Abstract
Autophagy is an important intracellular recycling system in eukaryotes that utilizes small vesicles to traffic cytosolic proteins and organelles to the vacuole for breakdown. Vesicle formation requires the conjugation of the two ubiquitin-fold polypeptides ATG8 and ATG12 to phosphatidylethanolamine and the ATG5 protein, respectively. Using Arabidopsis thaliana mutants affecting the ATG5 target or the ATG7 E1 required to initiate ligation of both ATG8 and ATG12, we previously showed that the ATG8/12 conjugation pathways together are important when plants encounter nutrient stress and during senescence. To characterize the ATG12 conjugation pathway specifically, we characterized a null mutant eliminating the E2-conjugating enzyme ATG10 that, similar to plants missing ATG5 or ATG7, cannot form the ATG12-ATG5 conjugate. atg10-1 plants are hypersensitive to nitrogen and carbon starvation and initiate senescence and programmed cell death (PCD) more quickly than wild type, as indicated by elevated levels of senescence- and PCD-related mRNAs and proteins during carbon starvation. As detected with a GFP-ATG8a reporter, atg10-1 and atg5-1 mutant plants fail to accumulate autophagic bodies inside the vacuole. These results indicate that ATG10 is essential for ATG12 conjugation and that the ATG12-ATG5 conjugate is necessary to form autophagic vesicles and for the timely progression of senescence and PCD in plants.
AS with other eukaryotes, plants have developed sophisticated mechanisms to recycle intracellular proteins. Most selective protein turnover occurs by the ubiquitin (Ub)/26S proteasome pathway, which directs the correct removal of short-lived regulatory and abnormal proteins (Smalle and Vierstra 2004). Conversely, autophagy is a catabolic process that is largely responsible for nonselective bulk turnover of cytosolic components from individual proteins and protein complexes to the removal of whole organelles (Thompson and Vierstra 2005; Bassham 2007). It involves the engulfment of cytoplasm in small vesicles followed by their deposition into the lytic vacuole (lysosome in animals) where the vesicles and cargo are quickly degraded by a cache of vacuolar proteases, peptidases, lipases, and other hydrolytic enzymes.
Thus far, primarily using the yeasts Saccharomyces cerevisiae and Pichia pastoris as models, at least two autophagic routes have been identified (for reviews see Ohsumi 2001; Thompson and Vierstra 2005; Klionsky 2007). Microautophagy proceeds by forming tubular invaginations of cytoplasm into the vacuole, which pinch off and release vesicles called autophagic bodies into the vacuolar lumen. In contrast, macroautophagy involves the de novo formation of small double-membrane-bound vesicles called autophagosomes within the cytoplasm, which sequester cytosolic constituents. These vesicles dock with the vacuole, where the outer membrane fuses with the tonoplast to release the inner compartment into the vacuolar lumen as an autophagic body. In addition, a derivative of macroautophagy called the cytoplasm-to-vacuole targeting (CVT) pathway exists to encapsulate and deliver functional proteins such as preaminopeptidase to the vacuole (Klionsky 2007). While the CVT pathway has been confirmed only in S. cerevisiae, it is possible that a similar vacuolar transport pathway is active in plants (Thompson and Vierstra 2005; Seay et al. 2006). Both micro- and macroautophagy are essential in yeast for maintaining nitrogen (N) and carbon (C) pools, recycling amino acids, removing unwanted or damaged organelles, and survival during starvation. Additional roles in programmed cell death (PCD) and various pathologies have been observed in animals (Bursch 2001; Levine and Klionsky 2004; Ueno et al. 2004; Juhasz et al. 2007).
Through genetic dissection of autophagy in yeasts over the past decade, several groups have discovered a set of autophagy (ATG) proteins common to both micro- and macroautophagy (Tsukada and Ohsumi 1993; Thumm et al. 1994; Harding et al. 1995). In particular, two Ub-like conjugation pathways were identified as essential for proper formation of autophagic vesicles (Ohsumi 2001). These pathways employ two Ub-fold proteins, ATG8 and ATG12, as tags, which through an ATP-dependent reaction cascade become conjugated to their respective targets, the lipid phosphatidylethanolamine (PE) and the ATG5 protein. Both tags are first activated by the common E1-activating enzyme ATG7, which couples ATP hydrolysis to the formation of ATG8-ATG7 and ATG12-ATG7 thioester intermediates. Activated ATG8 and ATG12 are then donated by transesterification to their respective conjugating enzymes (or E2s), ATG3 and ATG10, which then form covalent adducts with the targets via an amide bond between the C-terminal glycines of ATG8 and ATG12 and the ethanolamine moiety of PE and a specific Lys in ATG5, respectively (Mizushima et al. 1998; Ichimura et al. 2000). For ATG8, this Gly becomes exposed after processing of the initial translation product by the ATG4 protease that removes the amino acids C-terminal to this residue (Kirisako et al. 2000).
In yeasts, the ATG8-PE conjugate binds to the autophagic membrane via the lipid moiety and appears to help the membrane to expand during vesicle formation (Kirisako et al. 1999). The ATG12-ATG5 conjugate assembles with ATG16 to form a hetero-octomeric structure that is peripherally associated with the autophagic membrane (Mizushima et al. 1999; Suzuki et al. 2001; Kuma et al. 2002). Assembly of this ATG12/5/16 protein complex appears to precede formation of ATG8-PE and may enhance this lipidation reaction. Through the concerted action of both conjugates and other ATG components, an autophagic body is eventually deposited in the vacuole.
Orthologous autophagic systems have been identified in numerous eukaryotes, including Drosophila melanogaster, Caenorhabditis elegans, mice, and humans, as well as several members of the plant kingdom (Thompson and Vierstra 2005; Klionsky 2007). For example, genes encoding many ATG proteins have been detected in Arabidopsis thaliana, rice (Oryza sativa), and maize (Zea mays), including most components of the ATG8 and ATG12 conjugation pathways (Doelling et al. 2002; Hanaoka et al. 2002; Bassham 2007; A. Suttangkakul, T. Chung and R. D. Vierstra, unpublished results). Whereas the E1, ATG7, the E2s, ATG3 and ATG10, and the ATG5 target are encoded by single genes in Arabidopsis, the polypeptide tags ATG8 and ATG12 are encoded by gene families containing nine and two members, respectively. This increased complexity coupled with our failure to detect obvious Arabidopsis orthologs for other yeast ATG genes (e.g., ATG16 and several encoding components of the yeast ATG1/13 kinase complex) suggests that the plant autophagic system is not identical to that in yeasts and even may have evolved new components and functions.
Reverse genetic analyses of Arabidopsis mutants affecting ATG5 and ATG7 recently revealed that plants defective in ATG8/ATG12 conjugation senesce earlier than wild type and are also hypersensitive to N starvation and limiting light that depresses fixed C availability (referred to here as C limitation) (Doelling et al. 2002; Thompson et al. 2005). Combined with analyses of other autophagic proteins, such as ATG4a/4b, ATG6, ATG9, ATG18, and VTI12, it appears that autophagy is essential for appropriate C and N recycling in plants (Hanaoka et al. 2002; Surpin et al. 2003; Yoshimoto et al. 2004; Xiong et al. 2005; Fujiki et al. 2007; Qin et al. 2007). Additionally, Liu et al. (2005) found from analysis of an atg6 mutant that autophagy helps to restrict PCD triggered by the hypersensitive response (HR) close to the site of pathogen invasion, although the relationship between PCD and autophagy is unclear. Surprisingly, no genetic connections to plant development have been observed despite the predicted need for autophagy in processes such as xylogenesis, sclereid, fiber and aerenchyma maturation, organ abscission, anther dehiscence, and female gametogenesis and embryogenesis, all of which likely involve the wholesale turnover of cellular constituents by PCD mechanisms (Bursch et al. 2004; van Doorn and Woltering 2005).
To further describe the functions of the plant ATG system during autophagy and its potential involvement in PCD and to define the role(s) of the ATG12 conjugation pathway more specifically, we initiated a reverse genetic analysis of Arabidopsis ATG10, the E2 predicted to be responsible for ATG12 conjugation. Key questions included the following: Can ATG5 function in the absence of ATG12 modification? Is ATG5 the sole target of ATG12 and is ATG10 the sole E2? Is the ATG12 conjugation pathway individually essential to form autophagic bodies decorated with ATG8? Does inactivation of ATG12 conjugation have the same phenotypic consequences as does inactivation of ATG8 conjugation? Here, we show that a T-DNA insertion allele disrupting the ATG10 gene affects the normal response of seedlings exposed to N- or C-limiting environments. These mutant plants cannot form the ATG12-ATG5 conjugate and fail to accumulate autophagic bodies inside the vacuole during nutrient starvation. The plants also appear to carry out senescence and PCD much more quickly than wild-type plants, as indicated by elevated levels of a collection of senescence- and PCD-related transcripts and proteins under C-limiting conditions. These results indicate that ATG12 conjugation is essential for the proper formation of autophagic vesicles and that the defects in the ATG system upregulate PCD in addition to attenuating N and C recycling during starvation.
MATERIALS AND METHODS
Sequence analysis of ATG10 proteins:
ATG10 protein sequences were identified in the A. thaliana ecotype Columbia (Col-0) (http://www.Arabidopsis.org), rice (O. sativa) (http://www.tigr.org), poplar (Populus trichocarpa) (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home), Physcomitrella (Physcomitrella patens) (http://moss.nibb.ac.jp), Drosophila (D. melanogaster), and mouse (Mus musculus) (http://www.ncbi.nlm.nih.gov) databases using the yeast ATG10 protein sequence as the query (Ichimura et al. 2000). Intron/exon junctions in A. thaliana ATG10 were determined by alignment with the full-length cDNA sequence from The Arabidopsis Information Resource (TAIR; http://www.Arabidopsis.org). Coding regions for the other plant ATG10 genes were deduced by comparison to Arabidopsis ATG10 and alignments of genomic sequences to those available for cDNAs. Amino acid sequence comparisons were performed using CLUSTALX and MACBOXSHADE (Institute of Animal Health, Pirbright, UK). GenBank, TAIR, and The Institute for Genomic Research accession numbers for the sequences described in this article are At3g07525 (AtATG10), Os04g41990 (OsATG10a), Os12g32210 (OsATG10b), eugene3.00141226 (PtATG10), YLL042C (ScATG10), FBpp0087919 (DmATG10), and Q8R1P4 (MmATG10).
Isolation and complementation of atg10-1:
The atg10-1 T-DNA insertion mutant (SALK_084434) was obtained from the SIGnAL T-DNA collection generated in the A. thaliana Col-0 ecotype (Alonso et al. 2003). Homozygous mutant plants were identified by PCR using the 5′- and 3′-gene-specific primers ATGGATTCAGCTCGAGAGGTCAGCG and ACAGGGATGTAGCTTGAACCATGGCCTGTT, respectively, in combination with the left border T-DNA-specific primer TGGTTCACGTAGTGGGCCATCG (Alonso et al. 2003), and by kanamycin resistance conferred by the T-DNA. The mutant was backcrossed three times to wild-type Col-0 to help remove extraneous mutations.
For complementation, the full-length coding region of the ATG10 cDNA was amplified by PCR using the primers GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGATTCAGCTCGAGAGGTCA and GGGGACCACTTTGTACAAGAAAGCTGGGTTCTAATTCAGCATCTCAAGAGGG designed to introduce BP recombination sites at the 5′- and 3′-ends (underlined), respectively, for subsequent cloning into the Gateway pDONR221 vector (Invitrogen, Carlsbad, CA). Using the primer pair CTACATCCCTCTGGGACTGAGGACTG and CAGTCCTCAGTCCCAGAGGGATGTAG (altered nucleotides underlined), the active-site Cys178 codon was changed to that for serine by the Quickchange method (Stratagene, La Jolla, CA). The ATG10 and ATG10C-S coding regions were transferred to the Gateway pEARLEY201 vector (Earley et al. 2006) by an LR recombination reaction to append the cauliflower mosaic virus (CaMV) 35S promoter and codons for a HA epitope tag to the 5′-end.
The resulting 35S:ATG10 and 35S:ATG10C-S transgenes were introduced into Agrobacterium tumefaciens strain GV3101 and then transformed into homozygous atg10-1 plants by the floral dip method (Clough and Bent 1998). T2 plants homozygous for the atg10-1 mutation were confirmed to contain the transgenes by PCR using primers TGACGTAAGGGATGACGCACAAT and ACTAGTCCCGGGTCTTAATTAACTCTC. PCR products from 35S:ATG10C-S plants were sequenced to confirm the Cys178-Ser mutation. Transgene expression was demonstrated by reverse transcription–PCR (RT–PCR) analysis using 28 amplification cycles with Ex-Taq polymerase (TaKaRa, Madison, WI) and the 5′- and 3′-ATG10 gene-specific primers ATGGATTCAGCTCGAGAGGTCAGCGAT and CAGTCCTCAGTCCCACAGGGATGTAG. The ATG8e 5′- and 3′-gene-specific primers, GCATCTTTAAGATGGACGACGATTTCGAA and ATGTGTTCTCGCCACTGTAAGTGATGTAA, were used as an internal RT–PCR control. Because the 5′-ATG8e primer spans an intron, ATG8e genomic DNA is not amplified by this primer set.
Plant growth conditions:
Arabidopsis seeds were vapor-phase sterilized (Clough and Bent 1998), incubated in water at 4° for 2 days, and germinated on solid Gamborg's B5 (Sigma, St. Louis) medium containing 0.7% agar or in liquid growth medium (GM; Sigma) containing 2% sucrose. The plates and liquid cultures were incubated at 21° in a 16-hr light/8-hr dark photoperiod for long day (LD; fluence rate = 95 μmol m−2 sec−1), an 8-hr light/16-hr dark photoperiod for short day (SD; fluence rate = 95 μmol m−2 sec−1), or in continuous light (fluence rate = 65 μmol m−2 sec−1).
For exposure to N-starvation conditions, 1-week-old seedlings grown in the LD were transferred to N-deficient liquid or solid media containing Murashige and Skoog micronutrient salts (Sigma), 3 mm CaCl2, 1.5 mm MgSO4, 1.25 mm KH2PO4, 5 mm KCl, and 2 mm 2-(N-morpholino)ethanesulfonic acid (pH 5.7). After various amounts of time on the N-deficient solid medium, seedlings were transferred back to GM agar. For exposure to C-limiting conditions, seedlings grown in solid GM for 3 weeks in SD were transferred to soil and grown for 3 more weeks. The plants were then transferred to continuous darkness for various lengths of time and either collected immediately or returned to SD for a 1-week recovery. For confocal microscopy, seeds were germinated in liquid GM. After 1 week, the seedlings were transferred to N-deficient medium for 2 days. Twelve to 16 hr prior to examination by fluorescence confocal microscopy, concanamycin A (Sigma) was added to the medium to a final concentration of 0.5 μm. Plants stably expressing 35S:GFP-ATG8a in the wild-type and atg7-1 backgrounds were as described in Thompson et al. (2005). 35S:GFP-ATG8a was introduced into the atg10-1 and atg5-1 mutant backgrounds by crossing. Homozygous atg10-1 and atg5-1 seedlings expressing the GFP-ATG8a transgene were identified by Basta resistance and verified by fluorescence microscopy and PCR.
DNA/RNA gel-blot analyses:
Total genomic DNA was isolated from 1 g of leaf tissue as described (Balk and Leaver 2001). Twenty micrograms of DNA per sample was subjected to gel electrophoresis using 1.5% agar, the DNA was stained with ethidium bromide and then transferred to Hybond XL membrane (GE Healthcare, Piscataway, NJ) for DNA gel-blot analysis. The 32P-labeled 18S rRNA riboprobe was synthesized with SP6 RNA polymerase using a linearized pGEMT (Promega, Madison, WI) cDNA construction and the Riboprobe system (Promega). Membranes were hybridized overnight at 68° and washed as described (Smalle et al. 2002) prior to autoradiography.
RNA was isolated from liquid-grown and soil-grown plants using the Trizol reagent (Invitrogen). RNA for RT–PCR was treated with DNase RQI (Promega) prior to the synthesis of first-strand cDNA by Superscript II-reverse transcriptase (Invitrogen). The first-strand synthesis primers were the ATG10 gene-specific primers AAGCCACTCATATGTTAATGAAACTCAAGTT and AGAGATTCATCCTCTGGAATTTCCTC (primers 2 and 3, respectively; Figure 2B) or the H2A 3′ gene-specific primer GCAACTTGCTTAGCTCCTCATCATTCCTC (control; Figure 2B). RT–PCR included 35 cycles with Ex-Taq polymerase, the first-strand synthesis primer, and either the ATG10 5′ gene-specific primer pair ATGGATTCAGCTCGAGAGGTCAGCGAT and TAGTTTACAGTGCATCATACAAGGTTCCTG (primers 1 and 4, respectively; Figure 2B).
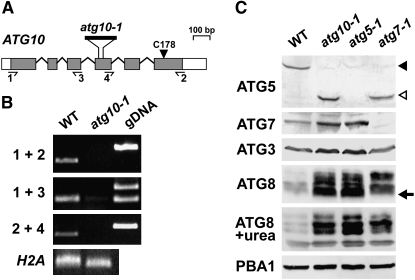
Figure 2.—
Description of the Arabidopsis atg10-1 mutant. (A) Diagram of the Arabidopsis ATG10 gene. Lines indicate introns and boxes indicate exons with coding regions shaded and 5′ and 3′-UTRs as open boxes. The arrowhead locates the active-site cysteine (Cys178). The position of the T-DNA in the atg10-1 mutant is shown. Half-arrows locate the primer binding sites used in B. (B) RT–PCR analysis of the atg10-1 mutant. Total RNA isolated from wild-type (WT) and atg10-1 seedlings was subjected to RT–PCR using the 1 + 2, 1 + 3, or 2 + 4 primer pairs. PCR amplification of genomic DNA (gDNA) and RT–PCR using a primer pair specific for the histone H2A gene were included as controls. (C) Immunoblot detection of the ATG12-ATG5 conjugate. Crude protein extracts from 10-day-old wild-type, atg10-1, atg5-1, and atg7-1 seedlings were subjected to SDS–PAGE with or without 6 m urea and immunoblot analysis with anti-ATG5, anti-ATG7, anti-ATG3, and anti-ATG8 antibodies. Equal protein loads were confirmed by immunoblot analysis with anti-PBA1 antibodies. Solid and open arrowheads identify the ATG12-ATG5 conjugate (50 kDa) and free ATG5 (40 kDa), respectively. Arrow identifies possible ATG8-PE conjugates.
For RNA gel-blot analysis, total RNA was isolated according to Smalle et al. (2002). 32P-labeled riboprobes were synthesized with T7, SP6, or T3 RNA polymerase using the Riboprobe system (Promega) and the linearized pGEMT (Promega) or pBluescript (Stratagene) cDNA constructions for ATG8e, ATG12a, ATG12b, SAG12, PED1, GPX2, CSD1, CAT3, NYE1, TUB4, and 18S rRNA. The CAB, SEN1, and ATG8a probes were from Doelling et al. (2002). Membranes were hybridized overnight at 68° and washed as described (Smalle et al. 2002) prior to autoradiography.
Protein isolation and immunoblot analysis:
Total protein was isolated from liquid- or soil-grown plants by homogenization in 2:1 (volume to gram fresh weight) SDS–PAGE sample buffer [125 mm Tris–HCl (pH 6.8), 5% SDS, 20% glycerol, and 10% 2-mercaptoethanol] and extracts were clarified by centrifugation at 10,000 × g. Proteins were subjected to SDS–PAGE in 12–16% acrylamide gels with or without 6 m urea in the separating gel and either stained with silver or electrophoretically transferred onto PVDF membranes (Millipore, Bedford, MA) for immunoblot analysis using alkaline phosphatase-labeled or peroxidase-labeled goat anti-mouse or goat anti-rabbit immunoglobulins (Kirkegaard & Perry Laboratories, Gaithersburg, MD) for detection. Sample sizes were adjusted to reflect either equal protein or equal fresh weight as indicated. Changes in total protein content during C starvation were measured by spotting the SDS-containing crude extracts directly on PVDF membranes, staining the membranes with Ponceau S, and quantifying the amount of protein densitometrically using bovine serum albumin as the standard.
Antibodies against Arabidopsis ATG3 were produced in rabbits (Harlan Polyclonal Antibody Service, Madison, WI) using recombinant protein expressed with N-terminal His6 and maltose-binding protein (MBP) tags. The full-length ATG3 coding region was inserted into the Gateway pDONR221 vector (Invitrogen), transferred to pVP13 (Center for Eukaryotic Structural Genomics; http://www.uwstructuralgenomics.org) by an LR reaction, and introduced into Escherichia coli BL21 Codon Plus cells (Novagen, Madison, WI). Following a 3-hr induction of log-phase cultures by the addition of 1 mm isopropyl-β-d-thiogalactoside, soluble His6-MBP-ATG3 was purified by NiNTA chromatography (QIAGEN Sciences, Germantown, MD). The His6 and MBP tags were removed by tobacco etch virus protease (Invitrogen) cleavage and the digested protein was further purified by SDS–PAGE. Gel fragments were injected directly into rabbits. The anti-vacuolar processing enzyme (anti-VPEγ) and anti-SAG2 antibodies were as described (Grbic 2003; Rojo et al. 2003). The anti-PBA1, -ATG7, -ATG5, and -ATG8a antibodies were from Doelling et al. (2002), Smalle et al. (2002), and Thompson et al. (2005). Anti-H3A antibodies were supplied by Abcam (Cambridge, MA). Antibodies against the large subunit of spinach RUBISCO were provided by Archie Portis (University of Illinois, Champagne, IL).
Leaf staining and fluorescence confocal microscopy:
Lactophenol blue was used to discriminate between live and dead cells according to Rate et al. (1999). The seventh leaf from individual plants was harvested at various times during the dark treatment and immediately boiled in a lactophenol blue solution [10 ml lactic acid, 10 ml glycerol, 10 ml liquid phenol, 10 ml water, and 10 mg trypan blue (Sigma) for 1 min], cleared in alcoholic lactophenol (2:1 95% ethanol:lactophenol) for 2 min, and incubated in 50% ethanol for 1 day. Leaves were washed in water before imaging on a Leica MZFLIII microscope equipped with an Optronics digital camera.
Fluorescence confocal microscopy of hypocotyl cells expressing either free GFP or GFP-ATG8a was conducted with a Zeiss 510-Meta scanning laser confocal microscope using a 488-nm light excitation (Thompson et al. 2005). Fluorescence was monitored 12–16 hr after treatment with 0.5 μm concanamycin A (Wako Chemicals, Richmond VA) using the BP505–530 (excitation 488 nm, emission 505–530 nm) filter set. Images were processed with LSM510 software and National Institutes of Health ImageJ (http://rsb.info.nih.gov/ij/). The density of fluorescent vesicles within the vacuoles of each genetic background was determined by counting their number within a 20- × 20-μm2 section of the central vacuole from representative cells. The data for wild type represent the average number in a 100-μm2 area (±SE) from three independent experiments that each analyzed images captured from 9 to 36 different cells. The data for the atg7-1, atg5-1, and atg10-1 lines represent the average number in a 100-μm2 area (±SD) from one experiment that analyzed images captured from 12 to 30 different cells.
RESULTS
Isolation of a mutant affecting ATG10:
To more specifically define the functions of ATG12, we searched for mutants affecting the cognate E2 ATG10 (Ichimura et al. 2000). Genomic database searches by BLASTP identified single ATG10 genes in Arabidopsis (ecotype Col-0; At3g07525) and poplar (P. trichocarpa; eugene3.00141226), and two ATG10 genes in rice (OsATG10a, Os04g41990; OsATG10b, Os12g32210) (Figure 1). We also detected several genomic fragments predicted to encode ATG10 in the moss P. patens genome, which likely were derived from the same locus. By analysis of genomic and full-length cDNA sequences, the Arabidopsis ATG10 gene was determined to encode a 225-amino-acid protein with 49 and 61% similarity to its rice and poplar orthologs, respectively. In contrast, the Arabidopsis protein shares only 22, 29, and 35% similarity with its nonplant counterparts in S. cerevisiae, Drosophila, and mice (Figure 1). However, several regions with strong amino acid conservation are apparent among the group, including a block bracketing the presumed active-site cysteine (residue 178 in AtATG10) that forms the thioester intermediate with ATG12 prior to its transfer to ATG5 (Figure 1).
Figure 1.—
Amino acid sequence comparison of the ATG10 protein among eukaryotes. The alignment includes sequences from Arabidopsis (At), rice (Os), poplar (Pt), yeast (Sc), Drosophila (Dm), and mice (Mm). Identical and similar amino acids are shown against solid and shaded backgrounds, respectively. Dots denote gaps. Numbers at the end of each sequence indicate the amino acid length of the protein. The arrowhead marks the active-site cysteine (Cys178 in At) involved in forming the thioester intermediate with ATG12. The bracket locates the T-DNA insertion site in the atg10-1 mutant.
In a screen of the available Arabidopsis T-DNA insertion populations prepared with the Col-0 background, we identified a mutant allele of ATG10 designated atg10-1 in the SIGnAL collection (Figure 2A; Alonso et al. 2003). The mutant was backcrossed three times to the wild-type Col-0 ecotype to eliminate possible extraneous secondary mutations, using kanamycin resistance associated with the T-DNA to track the mutation and then self-fertilized to generate homozygous individuals. Genomic PCR of atg10-1 plants with 5′ and 3′ gene-specific primer pairs alone or in combination with the T-DNA left border primer confirmed disruption of the wild-type ATG10 gene and the presence of the introduced T-DNA (see Figure 7A below). Sequencing the region flanking the T-DNA revealed that it was inserted as a tandem duplication in the fourth exon and simultaneously created a 28-bp deletion in the ATG10 coding region. RT–PCR analysis of homozygous atg10-1 seedlings failed to amplify the full-length ATG10 mRNA (primers 1 and 2) (Figure 2B). Although a slight amount of RT–PCR product encoding the region upstream of the T-DNA was generated from atg10-1 transcripts (primers 1 and 3), amplification of the downstream region was not detected (primers 2 and 4) (Figure 2B). Given that the missing downstream sequence encodes Cys178, it is highly likely that atg10-1 is a functionally null allele.
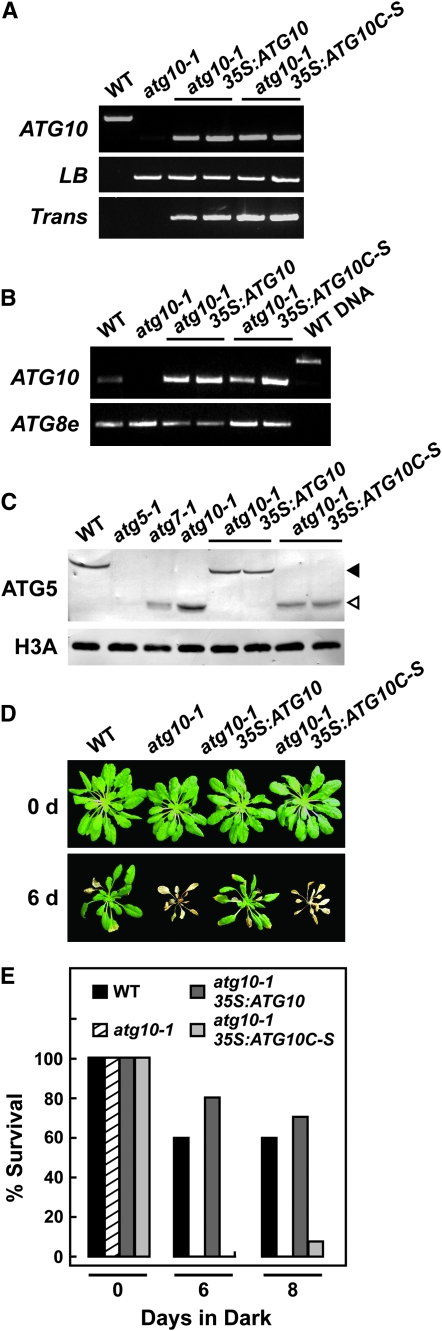
Figure 7.—
Attempted rescue of the atg10-1 phenotype with the 35S:ATG10 and 35S:ATG10C-S transgenes. (A) PCR analysis of the atg10-1 mutant and complemented plants. Total genomic DNA was subjected to PCR using either the ATG10 5′- and 3′-gene-specific primers (ATG10) or the T-DNA left border and ATG10 3′-primers (LB) or primers specific to the transgene (Trans). (B) Semiquantitative RT–PCR of the atg10-1 mutant and complemented plants. Total RNA was subjected to RT followed by 28 cycles of PCR using ATG10 5′- and 3′-gene-specific primers. A primer pair specific for ATG8e was used as an internal control. (C) Immunoblot detection of the ATG12-ATG5 conjugate in atg10-1 mutants complemented with 35S:ATG10 or the 35:ATG10C-S transgene. Tissue was collected from wild-type (WT), atg5-1, atg7-1, atg10-1, atg10-1/35S:ATG10, and atg10-1/35S:ATG10C-S seedlings and subjected to SDS–PAGE followed by immunoblot analysis with anti-ATG5 antibodies. Equal protein loads were confirmed by immunoblot analysis with anti-H3A antibodies. Open and solid arrowheads identify free ATG5 (40 kDa) and the ATG12-ATG5 conjugate (50 kDa), respectively. (D and E) Survival under C-limiting growth conditions induced by extended darkness. Six-week-old plants were grown under SD, exposed to 6 or 8 days of continuous darkness, and then transferred back to SD. (D) Representative plants after a 1-week recovery from darkness. (E) Percentage of plants that survived 6 or 8 days of continuous darkness as determined by resumption of growth after 1 week in SD. Each bar represents the analysis of 10 seedlings.
To demonstrate that ATG10 is the sole E2 that assembles the ATG12-ATG5 conjugate, we performed immunoblot analysis on crude extracts from homozygous atg10-1 seedlings using antibodies against ATG5 (Thompson et al. 2005). As shown in Figure 2C, the 50-kDa presumed ATG12-ATG5 conjugate (solid arrowhead) was detected in the wild-type extracts, the 40-kDa free form of ATG5 (open arrowhead) was detected in atg7-1 extracts, and neither was detected in the atg5-1 extracts. Only the free form of ATG5 was found in the atg10-1 extracts, indicating that ATG10 is essential to form the ATG12-ATG5 adduct. A similar result was recently reported by Suzuki et al. (2005) with anti-ATG12 antibodies and the same atg10-1 allele but confirmation that the ATG5 protein was the target was not confirmed immunologically.
We previously showed, using the atg5-1 and atg7-1 mutants, that defects in the ATG8/12 conjugation pathways substantially increase the levels of ATG8 even when the plants are grown under normal (nonstressed) conditions (Thompson et al. 2005). To see if a similar effect occurs when ATG10 is absent, we examined by immunoblot analysis the abundance of the various ATG8 isoforms, the E1 ATG7, and their cognate E2 ATG3 in whole seedlings grown under LD with N. (The anti-ATG3 antiserum was prepared using recombinant His6-tagged antigen, which was purified by nickel chelate affinity chromatography before injection.) We observed small increases in ATG3 and ATG7 and a large increase in ATG8 in atg10-1 plants as compared to wild type, indicating that all three components were upregulated (Figure 2C). The anti-ATG8a antibodies detected a mixture of ATG8 proteins that likely represent different isoforms from the nine-member ATG8 family with or without modification with PE.
Increased levels of numerous ATG8 species in the atg5-1 and atg7-1 backgrounds are consistent with previous studies (Thompson et al. 2005); however, it appeared that several forms differentially accumulated in the three mutants on the basis of variable banding patterns. In particular, the form in the atg10-1 and atg5-1 seedlings with the fastest SDS–PAGE migration was absent in the wild-type and atg7-1 seedlings (see arrow in Figure 2C). This species may represent the ATG8-PE conjugate, given that the atg7-1 mutation should completely abolish this lipidation reaction while the atg10-1 and the atg5-1 mutations may not, since functional ATG3 and ATG7 are still present and PE remains available. In the wild-type background, the ATG8-PE conjugate should be formed, but may be rapidly consumed during autophagy. SDS–PAGE in the presence of urea has been shown previously to separate free ATG8 from its lipidated derivative (Kirisako et al. 1999; Yoshimoto et al. 2004). However, in our hands, this method failed to conclusively identify the lipidated species, although differences in the abundance of the various ATG8 species were again seen between the mutants and wild type (Figure 2C).
atg10-1 seedlings are hypersensitive to N- and C-limiting growth conditions:
Several Arabidopsis atg mutants, including atg7-1 (Doelling et al. 2002), atg9-1 (Hanaoka et al. 2002), atg4a/b (Yoshimoto et al. 2004), and atg5-1 (Thompson et al. 2005), have been described phenotypically. These mutant plants are slightly smaller and flower later than wild type, have reduced seed set, senesce earlier, and are sensitive to both N starvation and C-limiting growth conditions especially under a SD photoperiod (8 hr light/16 hr dark). Disruption of ATG10 generated a similar set of phenotypes. atg10-1 rosettes developed slightly slower, bolted later, and produced ∼70% less seed as compared to wild-type Col-0 (data not shown).
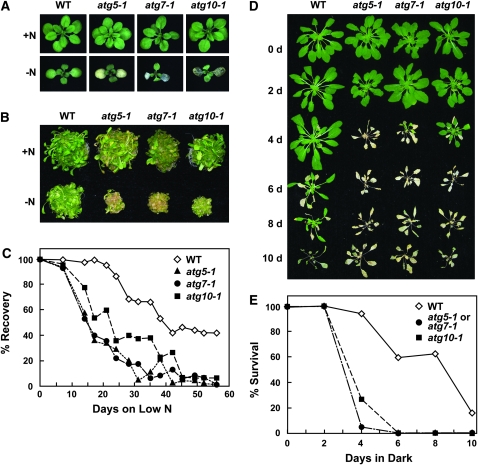
As with other autophagy mutants, atg10-1 seedlings were hypersensitive to N-deficient growth conditions. Wild-type, atg5-1, atg7-1, and atg10-1 seedlings were grown under a LD photoperiod (16 hr light/8 hr dark) or continuous light in medium containing sucrose and N for 1 week and then were transferred to N-deficient medium for increasing lengths of time. As shown in Figure 3, A and B, 2 weeks of such N starvation slowed leaf emergence and expansion and enhanced cotyledon chlorosis of atg10-1 seedlings similar to that previously described for atg7-1 and atg5-1 (Doelling et al. 2002; Thompson et al. 2005). When the plants were exposed to various durations of N starvation and then returned to N-rich medium, the three homozygous mutant populations had dramatically impaired recovery (Figure 3C). Whereas nearly all of the wild-type plants resumed growth after 17 days on N-deficient medium, ∼50% of atg10-1 and 60% of atg5-1 and atg7-1 plants failed to recover. After 45 days on N-deficient medium, almost all of the mutant seedlings died, while >40% of wild-type plants resumed growth.
Figure 3.—
Enhanced sensitivity of atg10-1 plants to N- and C-limiting conditions. Lines include wild-type Col-0 (WT) and homozygous atg5-1, atg7-1, and atg10-1 mutants. (A and B) Representative plants grown for 1 week on N-rich solid (A) or liquid (B) media and transferred to N-rich (+N) or N-deficient (−N) media for 2 weeks. (C) Survival on N-deficient medium. One-week-old seedlings were sown on N-rich solid medium and transferred to N-deficient medium for various lengths of time before transfer back to N-rich medium, all under SD. The graph plots the percentage of plants that resumed growth after exposure to N-deficient medium. Each point represents the analysis of 45 seedlings. (D and E) Survival under C-limiting growth conditions induced by extended darkness. Six-week-old plants were grown under SD, transferred to darkness for various lengths of time, and then transferred back to SD. (D) Representative plants after 1-week recovery from 0-, 2-, 4-, 6-, 8-, or 10-day dark treatments. (E) Percentage of plants that survived increasing days in the dark as determined by resumption of growth after 1 week in SD. Each point represents the analysis of 15 seedlings.
To test the effects of C limitation, plants were grown in SD for 6 weeks to maintain a low level of fixed C, transferred to the dark for various lengths of time, and then allowed to recover in SD for 1 week. As shown in Figure 3, D and E, the atg10-1 homozygous mutants were hypersensitive to C limitation. Similar to atg5-1 and atg7-1, atg10-1 plants were more chlorotic and more wilted than wild type right after the extended dark treatments and showed poor recovery after transfer back to the light. The survival profiles for all three mutants were similar. Whereas a majority of the wild-type plants survived up to 8 days of darkness, most of the mutant plants died following only 4 days (Figure 3, D and E).
Molecular defects of atg10-1 seedlings under C-limiting conditions:
To examine the effects of C limitation at the molecular level, we collected plants immediately after various days of dark treatment and assessed the levels of several autophagy and senescence-related proteins and transcripts. Both wild-type and atg10-1 seedlings progressively lost total protein during extended darkness, with ∼75% of the total remaining in each after 8 days of dark treatment (Figure 4A). However, examination of the protein profiles by SDS–PAGE revealed differences, with the atg10-1 plants losing high-molecular-mass polypeptides and specific lower-molecular-mass species more rapidly than wild type as the dark treatment continued (Figure 4B). Notable were the more rapid declines of the large and small subunits of RUBISCO (∼50 and 13 kDa), further indicative of reduced photosynthetic capacity. By immunoblot analyses, we also confirmed an accelerated loss of the large subunit of RUBISCO and VPEγ in the atg10-1 background (Figure 4C), the latter of which is enriched in the lytic vacuoles of senescing vegetative organs (Kinoshita et al. 1999). By comparison, histone 3A (H3A) disappeared at similar rates in wild-type and atg10-1 seedlings (Figure 4C).
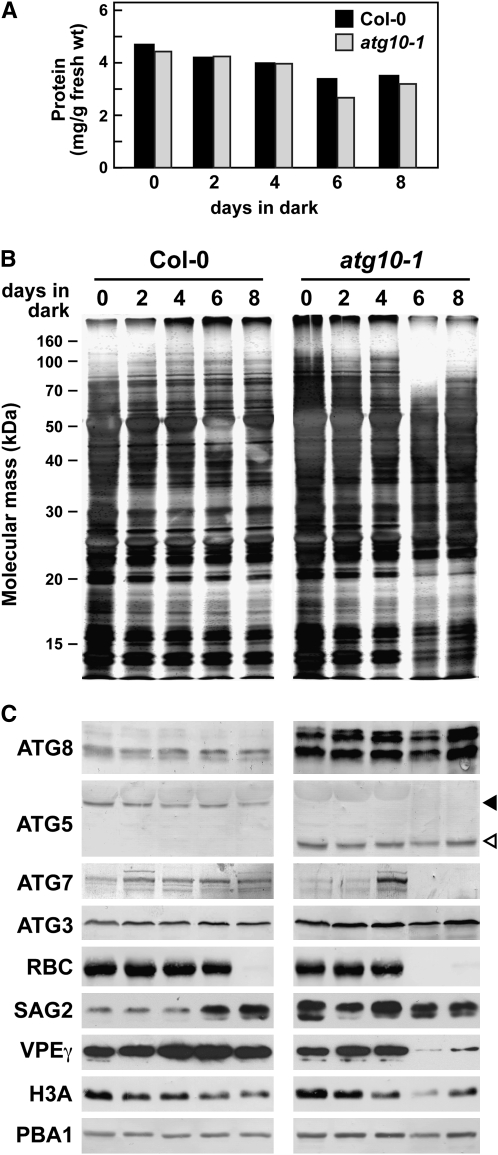
Figure 4.—
Protein profile of atg10-1 plants exposed to C-limiting conditions induced by extended darkness. Tissue was collected from wild-type (wt) and atg10-1 seedlings just after the indicated days of extended darkness (see Figure 3). (A) Quantification of total protein from wild-type and atg10-1 extracts. (B) Profile of total protein separated by SDS–PAGE and stained with silver. (C) Immunoblot analysis with antibodies against ATG8, ATG5, ATG7, ATG3, the large subunit of RUBISCO (RBC), SAG2, VPEγ, H3A, and the β1-subunit of the 26S proteasome (PBA1). Solid and open arrowheads identify the ATG12-ATG5 conjugate (50 kDa) and free ATG5 (40 kDa), respectively. Equivalent amounts of tissue fresh weight were analyzed in each lane.
Not all proteins decreased in abundance during extended darkness. For example, the level of the SENESCENCE-ASSOCIATED GENE (SAG)-2 cysteine protease (Grbic 2003) increased in the wild-type background as the plants remained in the dark for longer periods of time, implying that wild-type plants activated their senescence program (Greenberg 1996; Pennell and Lamb 1997; Figure 4C). In the atg10-1 seedlings, SAG2 levels were high even before dark treatment and remained high during prolonged darkness, suggesting that inactivation of autophagy constitutively induces the senescence program. Likewise, levels of various ATG8 isoforms were constitutively upregulated in the atg10-1 seedlings as compared to wild type (Figure 4C). These high levels were retained throughout the extended darkness, similar to that observed for the atg5-1 and atg7-1 mutants (Thompson et al. 2005; A. R. Phillips, unpublished data). Levels of ATG3, ATG5, and the 26S proteasome subunit PBA1 also remained high during the dark treatments in both the wild-type and atg10-1 plants, which likely reflects an attempt to maintain protein recycling systems during C limitation (Figure 4C). For ATG5, its ATG12 conjugate was retained in wild type, while the free form was retained in the atg10-1 background. The ATG7 protein remained high in wild type but decreased in atg10-1 seedlings during the dark treatment. (We note that an increase in the ATG7 level at day 4 is apparent in Figure 4C for the experiment involving atg10-1 plants, but this effect was not seen in other trials.)
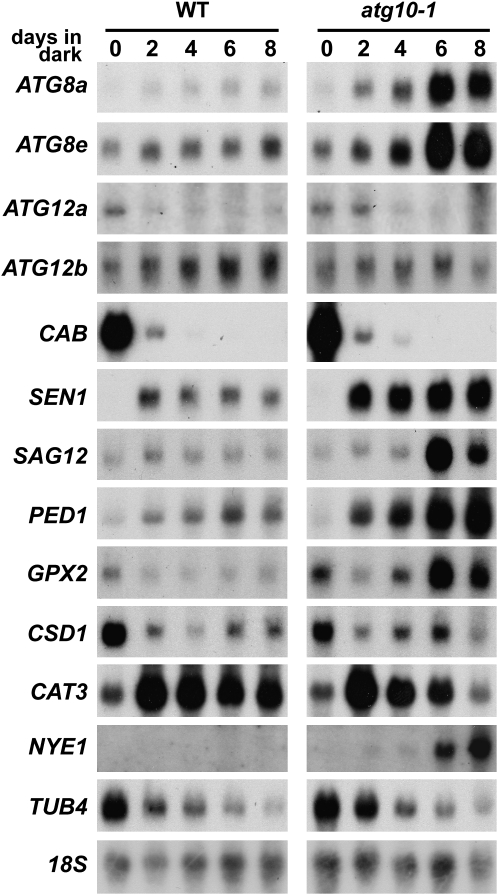
We then exploited RNA gel-blot analyses to investigate changes at the transcript level using 18S rRNA as a marker for equal RNA loading (Figure 5). Similar to previous studies (Weaver and Amasino 2001; Doelling et al. 2002; Thompson et al. 2005), chlorophyll a/b-binding protein (CAB) mRNA levels dropped rapidly following incubation in the dark (between 0 and 2 days) in both wild-type and atg10-1 plants, consistent with the instability of the CAB mRNA in the dark and its light-induced transcription (Figure 5; Giuliano et al. 1988). Levels of β4-tubulin (TUB4) mRNA also decreased in both backgrounds, which is similar to the reported decrease of β9-tubulin mRNA levels during senescence (Swidzinski et al. 2002). The abundance of several ATG8 transcripts were previously shown to increase in response to limited nutrient levels (Contento et al. 2004; Thompson et al. 2005; Rose et al. 2006; Osuna et al. 2007). Here, we found that mRNA levels for two different isoforms, ATG8a and ATG8e, were even more increased by darkness in the atg10-1 mutant plants. By contrast, abundance of the two ATG12 transcripts appears to be differentially regulated by darkness. Whereas the ATG12a mRNA dropped soon after the dark treatment (days 2–4), the ATG12b mRNA slowly increased in abundance over the course of prolonged darkness in both the wild-type and atg10-1 backgrounds (Figure 5).
Figure 5.—
RNA profile of atg10-1 plants exposed to C-limiting conditions induced by extended darkness. Tissue was collected from wild-type (WT) and atg10-1 seedlings just after the indicated days of extended darkness (see Figure 3). Equal amounts of total RNA (10 μg) were subjected to gel-blot analysis using probes for ATG8a, ATG8e, CAB, SEN1, SAG12, PED1, GPX2, CSD1, CAT3, NYE1, and TUB4. Near equal loading of total RNA was confirmed by RNA gel-blot analysis of 18S rRNA (18S) and staining for total rRNA with methylene blue (data not shown).
The abundance of several senescence- and PCD-related transcripts were also consistently increased in atg10-1 seedlings as compared to wild type (Figure 5). These included SENESCENCE (SEN)-1 and SAG12 genes, both markers for dark-induced senescence; the PEROXOSOME DEFECTIVE (PED)-1, which encodes a thiolase involved in fatty acid β-oxidation during germination and senescence; and glutathione peroxidase 2 (GPX2), which responds to oxidative stress (Oh et al. 1996; Hayashi et al. 1998; Weaver et al. 1998; Mullineaux et al. 2000; Swidzinski et al. 2002; van der Graaff et al. 2006). However, not all senescence/PCD mRNAs were affected by the atg10-1 mutation. Transcripts from the Cu/Zn superoxide dismutase 1 (CSD1) and the CAT3 catalase genes, which have previously been associated with senescence and oxidative stress (Swidzinski et al. 2002; Contento et al. 2004), were not upregulated in the atg10-1 background. Whereas the CSD1 mRNA decreased soon after dark treatment in both backgrounds, the CAT3 mRNA first increased at the beginning of darkness and then decreased, with the drop even more rapid in the atg10-1 background. [It should be noted that CAT3 expression is coordinately regulated by the circadian clock, the rhythm of which may cease after several days in the dark (Zhang et al. 2007).] These results, combined with previous studies (Contento et al. 2004; Liu et al. 2005; Thompson et al. 2005; Xiong et al. 2007), suggest that autophagy is connected with some, but not all, aspects of the senescence and PCD programs.
Presumably, the more rapid chlorosis of plants missing ATG7, ATG5, and ATG10 (Doelling et al. 2002; Thompson et al. 2005; this report) in the dark is caused, in part, by increased chlorophyll degradation. This breakdown requires pheophorbide a oxygenase, which is regulated in turn by the NON-YELLOWING (NYE)-1 nuclear-encoded chloroplast protein (Ren et al. 2007). Levels of NYE1 positively correlate with chlorophyll turnover, with the abundance of the NYE1 transcript increasing markedly during senescence, suggesting that the NYE1 protein is a major regulator of chlorophyll turnover. Here, we found that the amount of the NYE1 mRNA is dramatically affected by the atg10-1 mutation (Figure 5). Whereas the NYE1 transcript was barely detectable in wild-type seedlings even after prolonged darkness, it rose substantially following extended darkness in the atg10-1 seedlings, coinciding with increased chlorosis of the leaves.
Disruptions of ATG10, ATG5, and ATG7 enhance programmed cell death:
As shown in Figures 3–5, the survival of atg10-1 seedlings is severely compromised when exposed to extended darkness and many senescence and PCD-associated factors are upregulated over the course of the treatment. It is also likely that these plants are impaired in normal cell death pathways that involve autophagy (autophagic PCD: Bursch et al. 2004; van Doorn and Woltering 2005) due to their inability to conjugate ATG12 to ATG5. To further investigate how atg mutants undergo PCD in the absence of ATG-dependent autophagy, we tested for the involvement of other PCD types, such as apoptosis (thought not to occur in plants), nonlysosomal PCD, and necrosis, which may work exclusively or together (mixed-type PCD: Bursch et al. 2004; van Doorn and Woltering 2005).
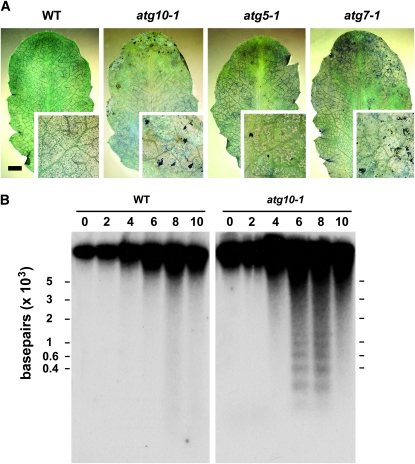
Necrosis was observed by staining wild-type, atg7-1, atg5-1, and atg10-1 leaves immediately after extended darkness with lactophenol blue, a dye excluded from live cells (Rate et al. 1999). After 6 days of extended darkness, which is sufficient to kill mutant but not wild-type plants during their SD light recovery, we saw little evidence for massive cell death in each plant line (Figure 6A). However, some punctate staining was observed in the leaf margins of the mutants, a site where senescence usually begins. Consequently, it appears that extensive necrosis had not yet occurred in the atg mutants even though PCD and senescence components were upregulated (Figure 5).
Figure 6.—
DNA fragmentation is accelerated in atg10-1 plants upon exposure to C-limiting conditions induced by extended darkness. Individual leaves were collected from wild-type (WT) and homozygous atg10-1, atg5-1, and atg7-1 plants after increasing days in continuous darkness. (A) Representative seventh rosette leaves following 6 days in the dark stained with lactophenol blue to identify dead cells. Bar, 2 mm. The insets are three-fold magnifications. (B) Twenty micrograms of total genomic DNA were separated by gel electrophoresis using 1.5% agar and subjected to DNA gel-blot analysis using an 18S rRNA antisense probe.
We next examined the extent of DNA fragmentation during darkness by isolating chromosomal DNA from the wild-type and mutant plants just after various days of dark treatment. The appearance of high-molecular-weight (HMW) DNA fragments (>10 kbp) is a characteristic of autophagic PCD, while their appearance, along with low-molecular-weight (LMW) oligonucleosome-sized DNA pieces (∼200-bp laddering), is characteristic of apoptotic PCD (Pennell and Lamb 1997; Bursch et al. 2004). As can be seen in Figure 6B, some HMW DNA fragmentation, as observed by smearing of the DNA near the top of the gel, was apparent in the wild-type plants after 8 and 10 days in the dark. This fragmentation was accentuated in the atg10-1 plants with the mutant plants also accumulating a ladder of LMW DNA fragments differing by ∼200 bp, the expected size of oligonucleosomal DNA (Brown et al. 1993). Coupled with the upregulation of senescence-related proteins and transcripts, including the cysteine proteases SAG2 and SAG12, we propose that the atg mutants enhance PCD by activating additional cell death pathways (mixed-type PCD).
Complementation of atg10-1:
To verify that the loss of the ATG12-ATG5 conjugate and the N- and C-limiting phenotypes were directly caused by the loss of ATG10, we attempted to rescue the defects by introducing a transgene encoding the full-length ATG10 protein in the homozygous atg10-1 background. The transgene was modified to encode the ATG10 protein with an N-terminal HA epitope tag and expressed under the control of the CaMV 35S promoter. To confirm that the E2 activity of ATG10 was necessary, we also attempted to rescue the atg10-1 plants with an active-site mutant in which Cys178 was replaced with a serine (ATG10C-S). Several groups have demonstrated using yeast and mouse orthologs that such an ATG10 mutant protein can still form a stable ester adduct with ATG12 but cannot transfer the tag to its target ATG5 (Shintani et al. 1999; Mizushima et al. 2002; Nemoto et al. 2003). The presence of the atg10-1 mutation and the 35S:ATG10 and 35S:ATG10C-S transgenes and the absence of the wild-type ATG10 locus were tracked by genomic PCR in progeny from independent transformants (Figure 7A). As can be seen in Figure 7B, homozygous T2 atg10-1 seedlings carrying either the 35S:ATG10 or 35S:ATG10C-S transgene expressed comparable transcript levels as determined by semiquantitative RT–PCR.
Introduction of the functional 35S:ATG10 transgene in turn fully rescued formation of the ATG12-ATG5 conjugate and the atg10-1 mutant phenotypes. While atg10-1 plants contained only the free form of ATG5 at 40 kDa, only the conjugate at 50 kDa was detected in the atg10-1/35S:ATG10 plants similar to that observed in wild type (Figure 7C). The 35S:ATG10C-S transgene, in contrast, failed to restore formation of the 50-kDa species, confirming that the E2 activity of ATG10 depends on Cys178. Likewise, a functional ATG10 transgene was required to rescue the atg10-1 phenotypic defects. As can be seen in Figure 7, D and E, the atg10-1 plants harboring the 35S:ATG10 but not the 35S:ATG10C-S transgene were restored in their ability to survive extended darkness to the level seen with wild-type plants.
Accumulation of ATG8-labeled vesicles is dependent on ATG10 and ATG5:
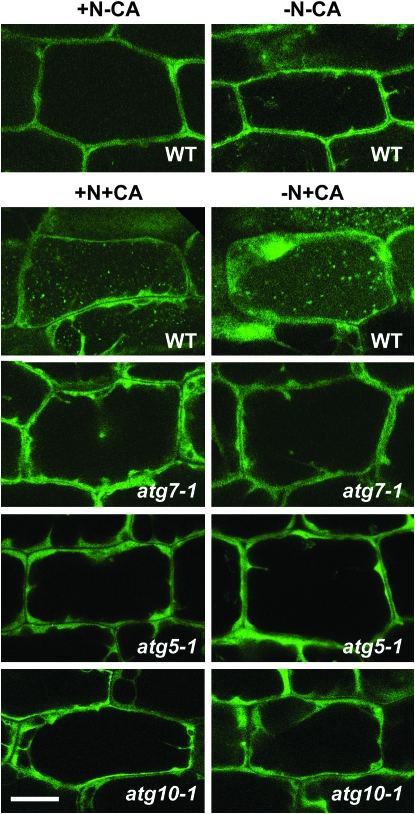
Previous studies with yeast, mammalian, and plant cells demonstrated that GFP-ATG8 fusions are excellent markers for visualizing autophagosomes and autophagic bodies in vivo (Suzuki et al. 2001; Mizushima et al. 2004; Yoshimoto et al. 2004; Contento et al. 2005; Slavikova et al. 2005; Thompson et al. 2005). For example, from analysis of Arabidopsis atg7-1 and atg4a-1 atg4b-1 plants stably expressing GFP-ATG8a (Yoshimoto et al. 2004; Thompson et al. 2005), it was previously shown that inhibition of ATG8 conjugation specifically or a block affecting both ATG8 and ATG12 conjugation abrogate autophagic body accumulation. To examine the importance of the ATG12-ATG5 conjugate specifically, we introduced the GFP-ATG8a fusion transgene into atg5-1 and atg10-1 plants. Seedlings expressing the fusion protein were grown in liquid medium with or without N, treated with concanamycin A, and then observed by fluorescence confocal microscopy for autophagic body accumulation. By raising vacuolar pH, concanamycin A helps stabilize autophagic bodies by slowing their breakdown by luminal hydrolases (Drose et al. 1993).
Similar to previous studies (Yoshimoto et al. 2004; Thompson et al. 2005), small punctuate structures likely representing autophagic bodies appeared in the vacuolar lumen of wild-type hypocotyl cells exposed to concanamycin A with their numbers increased ∼1.6-fold by N starvation (Figure 8; Table 1). As with the atg7-1 mutant (Thompson et al. 2005), the atg5-1 and atg10-1 mutants failed to accumulate these GFP-ATG8a-labeled vesicles, even after treatment in low-N medium containing concanamycin A (Figure 8; Table 1; data not shown). Instead, the GFP signal remained diffuse in the cytoplasm, similar to that observed for free GFP (data not shown). These results support the assertion that the GFP-ATG8a-labeled structures are bona fide autophagic bodies and show that their assembly and delivery to the vacuole requires formation of the ATG12-ATG5 conjugate.
Figure 8.—
Visualization of autophagic vesicles in wild-type and various atg mutants using a GFP-ATG8 fusion. Eight-day-old wild-type, atg10-1, atg5-1, and atg7-1 seedlings expressing a GFP-ATG8a fusion were grown in N-rich liquid medium for 6 days, transferred to N-rich (+N) or N-deficient (−N) liquid media for ∼1.5 days, and then incubated for an additional 12–16 hr with 0.5 μm concanamycin A (+CA) or an equal volume of DMSO (−CA). Hypocotyls were visualized by fluorescence confocal microscopy of GFP. Bar, 50 μm.
TABLE 1.
Autophagic body accumulation in the vacuoles of wild-type and atg mutant seedlings
| +N−CA | −N−CA | +N+CA | −N+CA | |
|---|---|---|---|---|
| Wild-type Col-0a | 0.09 ± 0.01 | 0.05 ± 0.01 | 2.46 ± 0.07 | 3.88 ± 0.33 |
| atg10-1b | 0.08 ± 0.13 | 0.11 ± 0.14 | 0.03 ± 0.08 | 0.11 ± 0.18 |
| atg5-1b | 0.08 ± 0.13 | 0.08 ± 0.15 | 0.03 ± 0.08 | 0.08 ± 0.12 |
| atg7-1b | 0.07 ± 0.14 | 0.10 ± 0.13 | 0.13 ± 0.13 | 0.11 ± 0.13 |
Each line expressed GFP-ATG8 under the control of the CaMV 35S promoter. Seedlings were grown in N-deficient liquid medium for 2 days and then treated with 0.5 μm concanamycin A (CA) for 12–16 hr prior to confocal fluorescence microscopy of the central vacuole.
Values are the average number of vacuolar vesicles per 100 μm2 of 9–32 cells each for three independent experiments (±SE).
Values are the average of number of vacuolar vesicles per 100 μm2 of 12–36 cells each for one experiment (±SD).
DISCUSSION
Prior genetic analyses of Arabidopsis ATG genes, including ATG7, which is required for the combined action of ATG8 and ATG12, have revealed several important roles for autophagy in plants. These include assisting in the remobilization of nutrients under N starvation and fixed C-limiting conditions and during senescence, removing oxidized proteins from the cytoplasm, and limiting the spread of necrosis during HR (Doelling et al. 2002; Hanaoka et al. 2002; Liu et al. 2005; Thompson et al. 2005; Xiong et al. 2007). In addition, cytological and molecular studies support the involvement of autophagy in PCD and HR (Liu et al. 2005; van Doorn and Woltering 2005; Bozhkov and Jansson 2007).
Here, we demonstrate the importance of the ATG12 conjugation pathway specifically through the reverse genetic analysis of the ATG10 gene encoding the E2 responsible for the ligation of this polypeptide tag. A T-DNA insertion mutant preventing ATG10 accumulation fails to form the ATG12-ATG5 conjugate, demonstrating that this E2 directs ATG12 ligation. Phenotypically, the atg10-1 mutant plants resemble atg5-1 and atg7-1 plants previously characterized (Doelling et al. 2002; Thompson et al. 2005). Under standard growth conditions, the atg10-1 plants germinate and develop normally, but in SD they grow slower, flower later, senesce earlier, and produce less seed. More importantly, they display enhanced chlorosis and die more rapidly during N starvation and when exposed to extended darkness that substantially reduces fixed C availability. These phenotypic defects can be rescued by transgenic expression of wild-type ATG10 but not by transgenic expression of an active-site mutant (Cys178-Ser), demonstrating that the enzymatic activity of ATG10 is required. The phenotypic similarity between the atg5-1 and atg10-1 mutants coupled with the detection of a single conjugate in wild-type plant extracts using either anti-ATG12 (Suzuki et al. 2005) or anti-ATG5 (this report) antibodies strongly suggests that ATG5 is the main, if not, sole target of ATG12 in plants.
The phenotypic similarity of atg10-1 and atg5-1 plants also supports expectations based on data from yeasts (Ohsumi 2001) that ATG5 functions optimally when conjugated to ATG12. However, for both N- and C-limiting growth conditions, we reproducibly observed a slight decrease in sensitivity for the atg10-1 plants as compared to atg7-1 and atg5-1. Since all three mutants appear to represent null alleles (Doelling et al. 2002: Thompson et al. 2005; this report), mutant strength is an unlikely explanation for this small difference. Other possibilities include (i) the action of free ATG5 by itself, (ii) the noncovalent association of ATG12 with ATG5, (iii) the direct transfer of activated ATG12 from ATG7 to ATG5 without an E2 intermediate, and/or (iv) the formation of the ATG12-ATG5 conjugate by another mechanism (e.g., using the E2 ATG3). However, the absence of an immunodetectable ATG12-ATG5 conjugate in the atg10-1 plants may preclude the last two scenarios (Figure 2C). We also note that almost all ATG5 in wild-type plants is present in the conjugated form regardless of the age of the plants or nutritional status. This constitutive conjugation combined with only small changes in ATG12a/b transcript abundance during C limitation imply that the formation of the ATG12-ATG5 conjugate, while necessary for autophagy, is not the trigger for this recycling pathway during senescence or under nutrient-limiting conditions. The ATG8-PE conjugate could be the trigger, given the substantial upregulation of various ATG8 mRNAs and proteins during C starvation of wild-type plants. The abundance of various ATG8 isoforms increases further in several atg mutant backgrounds (Thompson et al. 2005; this report). Since both the atg5-1 and atg10-1 mutations increase the steady-state levels of ATG8 transcripts, part of this increase likely reflects increased protein synthesis. However, it is also possible that atg defects indirectly raise the levels of ATG8 proteins by decreasing their breakdown during autophagy.
In yeast, formation of the autophagosomes and autophagic bodies requires both the ATG8-PE and ATG12-ATG5 conjugates (Suzuki et al. 2004, 2007). A similar scenario likely exists in plants. Prior studies using Arabidopsis mutants blocked in ATG8 processing (Yoshimoto et al. 2004) or expression of the ATG7 E1 (Thompson et al. 2005) together confirmed the importance of ATG8 conjugation. Here, we extend these observations to the ATG12-ATG5 conjugate specifically. Like atg7-1 plants (Thompson et al. 2005), atg5-1 and atg10-1 plants fail to accumulate GFP-ATG8-labeled vesicles in the vacuolar lumen upon treatment with concanamycin A under either N-rich or N-deficient conditions. The results with atg5-1 and atg10-1 plants in particular further support the notion that while formation of ATG8-PE conjugates may not be blocked by removal of the ATG12-ATG5 conjugate, their incorporation into autophagosomes/autophagic bodies is inhibited. However, we cannot discount the remote possibility that autophagic vesicles can be assembled without the ATG8 decoration.
One phenotypic conundrum for the collection of atg mutants is that, compared to wild-type plants, they senesce earlier and die more rapidly under N- and C-limiting environments despite their inability to direct autophagic breakdown. One probable scenario is that defects in autophagy under starvation conditions irreversibly trigger other stress-activated PCD pathways that then compromise cell viability more quickly than normal. Possibilities include apoptosis [although true apoptosis involving phagocytosis does not occur in plants (van Doorn and Woltering 2005)], nonlysosomal PCD, necrosis, and/or an ATG-independent autophagic system. In support, we detected several hallmarks of various cell death pathways when atg mutants were exposed to darkness, including the fragmentation of genomic DNA into HMW and LMW species, loss of turgor in leaf tissue, and death of the shoot meristem. This was accompanied by more rapid leaf chlorosis and a faster loss of specific proteins from most, if not, all cellular compartments, including chloroplasts, mitochondria, and cytoplasm, which suggests a severe disruption of cellular homeostasis (Thompson et al. 2005; this report).
Prior to their more rapid death, the atg mutant plants dramatically increase mRNA abundance for a suite of genes often associated with senescence-induced PCD, including SEN1, SAG12, PED1, GPX2, and NYE1, which implies that one or more PCD pathways are activated by extended darkness that in turn accelerate cell death (Swidzinski et al. 2002). This upregulation could reflect a direct connection between autophagy and stress pathways or an indirect result of atg mutants attempting to cope with acute stress. However, not all factors associated with PCD and senescence were increased by autophagic defects. Most notably, levels of the VPEγ protein were not retained despite the proposed role of this caspase-like protease in activating hydrolyases needed during senescence and HR (Hatsugai et al. 2004).
With respect to the mechanism(s) of accelerated death, the absence of large-scale patches of dead cells just after the atg plants exited darkness would preclude necrotic mechanisms. However, it remains possible that necrosis occurs only after returning the plants to full light when oxidative damage induced by light would become challenging. The subsequent failure to upregulate CSD1 and CAT3 expression, both of which help scavenge oxidative species, could then accentuate the problem. One likely contributor to the enhanced chlorosis is premature activation of an autophagy-independent chlorophyll catabolic pathway involving NYE1, which is dramatically upregulated when atg plants encounter prolonged darkness. Coupled with the more rapid loss of RUBISCO, we propose that nonautophagic chloroplast breakdown represents an important source of nutrients when autophagy is compromised.
It remains unclear what signaling pathway(s) trigger PCD in the atg mutants and what are the molecular consequences of this upregulation. One likely effector could be reactive oxygen species (ROS). Xiong et al. (2007) reported that autophagy is enhanced when plants encounter severe oxidative stress, which then works to eliminate oxidized proteins and potentially dampen cytosolic ROS accumulation. In addition, Liu et al. (2005) endorsed a role for autophagy in protecting cells from damage by reactive oxygen intermediates during HR. One direct target of ROS could be ATG4. Recent studies with the human autophagy system showed that this protease is activated by starvation-induced oxidation, thus providing a mechanism to directly regulate ATG8 availability (Scherz-Shouval et al. 2007). Considering that ROS activate PCD, it is possible that levels of ROS are abnormally high in the absence of autophagy and are further increased during dark-induced senescence to accelerate PCD. Clearly, a more complete picture of genes affected by the inhibition of autophagy, coupled with genetic and biochemical analyses of ROS accumulation in various atg backgrounds, is now needed to test this connection.
Acknowledgments
We thank Archie Portis, Natasha Raikhel, and Sara Patterson for the supply of the RUBISCO, VPEγ, and SAG2 antibodies, respectively, and Joseph Walker, Taijoon Chung, and Scott Saracco for technical assistance and helpful discussions. This project was supported by a grant from the National Research Initiative of the U. S. Department of Agriculture Cooperative State Research, Education and Extension Service (2005-35301-15768) to R.D.V.; a Thailand Predoctoral Fellowship to A.S.; and Wisconsin Alumni Research Foundation and Louis and Elsa Thomsen Wisconsin Distinguished Predoctoral Fellowships to A.R.P.
References
- Alonso, J. M., A. N. Stepanova, T. J. Leisse, C. J. Kim, H. M. Chen et al., 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Balk, J., and C. J. Leaver, 2001. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham, D. C., 2007. Plant autophagy: more than a starvation response. Curr. Opin. Plant Biol. 10 587–593. [DOI] [PubMed] [Google Scholar]
- Bozhkov, P., and C. Jansson, 2007. Autophagy and cell-death proteases in plants: two wheels of a funeral cart. Autophagy 3 136–138. [DOI] [PubMed] [Google Scholar]
- Brown, D. G., X. M. Sun and G. M. Cohen, 1993. Dexamethasone-induced apoptosis involves cleavage of DNA to large fragments prior to internucleosomal fragmentation. J. Biol. Chem. 268 3037–3039. [PubMed] [Google Scholar]
- Bursch, W., 2001. The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ. 8 569–581. [DOI] [PubMed] [Google Scholar]
- Bursch, W., A. Ellinger, C. Gerner and R. Schulte-Hermann, 2004. Autophagocytosis and programmed cell death, pp. 287–303 in Autophagy, edited by D. J. Klionsky. Eurekah.com/Landes Bioscience, Georgetown, TX.
- Clough, S. J., and A. F. Bent, 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Contento, A. L., S. J. Kim and D. C. Bassham, 2004. Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol. 135 2330–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento, A. L., Y. Xiong and D. C. Bassham, 2005. Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 42 598–608. [DOI] [PubMed] [Google Scholar]
- Doelling, J. H., J. M. Walker, E. M. Friedman, A. R. Thompson and R. D. Vierstra, 2002. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277 33105–33114. [DOI] [PubMed] [Google Scholar]
- Drose, S., K. U. Bindseil, E. J. Bowmama, A. Siebers, A. Zeeck et al., 1993. Inhibitory effect of modified bafilomycins and concanamycins and P- and V-type adenosinetriphosphatases. Biochemistry 32 3902–3906. [DOI] [PubMed] [Google Scholar]
- Earley, K. W., J. R. Haag, O. Pontes, K. Opper, T. Juehne et al., 2006. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. [DOI] [PubMed] [Google Scholar]
- Fujiki, Y., K. Yoshimoto and Y. Ohsumi, 2007. An Arabidopsis homolog of yeast ATG6/VPS30 is essential for pollen germination. Plant Physiol. 143 1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano, G., N. E. Hoffman, K. Ko, P. A. Scolnik and A. R. Cashmore, 1988. A light-entrained circadian clock controls transcription of several plant genes. EMBO J. 7 3635–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic, V., 2003. SAG2 and SAG12 protein expression in senescing Arabidopsis plants. Physiol. Plant. 119 263–269. [Google Scholar]
- Greenberg, J. T., 1996. Programmed cell death: a way of life for plants. Proc. Natl. Acad. Sci. USA 93 12094–12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka, H., T. Noda, Y. Shirano, T. Kato, H. Hayashi et al., 2002. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, T. M., K. A. Morano, S. V. Scott and D. J. Klionsky, 1995. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai, N., M. Kuroyanagi, K. Yamada, T. Meshi, S. Tsuda et al., 2004. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305 855–858. [DOI] [PubMed] [Google Scholar]
- Hayashi, M., K. Toriyama, M. Kondo and M. Nishimura, 1998. 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell 10 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura, Y., T. Kirisako, T. Takao, Y. Satomi, Y. Shimonishi et al., 2000. A ubiquitin-like system mediates protein lipidation. Nature 408 488–492. [DOI] [PubMed] [Google Scholar]
- Juhasz, G., B. Erdi, M. Sass, and T. P. Neufeld, 2007. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 21 3061–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., K. Yamada, N. Hiraiwa, M. Kondo, M. Nishimura et al., 1999. Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J. 19 43–53. [DOI] [PubMed] [Google Scholar]
- Kirisako, T., M. Baba, N. Ishihara, K. Miyazawa, M. Ohsumi et al., 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako, T., Y. Ichimura, H. Okada, Y. Kabeya, N. Mizushima et al., 2000. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., 2007. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 11 931–937. [DOI] [PubMed] [Google Scholar]
- Kuma, A., N. Mizushima, N. Ishihara and Y. Ohsumi, 2002. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 277 18619–18625. [DOI] [PubMed] [Google Scholar]
- Levine, B., and D. J. Klionsky, 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6 463–477. [DOI] [PubMed] [Google Scholar]
- Liu, Y., M. Schiff, K. Czymmek, Z. Talloczy, B. Levine et al., 2005. Autophagy regulates programmed cell death during the plant innate immune response. Cell 121 567–577. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., T. Noda, T. Yoshimori, Y. Tanaka, T. Ishii et al., 1998. A protein conjugation system essential for autophagy. Nature 395 395–398. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., T. Noda and Y. Ohsumi, 1999. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 18 3888–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N., T. Yoshimori and Y. Ohsumi, 2002. Mouse Apg10 as an Apg12-conjugating enzyme: analysis by the conjugation-mediated yeast two-hybrid method. FEBS Lett. 532 450–454. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., A. Yamamoto, M. Matsui, T. Yoshimori and Y. Ohsumi, 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux, P., L. Ball, C. Escobar, B. Karpinska, G. Creissen et al., 2000. Are diverse signalling pathways integrated in the regulation of Arabidopsis antioxidant defence gene expression in response to excess excitation energy? Philos. Trans. R. Soc. Lond. B Biol. Sci. 355 1531–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto, T., I. Tanida, E. Tanida-Miyake, N. Minematsu-Ikeguchi, M. Yokota et al., 2003. The mouse APG10 homologue, an E2-like enzyme for Apg12p conjugation, facilitates MAP-LC3 modification. J. Biol. Chem. 278 39517–39526. [DOI] [PubMed] [Google Scholar]
- Oh, S. A., S. Y. Lee, I. K. Chung, C. H. Lee and H. G. Nam, 1996. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 30 739–754. [DOI] [PubMed] [Google Scholar]
- Ohsumi, Y., 2001. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2 211–216. [DOI] [PubMed] [Google Scholar]
- Osuna, D., B. Usadel, R. Morcuende, Y. Gibon, O. E. Blasing et al., 2007. Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 49 463–491. [DOI] [PubMed] [Google Scholar]
- Pennell, R. I., and C. Lamb, 1997. Programmed cell death in plants. Plant Cell 9 1157–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, G., Z. Ma, L. Zhang, S. Xing, X. Hou et al., 2007. Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 17 249–263. [DOI] [PubMed] [Google Scholar]
- Rate, D. N., J. V. Cuenca, G. R. Bowman, D. S. Guttman and J. T. Greenberg, 1999. The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, G., K. An, Y. Liao, X. Zhou, Y. Cao et al., 2007. Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol. 144 1429–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, E., J. Zouhar, C. Carter, V. Kovaleva and N. V. Raikhel, 2003. A unique mechanism for protein processing and degradation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100 7389–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, T. L., L. Bonneau, C. Der, D. Marty-Mazars and F. Marty, 2006. Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol. Cell. 98 53–67. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval, R., E. Shvets, E. Fass, H. Shorer, L. Gil et al., 2007. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26 1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seay, M., S. Patel and S. P. Dinesh-Kumar, 2006. Autophagy and plant innate immunity. Cell. Microbiol. 8 899–906. [DOI] [PubMed] [Google Scholar]
- Shintani, T., N. Mizushima, Y. Ogawa, A. Matsuura, T. Noda et al., 1999. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 18 5234–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavikova, S., G. Shy, Y. Yao, R. Glozman, H. Levanony et al., 2005. The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56 2839–2849. [DOI] [PubMed] [Google Scholar]
- Smalle, J., and R. D. Vierstra, 2004. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590. [DOI] [PubMed] [Google Scholar]
- Smalle, J., J. Kurepa, P. Yang, E. Babiychuk, S. Kushnir et al., 2002. Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin, M., H. Zheng, M. T. Morita, C. Saito, E. Avila et al., 2003. The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15 2885–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., T. Kirisako, Y. Kamada, N. Mizushima, T. Noda et al., 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20 5971–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., T. Noda and Y. Ohsumi, 2004. Interrelationships among ATG proteins during autophagy in Saccharomyces cerevisiae. Yeast 21 1057–1065. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., Y. Kubota, T. Sekito and Y. Ohsumi, 2007. Hierarchy of ATG proteins in pre-autophagosomal structure organization. Genes Cells 12 209–218. [DOI] [PubMed] [Google Scholar]
- Suzuki, N. N., K. Yoshimoto, Y. Fujioka, Y. Ohsumi and F. Inagaki, 2005. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy 1 119–126. [DOI] [PubMed] [Google Scholar]
- Swidzinski, J. A., L. J. Sweetlove and C. J. Leaver, 2002. A custom microarray analysis of gene expression during programmed cell death in Arabidopsis thaliana. Plant J. 30 431–446. [DOI] [PubMed] [Google Scholar]
- Thompson, A. R., and R. D. Vierstra, 2005. Autophagic recycling: lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 8 165–173. [DOI] [PubMed] [Google Scholar]
- Thompson, A. R., J. H. Doelling, A. Suttangkakul and R. D. Vierstra, 2005. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm, M., R. Egner, B. Koch, M. Schlumpberger, M. Straub et al., 1994. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349 275–280. [DOI] [PubMed] [Google Scholar]
- Tsukada, M., and Y. Ohsumi, 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333 169–174. [DOI] [PubMed] [Google Scholar]
- Ueno, T., I. Tanida and E. Kominami, 2004. Autophagy and neuromuscular diseases, pp. 264–286 in Autophagy, edited by D. J. Klionsky. Eurekah.com/Landes Bioscience, Georgetown, TX.
- van der Graaff, E., R. Schwacke, A. Schneider, M. Desimone, U. I. Flugge et al., 2006. Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141 776–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn, W. G., and E. J. Woltering, 2005. Many ways to exit? Cell death categories in plants. Trends Plant Sci. 10 117–122. [DOI] [PubMed] [Google Scholar]
- Weaver, L. M., and R. M. Amasino, 2001. Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol. 127 876–886. [PMC free article] [PubMed] [Google Scholar]
- Weaver, L. M., S. Gan, B. Quirino and R. M. Amasino, 1998. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37 455–469. [DOI] [PubMed] [Google Scholar]
- Xiong, Y., A. L. Contento and D. C. Bassham, 2005. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42 535–546. [DOI] [PubMed] [Google Scholar]
- Xiong, Y., A. L. Contento, P. Q. Nguyen and D. C. Bassham, 2007. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, K., H. Hanaoka, S. Sato, T. Kato, S. Tabata et al., 2004. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Y. Chen, Z. Y. Wang, Z. Chen, H. Gu et al., 2007. Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J. 51 512–525. [DOI] [PubMed] [Google Scholar]