increase in instability compared with the heterozygous counterpart increasing from 4.03E-03 to 1.94E-02 with loss of the second copy of MCM21.
TABLE 3.
Impact on genome stability due to deletions in COMA complex genes
| COMA mutation | Strain background | Sectoring (%) | Chromosome V instability rate (events/cell/generation) | Fold change over parental strain |
|---|---|---|---|---|
| — | Wild type | 0.07 | 7.86E-06 | |
| — | rad9Δ/rad9Δ | 0.27 | 3.37E-05 | |
| mcm21Δ/MCM21 | Wild type | 4.10 | 4.03E-03 | 512.72 |
| mcm21Δ/MCM21 | rad9Δ/rad9Δ | 0.65 | 4.21E-06 | 0.12 |
| mcm21Δ/mcm21Δ | Wild type | 0.00 | 1.94E-02 | 2468.19 |
| mcm21Δ/mcm21Δ | rad9Δ/rad9Δ | 0.00 | 1.87E-02 | 554.90 |
| ctf19Δ/CTF19 | Wild type | 6.80 | 2.54E-04 | 32.32 |
| ctf19Δ/CTF19 | rad9Δ/rad9Δ | 8.60 | 1.64E-04 | 4.87 |
| ctf19Δ/ctf19Δ | Wild type | 0.00 | 1.81E-02 | 2302.80 |
| ctf19Δ/ctf19Δ | rad9Δ/rad9Δ | 0.00 | 1.64E-02 | 486.65 |
| okp1Δ/OKP1 | Wild type | 5.00 | 2.72E-05 | 3.46 |
| okp1Δ/OKP1 | rad9Δ/rad9Δ | 3.33 | 1.29E-06 | 0.04 |
| ame1Δ/AME1 | Wild type | 6.60 | 1.47E-05 | 1.87 |
| ame1Δ/AME1 | rad9Δ/rad9Δ | 6.60 | 3.08E-05 | 0.91 |
Sectoring and chromosome V instability rates are shown for heterozygous and homozygous deletion strains for the four members of the COMA complex. Sectoring is shown as a percentage of the total number of sectors in ∼500 colonies. Chromosome V instability rate (events/cell/generation) as measured by conversion to canavanine-resistant cells was estimated via fluctuation analysis. The fold change in the chromosome V instability rate for the indicated strain compared to the respective parental strain rate is shown in column 4. OKP1 and AME1 are essential genes and data are available only for heterozygous strains.
The COMA complex is named for its four member proteins, Ctf19, Okp1, Mcm21, and Ame1, which are encoded by two nonessential genes, CTF19 and MCM21, and two essential genes, OKP1 and AME1. The ctf19Δ/CTF19 and ctf19Δ/ctf19Δ strains are also very unstable with chromosome V instability rates of 2.5E-04 and 1.8E-02, respectively, similar to the mcm21 mutant strains. Heterozygous deletion of the essential genes, OKP1 and AME1, results in increased instability, but not to the high level seen with deletion of the nonessential members of this complex. Similar to the mcm21Δ/MCM21 mutant strain, the combined rad9Δ/rad9Δ okp1Δ/OKP1 strain was more stable than okp1Δ/OKP1 alone.
DISCUSSION
In this study, we performed a genomewide screen in S. cerevisiae to successfully identify heterozygous mutations that cause a modest increase in genome instability in a checkpoint-deficient strain. The genes identified in this screen differ from prior screens, which were carried out in haploid strains and which typically identify mutant strains with much greater degrees of genomic instability (Begley et al. 2004; Yuen et al. 2007). We identified 398 heterozygous mutant strains with increased loss of a nonessential artificial chromosome, resulting from insertions into at least 172 different coding regions in this collection. Approximately 45% of these insertions also resulted in a statistically significant 3- to 25-fold increased instability of the endogenous chromosome V. Assays of genomic instability conducted in precise heterozygous deletions in nonmutagenized backgrounds demonstrated 80% reproducibility of the instability phenotype. These findings lend further support to the analysis of other heterozygous phenotypes in yeast as a potential model of human diseases that result from haplo-insufficiency.
Yuen et al. (2007) recently published work identifying 293 null mutant strains (either hemizygous deletions in haploids or homozygous deletions in diploids) in an otherwise wild-type background, which affected maintenance of genome structure. They utilized three different assays to identify these genes, one of which was similar to our initial assay using CF inheritance and colony sectoring to measure instability. When we compare our gene list with the 89 genes identified from this portion of their screen, we find 4 genes in both lists. Although we do not know the severity of sectoring required to be included in the Yuen et al. (2007) data set, as their scoring ranged from 1–3 (from subtle to prominent), the lack of significant overlap in the genes identified in the two screens may result from (1) our use of a rad9-deficient background and (2) the inability to a priori predict which genes will demonstrate both a null and heterozygous or dosage-sensitive phenotype.
Gene ontology analysis demonstrated that we identified insertions into genes implicated in a wide variety of functions but the list had statistically significant overrepresentations of groups such as cell cycle that have members known to participate in maintenance of genome stability. Further analysis of the genes in the cell cycle group revealed a predominance of G2, G2/M, and mitotic checkpoint genes and an absence of G1 cell-cycle-specific genes. The interactome demonstrated that many of these genes also genetically or biochemically interact with RAD9. However, we did not identify the genes encoding the checkpoint kinases MEC1, TEL1, or CHK1 presumably because enzymes are less likely to be dosage sensitive. Instead, we identified genes whose products are proposed to play more structural roles in the checkpoint response such as MRC1 and DBP11 (Kamimura et al. 1998; Osborn and Elledge 2003).
Further assays for sensitivity to genotoxic stress demonstrated that the majority of heterozygous deletions that increased spontaneous genomic instability did not significantly increase sensitivity to exogenous damage. Thus, the results of this screen are distinct from those that focused on mutants with increased sensitivity to specific agents (Birrell et al. 2001; Chang et al. 2002; Aouida et al. 2004). Most interesting was the observation that, for seven different deletions, the heterozygous strains were more resistant to the microtubule-depolymerizing agent benomyl in a rad9-deficient background and were more sensitive to benomyl in the wild-type background. In these strains, the heterozygous mutation may result in slowing during mitosis. In the wild-type case, this slowing could accentuate the mitotic delay that is induced by exposure to benomyl and result in poorer colony growth. In fact, experiments performed in conditions inducing slow growth, growth either at 18° or on low-nutrient plates, further accentuated sensitivity to benomyl (data not shown). Conversely, in the rad9-deficient background, this mitotic delay may compensate for the RAD9 checkpoint deficiency and result in relative resistance to benomyl. Consistent with this model, the rad9Δ/rad9Δ strain itself is more sensitive to benomyl exposure. Overlapping roles of Rad9p in both the DNA damage and spindle checkpoints have been previously reported by our lab and others (Garber and Rine 2002; Mikhailov et al. 2002; Scott and Plon 2003).
Although the initial screen was performed in a rad9Δ/rad9Δ background, additional testing of 65 precise gene deletions revealed that 85% of them were not exclusively RAD9 dependent and also increased genomic instability of strains in a wild-type background. However, the absolute sectoring rate of the majority of these heterozygous mutations in a wild-type background was still less than the rad9Δ/rad9Δ parental strain. Thus, use of the rad9Δ/rad9Δ background was necessary to bring the absolute instability of the strains to a visible level for the sectoring assay used in our initial screen. The majority of the genes identified demonstrate dosage-sensitive genomic instability independent of checkpoint status, and heterozygous mutations in homologous human genes might affect cancer susceptibility in the general population.
Twenty of the 65 precise deletions tested did increase genomic instability of a rad9Δ/rad9Δ strain at or above a multiplicative effect. The human orthologs of this set of genes would be good candidates for modifiers of the cancer phenotype in carriers of BRCA1 or other checkpoint mutations.
Included in the group of potential BRCA1-specific modifiers are the three genes that showed rad9 specificity for genome instability: SAM1, TMA17, and DML1. This rad9 specificity may reflect heterozygous mutations that induce instability through an intermediate that is normally detected and compensated for by the Rad9 checkpoint pathway and thus do not show instability in a wild-type background.
The budding yeast SAM1 gene encodes an enzyme required for production of S-adenosyl-l-methionine (SAM), availability of which has been associated with the development of cancer. Consistent with our findings in yeast, exogenous addition of SAM to primary human foreskin fibroblasts has recently been shown to prevent aneuploidy (Ramirez et al. 2007). While alterations in levels of SAM may be affected by diet or exposure to chemical agents, variation in levels may also result from polymorphisms in methyl metabolism genes.
The TMA17 gene was named for its translational machinery association (Fleischer et al. 2006), but further functional studies have not been performed. It is of particular interest that Mcm21 was one of five proteins found to interact with Tma17 through a two-hybrid analysis (Ito et al. 2001). Given this interaction and our finding of a CF and chromosome V instability phenotype for the heterozygous tma17 deletion strain, we hypothesize that Tma17p affects chromosome segregation as well as any putative function in translation.
DML1 was identified as the ortholog of the Drosophila melanogaster Misato gene, which functions in chromosome segregation (Miklos et al. 1997). Analysis of the haploid deletion of dml1 in S. cerevisiae by Gurvitz et al. (2002) revealed that it was an essential gene, although their work specifically implicated a defect in mitochondrial DNA inheritance but not chromosome segregation effects. In contrast, similar to the Drosophila work, our study of heterozygous dml1Δ strains demonstrates a role for Dml1 in chromosome segregation in checkpoint-deficient cells. Although recent studies of human Misato demonstrate a role in mitochondrial fusion (Kimura and Okano 2007), on the basis of our results in S. cerevisiae and the work in D. melanogaster, further analysis of the role of human Misato in maintenance of chromosome stability is warranted.
The mcm21Δ heterozygous deletion caused the greatest increase (∼500-fold) in genomic instability in a wild-type background. We also found that strains mutant for the other nonessential member of the COMA kinetochore complex, CTF19, have similar phenotypes. Thus, deficiency of either Mcm21 or Ctf19 (the nonessential COMA complex members) results in very high levels of chromosome instability. This is consistent with the results of synthetic lethal screens, which demonstrated that combining mcm21Δ or ctf19Δ deletions with spindle checkpoint gene deletions results in lethality (Lee and Spencer 2004; Tong et al. 2004; Measday et al. 2005; Daniel et al. 2006). Heterozygous mutant strains for okp1 and ame1, the two essential members of the COMA complex, showed increases in genome instability but not to the level seen in the two nonessential members. We propose that when either of the nonessential members (Mcm21 or Ctf19) are deficient, Okp1 and Ame1 perform their essential kinetochore function very inefficiently, resulting in very high levels of chromosome instability. Thus, the COMA complex appears to play a critical role in maintaining chromosome segregation fidelity and is encoded by dosage-sensitive genes.
Work ongoing at the time of this study identified a human homolog to Mcm21, called Mcm21R or hMcm21 or CENPO (Meraldi et al. 2006). Similar to our analysis, depletion of Mcm21R by siRNA in HeLa cells showed chromosome congression defects (McAinsh et al. 2006). Depletion of Mcm21R was also shown to allow bypass of mitotic arrest by preventing Mad2 binding to kinetochores, thus inactivating the spindle checkpoint. Interestingly, in our experiments, homozygous deletion of RAD9 reduced sectoring and chromosome V instability of the MCM21 heterozygous strains. Similar results were seen for OKP1 heterozygous strains. These phenotypes in yeast and human cells suggest that, in the presence of defective mitotic and spindle checkpoints, the apparent relative chromosome V stability may be due to reduced viability of the cells that mis-segregate chromosomes or chromosome fragments.
Evolutionary conservation of genes and/or pathways is paramount to the effective use of yeast to model human disease states. Therefore, extensive searches for homologs to the 172 unique genes identified in this screen were conducted using multiple algorithims, including Inparanoid, HomoloGene, OrthoMCL, BLASTP, and PubMed literature searches. This combined approach yielded a candidate human homolog for 50% of the yeast genes isolated from our screen (Table 4).
TABLE 4.
Identification of human homologs for 172 gene insertions identified in the screen
| Yeast protein | Human homolog | Yeast protein | Human homolog | Yeast protein | Human homolog |
|---|---|---|---|---|---|
| Ack1p | SEL1L (D) | Mcm21p | MCM21R (E) | Sec15p | SEC15L2 (A, B, C); EXOC6 (D) |
| Ape2p | NPEPPS (A, B, C, D) | Mes1p | MARS (A, B, C); SYMPK (D) | Sec6p | SEC6L1 (A); EXOC3 (B, D) |
| Apl4p | AP1G1 (A, C, D) | Mia40p | CHCHD4 (A, C, D, E) | Smc4p | SMC4 (A, B, C, D) |
| Atg26p | UGT3A2 (D) | Mnn4p | ANP32E (A) | Smy2p | TNNT1 (A); PERQ1 (D) |
| Aus1p | ABCG2 (D) | Mrc1p | CLSPN (E) | Snf3p | GTR2 (D) |
| Bfr2p | AATF (A, B, C) | Mrs6p | CHM (B, C, E); GDI1 (D) | Snt2p | AF17 (D) |
| Bph1p | NSMAF (A); WDFY4 (C); ABCC1 (D) | Msh5p | MSH5 (A, B, C, D, E) | Snu66p | SART1 (A, C) |
| Bpt1p | ABCC6 (C); WDFY3 (D) | Muc1p | MUC5B (A); MUC16 (C) | Sor1p | SORD (A, C, D) |
| Cdc9p | LIG1 (A, B, C, E); DNL1 (D) | Nab2p | ZC3H14 (D) | Spe1p | ODC1 (A, B, C, D) |
| Chs3p | HAS3 (D) | Nft1p | MRP2 (D) | Spe4p | SRM (C, D) |
| Cox13p | COX6A1 (A, C, D) | Ngr1p | TIAL1 (A, D); TRSPAP1 (C) | Spp2p | GPKOW (C) |
| Dbp5p | DDX19A (A, B, C, D) | Nmd2p | UPF2 (A, B, C, D, E) | Srv2p | CAP2 (A, D); CAP1 (B, C, E) |
| Dbp6p | DDX51 (A, B, C, D) | Npp1p | ENPP5 (A,B); ENPP3 (C,D,E) | Stu2p | CKAP5 (A, C, D) |
| Dbr1p | DBR1 (A, B, C, D, E) | Nup133p | hNUP133 (E) | Swe1p | PKMYT1 (C, D) |
| Did4p | CHMP2A (A, B, C, D) | Oac1p | SLC25A34 (A, B, C, D) | Swh1p | OSBP (A, B, C, D) |
| Dip5p | SLC7A4 (A) | Otu2 p | OTUD6B (A, B, C, D) | Swi1p | AR15B (D) |
| Dml1p | MSTO1 (C, D) | Pdc1p | HPCL (D) | Syf1p | XAB2 (A, C, D) |
| Dpb11p | TopBP1 (E) | Pet191p | MGC52110 (C) | Tom1p | HUWE1 (C, D) |
| Dst1p | TCEA1 (A, B, C, D) | Pfa4p | ZDHHC6 (C, D) | Tpm2p | TPM3 (A) |
| Dun1p | CHEK2 (B, C, D, E) | Ppa2p | IPYR (D) | Trx1p | TXN (A, B, D) |
| Ecm16p | DHX37 (A, B, C, D, E) | Pph21p | PPP2CB (A, D); PPP2C (A, B, C) | Tsc11p | RICTOR (C, D) |
| Elg1p | FRAG1 (E) | Prc1p | PPGB (A, C, D) | Tub2p | TUBB2C (A, B, C); TBB2 (D) |
| Emi2p | HKDC1 (C, D) | Pry1p | GAPR1 (B, C, D) | Ubp11p | UBP21 (D) |
| Ena1p | ATP2C1 (D) | Ptc3p | PPM1G (A, C); PP2CA (D) | Urm1p | C9orf74 (A, C, D) |
| Ena2p | ATP2C1 (D) | Pus9p | RPUSD2 (A, C, D) | Utp22p | NOL6 (A, B, C, D) |
| Ena5p | AT2A1 (D) | Pyc2p | PC (A, C, D) | Vma2p | ATP6V1B2 (A, B); ATP6V1B1 (C, D) |
| Eug1p | P4HB (A, C); PDIA3 (D) | Rad1p | ERCC4 (A, B, C, D, E) | Ybl104Cp | FLJ20323 (A, C, D) |
| Gda1p | ENTPD6 (A, C, D); ENTPD5 (B) | Rev7p | hREV7 (E) | Ydr066Cp | C1orf82 (A) |
| Hac1p | XBP1 (E) | Rfc4p | RFC2 (A, B, C, D) | Yel077Cp | LOC642662 (B) |
| Hop1p | HORMAD2 (A); HORMAD1 (C, D, E) | Rli1p | ABCE1 (A, B, C, D, E) | Yfr044Cp | CNDP2 (A, B, D) |
| Ime2p | ICK (A, C, D) | Rpl12Bp | RPL12 (A, C, D) | Ygl059Wp | PDK4 (A); BCKDK (D) |
| Lcb4p | SPHK2 (A, C); SPHK1 (D) | Rpo21p | POLR2A (A, B, C, D) | Yhr135Cp | CSNK1G1 (A, C, D) |
| Lhs1p | SH3KBP1 (A) HYOU1 (B, C, D) | Rrm3p | C15orf20 (A, C, D) | Yir035Cp | HSD11B1 (A, D) |
| Lsb1p | GRB2 (C); SH3D19 (D) | Sam1p | MAT1A (A, D); MAT2A (B, C) | Ypk1p | SGK2 (A, B, C, D) |
| Mch2p | SLC16A6 (A, D) | Sdc25p | SOS1 (D) |
Searches for human homologs to the 172 proteins encoded by the genes identified in the primary screen for CF loss were carried out using Inparanoid (A), HomoloGene (B), OrthoMCL (C), BLASTP (D), and PubMed (E). Any protein with an identified homolog is listed and the search program(s) that identified the human homolog are denoted by the letters in parentheses, which refer to the search programs above.
Consistent with our goal to use this screen to identify potential cancer predisposition loci, mutations or polymorphisms in human genes homologous to 15% of our mutants have already been associated with increased cancer risk, including TOPBP1, TXN, ERCC4, and CHEK2 (Meijers-Heijboer et al. 2002; Staalesen et al. 2004; Cebrian et al. 2006; Karppinen et al. 2006; Milne et al. 2006). For example, Karppinen et al. (2006) recently investigated TOPBP1's influence on hereditary predisposition to breast and/or ovarian cancer on the basis of its role in DNA damage and replication checkpoints and described a novel heterozygous variant in TOPBP1 associated with an increased breast and/or ovarian cancer risk. Similarly, a single nucleotide polypmorphism in the TXN gene has been associated with susceptibility to breast cancer by Cebrian et al. (2006) and surmised to account for 0.32% of the excess familial risk of breast cancer.
On the basis of their dosage-sensitive impact on genome instability, we believe that the human homologs of genes identified in our screen should be included in ongoing epidemiologic studies for cancer susceptibility loci as well as for modifiers of cancer susceptibility in mutation carriers. The impact of haplo-insufficiency of the mammalian orthologs could also be tested in heterozygous mouse models, including their impact on genome instability and cancer.
Acknowledgments
Special thanks go to Hannah Klein, Vicki Lundblad, and Phil Hieter for donation of strains. We extend thanks to Kenneth Scott, Chad Shaw, and Albert Ribes Zamora for technical support and helpful discussions. This work was supported by Department of Defense grant DAMD17-03-1-0464 to S.E.P.; E.D.S. and X.W. received support from the Dan L. Duncan Cancer Center at Baylor College of Medicine and Texas Children's Hospital. M.K. was supported in part by grant 3T11F01029 from the Polish Council of Scientific Research.
References
- Aouida, M., N. Page, A. Leduc, M. Peter and D. Ramotar, 2004. A genome-wide screen in Saccharomyces cerevisiae reveals altered transport as a mechanism of resistance to the anticancer drug bleomycin. Cancer Res. 64 1102–1109. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler et al., 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley, T. J., A. S. Rosenbach, T. Ideker and L. D. Samson, 2004. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol. Cell 16 117–125. [DOI] [PubMed] [Google Scholar]
- Birrell, G. W., G. Giaever, A. M. Chu, R. W. Davis and J. M. Brown, 2001. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl. Acad. Sci. USA 98 12608–12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork, P., K. Hofmann, P. Bucher, A. F. Neuwald, S. F. Altschul et al., 1997. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 11 68–76. [PubMed] [Google Scholar]
- Breitkreutz, B. J., C. Stark and M. Tyers, 2003. Osprey: a network visualization system. Genome Biol. 4 R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian, A., P. D. Pharoah, S. Ahmed, P. L. Smith, C. Luccarini et al., 2006. Tagging single-nucleotide polymorphisms in antioxidant defense enzymes and susceptibility to breast cancer. Cancer Res. 66 1225–1233. [DOI] [PubMed] [Google Scholar]
- Chang, M., M. Bellaoui, C. Boone and G. W. Brown, 2002. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl. Acad. Sci. USA 99 16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevix-Trench, G., R. L. Milne, A. C. Antoniou, F. J. Couch, D. F. Easton et al., 2007. An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA). Breast Cancer Res. 9 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, J. A., B. E. Keyes, Y. P. Ng, C. O. Freeman and D. J. Burke, 2006. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics 172 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge, S. J., 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274 1664–1672. [DOI] [PubMed] [Google Scholar]
- Fleischer, T. C., C. M. Weaver, K. J. McAfee, J. L. Jennings and A. J. Link, 2006. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 20 1294–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber, P. M., and J. Rine, 2002. Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics 161 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., and R. A. Woods, 2006. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol. Biol. 313 107–120. [DOI] [PubMed] [Google Scholar]
- Gurvitz, A., A. Hartig, H. Ruis, B. Hamilton and H. G. De Couet, 2002. Preliminary characterisation of DML1, an essential Saccharomyces cerevisiae gene related to misato of Drosophila melanogaster. FEMS Yeast Res. 2 123–135. [DOI] [PubMed] [Google Scholar]
- Hieter, P., C. Mann, M. Snyder and R. W. Davis, 1985. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell 40 381–392. [DOI] [PubMed] [Google Scholar]
- Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori et al., 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura, Y., H. Masumoto, A. Sugino and H. Araki, 1998. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell. Biol. 18 6102–6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karppinen, S. M., H. Erkko, K. Reini, H. Pospiech, K. Heikkinen et al., 2006. Identification of a common polymorphism in the TopBP1 gene associated with hereditary susceptibility to breast and ovarian cancer. Eur. J. Cancer 42 2647–2652. [DOI] [PubMed] [Google Scholar]
- Kimura, M., and Y. Okano, 2007. Human Misato regulates mitochondrial distribution and morphology. Exp. Cell Res. 313 1393–1404. [DOI] [PubMed] [Google Scholar]
- King, M. C., J. H. Marks and J. B. Mandell, 2003. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302 643–646. [DOI] [PubMed] [Google Scholar]
- Klein, H. L., 2001. Spontaneous chromosome loss in Saccharomyces cerevisiae is suppressed by DNA damage checkpoint functions. Genetics 159 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., S. Vidan and M. Snyder, 2002. Insertional mutagenesis: transposon-insertion libraries as mutagens in yeast. Methods Enzymol. 350 219–229. [DOI] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49 264–285. [DOI] [PubMed] [Google Scholar]
- Lee, M. S., and F. A. Spencer, 2004. Bipolar orientation of chromosomes in Saccharomyces cerevisiae is monitored by Mad1 and Mad2, but not by Mad3. Proc. Natl. Acad. Sci. USA 101 10655–10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., C. Yen, D. Liaw, K. Podsypanina, S. Bose et al., 1997. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275 1943–1947. [DOI] [PubMed] [Google Scholar]
- Lix, L. M., H. J. Keselman and A. M. Hinds, 2005. Robust tests for the multivariate Behrens-Fisher problem. Comput. Methods Programs Biomed. 77 129–139. [DOI] [PubMed] [Google Scholar]
- Maine, G. T., P. Sinha and B. K. Tye, 1984. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics 106 365–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh, A. D., P. Meraldi, V. M. Draviam, A. Toso and P. K. Sorger, 2006. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J. 25 4033–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday, V., K. Baetz, J. Guzzo, K. Yuen, T. Kwok et al., 2005. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc. Natl. Acad. Sci. USA 102 13956–13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijers-Heijboer, H., O. A. Van Den, J. Klijn, M. Wasielewski, S. A. De et al., 2002. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat. Genet. 31 55–59. [DOI] [PubMed] [Google Scholar]
- Meraldi, P., A. D. McAinsh, E. Rheinbay and P. K. Sorger, 2006. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7 R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov, A., R. W. Cole and C. L. Rieder, 2002. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 12 1797–1806. [DOI] [PubMed] [Google Scholar]
- Miklos, G. L., M. Yamamoto, R. G. Burns and R. Maleszka, 1997. An essential cell division gene of Drosophila, absent from Saccharomyces, encodes an unusual protein with tubulin-like and myosin-like peptide motifs. Proc. Natl. Acad. Sci. USA 94 5189–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne, R. L., G. Ribas, A. Gonzalez-Neira, R. Fagerholm, A. Salas et al., 2006. ERCC4 associated with breast cancer risk: a two-stage case-control study using high-throughput genotyping. Cancer Res. 66 9420–9427. [DOI] [PubMed] [Google Scholar]
- Nathanson, K. L., and B. L. Weber, 2001. “Other” breast cancer susceptibility genes: searching for more holy grail. Hum. Mol. Genet. 10 715–720. [DOI] [PubMed] [Google Scholar]
- Ohnishi, G., K. Endo, A. Doi, A. Fujita, Y. Daigaku et al., 2004. Spontaneous mutagenesis in haploid and diploid Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 325 928–933. [DOI] [PubMed] [Google Scholar]
- Ortiz, J., O. Stemmann, S. Rank and J. Lechner, 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13 1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn, A. J., and S. J. Elledge, 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, A., and P. L. Chen, 2003. NFBD1, like 53BP1, is an early and redundant transducer mediating Chk2 phosphorylation in response to DNA damage. J. Biol. Chem. 278 8873–8876. [DOI] [PubMed] [Google Scholar]
- Peng, A., and P. L. Chen, 2005. NFBD1/Mdc1 mediates ATR-dependent DNA damage response. Cancer Res. 65 1158–1163. [DOI] [PubMed] [Google Scholar]
- Phelan, C. M., T. R. Rebbeck, B. L. Weber, P. Devilee, M. H. Ruttledge et al., 1996. Ovarian cancer risk in BRCA1 carriers is modified by the HRAS1 variable number of tandem repeat (VNTR) locus. Nat. Genet. 12 309–311. [DOI] [PubMed] [Google Scholar]
- Ramirez, T., H. Stopper, R. Hock and L. A. Herrera, 2007. Prevention of aneuploidy by S-adenosyl-methionine in human cells treated with sodium arsenite. Mutat. Res. 617 16–22. [DOI] [PubMed] [Google Scholar]
- Rappold, I., K. Iwabuchi, T. Date and J. Chen, 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell Biol. 153 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck, T. R., 2002. Inherited predisposition and breast cancer: modifiers of BRCA1/2-associated breast cancer risk. Environ. Mol. Mutagen. 39 228–234. [DOI] [PubMed] [Google Scholar]
- Scott, K. L., and S. E. Plon, 2003. Loss of Sin3/Rpd3 histone deacetylase restores the DNA damage response in checkpoint-deficient strains of Saccharomyces cerevisiae. Mol. Cell. Biol. 23 4522–4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staalesen, V., J. Falck, S. Geisler, J. Bartkova, A. L. Borresen-Dale et al., 2004. Alternative splicing and mutation status of CHEK2 in stage III breast cancer. Oncogene 23 8535–8544. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303 808–813. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- Weaver, B. A., and D. W. Cleveland, 2006. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 18 658–667. [DOI] [PubMed] [Google Scholar]
- Weinert, T. A., and L. H. Hartwell, 1990. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol. Cell. Biol. 10 6554–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, B. L., 1938. The significance of the difference between two means when the population variances are unequal. Biometrika 29 350–362. [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901–906. [DOI] [PubMed] [Google Scholar]
- Yuen, K. W., C. D. Warren, O. Chen, T. Kwok, P. Hieter et al., 2007. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc. Natl. Acad. Sci. USA 104 3925–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Q., 2005. New algorithms for Luria-Delbruck fluctuation analysis. Math. Biosci. 196 198–214. [DOI] [PubMed] [Google Scholar]
- Zhou, B. B., and S. J. Elledge, 2000. The DNA damage response: putting checkpoints in perspective. Nature 408 433–439. [DOI] [PubMed] [Google Scholar]
- Zhu, J., and M. Q. Zhang, 1999. SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics 15 607–611. [DOI] [PubMed] [Google Scholar]