Abstract
Scans of the human genome have identified many loci as potential targets of recent selection, but exploration of these candidates is required to verify the accuracy of genomewide scans and clarify the importance of adaptive evolution in recent human history. We present analyses of one such candidate, enamelin, whose protein product operates in tooth enamel formation in 100 individuals from 10 populations. Evidence of a recent selective sweep at this locus confirms the signal of selection found by genomewide scans. Patterns of polymorphism in enamelin correspond with population-level differences in tooth enamel thickness, and selection on enamel thickness may drive adaptive enamelin evolution in human populations. We characterize a high-frequency nonsynonymous derived allele in non-African populations. The polymorphism occurs in codon 648, resulting in a nonconservative change from threonine to isoleucine, suggesting that the allele may affect enamelin function. Sequences of exons from 12 primate species show evidence of positive selection on enamelin. In primates, it has been documented that enamel thickness correlates with diet. Our work shows that bursts of adaptive enamelin evolution occur on primate lineages with inferred dietary changes. We hypothesize that among primate species the evolved differences in tooth enamel thickness are correlated with the adaptive evolution of enamelin.
WHOLE-GENOME scans make it possible to systematically identify regions of the human genome that have been subject to recent selection and are a first step in understanding the role of adaptive evolution in creating human phenotypic diversity (Sabeti et al. 2002; Clark et al. 2003; Akey et al. 2004; Bustamante et al. 2005; Carlson et al. 2005; Kelley et al. 2006; Nielsen et al. 2005; Voight et al. 2006; Wang et al. 2006). Scans for adaptive evolution have primarily used publicly available data, which can contain ascertainment and collection biases (Clark et al. 2005), and candidates identified by genome scans inevitably require verification. Here we present detailed analyses of one such candidate, enamelin (ENAM), a gene whose protein product is involved in tooth enamel formation. Population-level patterns of single nucleotide polymorphism (SNP) variation from the Perlegen data set identify enamelin as a target of recent selection among human populations (Hinds et al. 2005; Kelley et al. 2006). The enamelin locus is an outlier in the genomewide empirical distribution of Tajima's D values in both the Han Chinese (P = 0.006) and the European–American (P = 0.017) populations (supplemental Figure S1 at http://www.genetics.org/supplemental/) (Kelley et al. 2006). The signal of selection appears to be specific to enamelin; the genes on either side of enamelin do not show evidence of selection. To better understand the current and historical selective pressures that create divergence of enamelin, we sequenced this region in 10 human populations and 12 primate species.
enamelin is an essential gene in tooth enamel formation, encoding a 1142-amino-acid secretory protein with a 39-amino-acid signal peptide that is cleaved prior to secretion (Hu and Yamakoshi 2003; Hu et al. 2005). The secreted protein is proteolytically processed into several smaller functional products located in specific layers of the developing and mature enamel (Hu and Yamakoshi 2003). enamelin peptides compose ∼5% of the enamel matrix and are thought to influence the formation and elongation of enamel crystallites during tooth development (Hu et al. 2000, 2005; Paine et al. 2001; Mardh et al. 2002). Mutations in enamelin create variability in tooth enamel thickness and are responsible for heritable enamel development disorders (Amelogenesis imperfecta). The clinical phenotype is underdeveloped, thin, pitted (hypoplastic) enamel and mutations are responsible for autosomal dominant (Gutierrez et al. 2007; Pavlic et al. 2007) as well as recessive A. imperfecta (Rajpar et al. 2001; Kida et al. 2002; Mardh et al. 2002; Hart et al. 2003; Hu and Yamakoshi 2003; Kim et al. 2005). Dominant mutations occur mainly in the N-terminal region of the protein. Several C-terminal enamelin mutations responsible for A. imperfecta have observed dosage-dependent effects, and heterozygous individuals have an intermediate phenotype, which is not severe enough to be diagnosed as A. imperfecta (Ozdemir et al. 2005). The intermediate hypoplastic phenotype, possibly due to haplo-insuffiency of specific cleavage products, suggests that two functional enamelin copies are required for forming enamel (Ozdemir et al. 2005). The observed disease phenotypes and immunohistochemical analyses of the pattern of enamelin localization in the enamel matrix suggest that functional enamelin is involved in the control of enamel thickness and is necessary for proper enamel thickness formation (Hu et al. 1997; Hu and Yamakoshi 2003; Kim et al. 2005). Natural enamelin variation could therefore influence tooth enamel thickness.
Enamel thickness and tooth morphology often reflect dental adaptation to diet (Shellis et al. 1998), and among primate species, tooth enamel thickness differences are correlated with dietary differences (Fleagle 1988; Schwartz 2000). Within living primates, humans have the thickest enamel, which is thought to be due to a dietary shift from an ancestral diet that was composed mainly of leaves to harder objects found in savanna habitats (Gantt and Rafter 1998; Shellis et al. 1998). Tooth adaptations are specific to the physical properties of the diet (Fleagle 1988). Selection on enamel thickness is thought to be a consequence of mechanical (wear and crushing) and/or morphological (defined vs. reduced relief cusps) optimization to diets containing plant material or hard objects (Shellis et al. 1998). It has been suggested that moderate or low selective pressures over short evolutionary time periods could lead to measurable changes in enamel thickness (Hlusko et al. 2004). Enamel thickness is heritable and the genetic component explains observed population-level variation in baboons (Hlusko et al. 2004). In addition, population-specific differences in enamel thickness exist among human populations; specifically, African Americans have significantly thicker tooth enamel than European Americans (Harris et al. 2001).
Currently, the mechanisms underlying the observed differences in tooth enamel thickness between individuals and populations are unknown. In this study, we confirm that enamelin shows signs of positive selection as predicted by genomewide scans of adaptive evolution by sequencing enamelin in 100 human individuals from 10 populations. We also characterize the evolutionary history of enamelin in 12 extant primates. We correlate adaptive changes in enamelin with diet shifts along the primate lineage; to this point, there have been few studies correlating molecular evolutionary changes with ecological phenomena, with notable exceptions relating to diet such as RNase (Zhang 2003, 2006) and lysozyme (Messier and Stewart 1997).
MATERIALS AND METHODS
DNA samples:
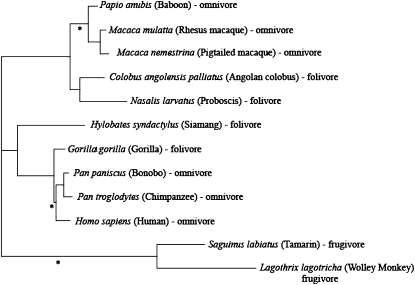
We sequenced enamelin exons in 11 nonhuman primate species that span a range of taxonomic relationships and dietary preferences (Figure 1). Dietary information was gathered from a variety of sources (Milton and May 1976; Richard 1985; Sussman 1987; Woodland Park Zoo, http://www.zoo.org). There is considerable evidence that chimpanzees hunt and eat meat and thus are classified as omnivores (Goodall 1986; Stanford 1998; Pickford 2005; Teelen 2007). PAUP* was used to infer ancestral dietary states on the basis of the known primate phylogeny (Boffelli et al. 2003; Swofford 2003). The following primates from the Coriell Cell Repositories were sequenced (Coriell ID numbers and base pairs surveyed are in parentheses): chimpanzee Pan troglodytes (NG06939, 3241), bonobo Pan paniscus (NG05253, 3116), gorilla Gorilla gorilla (NG05251, 3374), pigtailed macaque Macaca nemestrina (NG08452, 3060), rhesus monkey Macaca mulatta (NG07109, 3119), woolly monkey Lagothrix lagotricha (NG05356, 3056), and red-chested mustached tamarin Saguinus labiatus (NG05308, 2464). Four additional primate samples were obtained from Integrated Primate Biomaterial and Information Resource: baboon Papio anubis (PR00036, 2816), colobus Colobus angolensis palliatus (PR00099, 2908), proboscis Nasalis larvatus (PR00679, 3314), and siamang Hylobates syndactylus (PR00721, 3235). Sequences are deposited in GenBank under accession nos. EU482096–EU482107.
Figure 1.—
Unrooted species tree (Boffelli et al. 2003) for the primates sequenced at the enamelin locus. Corresponding dietary preferences are noted by each species name. Branches with inferred diet changes are indicated with an asterisk.
To survey human polymorphism in enamelin (ENAM), we used DNAs from 100 individuals composing the following panels from Coriell Cell Repositories Human Variation Collections (numbers in parentheses indicate individual accession numbers and number of individuals sequenced from the corresponding panel): Northern European HD01 (NA17002-17010, 9), Russian HD23 (NA13820, 13838, 13849, 13852, 13876-77, 13911-14, 10), Africans north of the Sahara HD11 (NA17378-17384, 7), Africans south of the Sahara HD12 (NA17341-17349, 9), Mbuti tribe from northeast Zaire (NA10492-10496, 5), Middle East HD05 (Version 1) (NA17041-17050, 10), Japanese HD07 (NA17051-17060, 10), Aboriginal tribe from Taiwan HD24 (NA13597-13606, 10), South America HD17 (NA17301-17310, 10), and Caucasian HD50CAU (NA17231-17250, 20). Sequences are deposited in GenBank under reference numbers EU482096–EU482107. To increase our statistical power, we combined our populations on the basis of the Rosenberg et al. (2002) analysis of population structure. The resulting five populations are European, north Saharan Africans, south Saharan Africans, Asians, and South Americans (Table 1).
TABLE 1.
Summary statistics of enamelin population differentiation
| Population | No. of chromosomes | Segregating sites | Singletons | πa | θa | Tajima's D |
|---|---|---|---|---|---|---|
| European | 98 | 13 | 5 | 0.6 | 2.6 | −2.1014** |
| Northern European | 18 | 1 | 1 | 0.1 | 0.3 | — |
| North American | 40 | 10 | 3 | 0.9 | 2.5 | −1.9269* |
| Russian | 20 | 7 | 7 | 0.7 | 2.1 | −2.1214* |
| Middle Eastern | 20 | 3 | 3 | 0.3 | 0.9 | −1.7233 |
| North Saharan Africa | 14 | 3 | 1 | 0.7 | 1.0 | −0.886 |
| South Saharan Africa | 28 | 23 | 9 | 6.2 | 6.2 | 0.01262 |
| Sub-Saharan Africans | 18 | 20 | 8 | 6.3 | 6.1 | 0.1549 |
| Mbuti tribe | 10 | 16 | 6 | 6.2 | 5.9 | 0.2272 |
| Asian | 40 | 2 | 1 | 0.2 | 0.5 | −1.29613 |
| Japanese | 20 | 2 | 1 | 0.3 | 0.6 | — |
| Abriginal Taiwanese | 20 | 0 | 0 | — | — | — |
| South American | 20 | 3 | 2 | 0.4 | 0.9 | −1.44071 |
| African | 42 | 24 | 10 | 5.7 | 5.9 | −0.05804 |
| Non-African | 158 | 15 | 5 | 0.5 | 2.8 | −2.1846** |
*P < 0.05; **P < 0.01.
×10−4.
PCR and sequencing:
enamelin is located on chromosome 4: 71859495–71877517, reference sequence NM_031889 (UCSC Genome Browser March 2006 Assembly). Primers were designed to amplify the exons, 3′-UTR and 5′-UTR from the known human sequence using PRIMER3 v 0.2 (Rozen and Skaletsky 2000). The enamelin locus spans 18.8 kbp; we sequenced 9521 bp. The sequences were concatenated for analysis. The primers and conditions for PCR and sequencing are available upon request. PCR products were diluted five times, cycle sequenced using BigDye v. 3.1, ethanol precipitated, and analyzed on an ABI 3100 automated sequencer.
Statistical analysis:
Primate enamelin sequences were manually assembled using Sequencher 4.2 (Gene Codes, Ann Arbor, MI). Multiple overlapping reads were aligned with the human reference sequence from the UCSC genome browser. Consensus sequences were exported, aligned using ClustalW (Higgins et al. 1996), and checked visually in Se-Al v.2.0 (Rambaut 1996). Maximum-likelihood-based methods (Yang et al. 2000) were used to detect the presence of adaptive evolution on the amino acid sequence of enamelin. These tests were implemented using CODEML in the PAML package (v. 3.15). A species tree (Boffelli et al. 2003) was used for PAML analyses.
Three likelihood-ratio tests were used to examine the data for evidence of positive selection, specifically by looking for codons and/or lineages with dN/dS ratios significantly >1. The tests are classified by the way in which the data are used to construct the likelihood ratios: sites, branch, and branch-site tests (Yang 1998; Yang et al. 2000). Significance for the three tests was determined by comparing the likelihood of a null model, without selection, to the likelihood of a selection model. The test statistic, the negative of twice the log-likelihood difference (−2Δl), is compared to the χ2 distribution to determine significance, and the degrees of freedom equal the number of parameter differences between the null and selection models. The log-likelihood difference asymptotically follows a χ2 distribution, which is conservative (Anisimova et al. 2001).
For the sites test, we used two null models, the first (M1) with two dN/dS estimates, one to be <1 and the other equal to 1, and the second (M7) with dN/dS values estimated between 0 and 1 from a beta distribution. Both corresponding selection models included an additional class of sites with an unrestricted dN/dS estimated from the data (M2 and M8). If the dN/dS ratio for the additional class of sites is estimated to be >1 we compare the M8 likelihood to that in M8a, the model in which the unrestricted dN/dS is set to 1 (Swanson et al. 2003). For the M8a vs. M8 comparison, the appropriate test statistic is unknown; however, our test statistic meets the 1% critical value for two different distributions (Wong et al. 2004). We also implemented a test for variation between sites; the test compares a model with no variation between sites (M0) to a model that allows variation between sites (M3). For the branch test, the null model, a phylogenetic tree without selection, a single dN/dS for the entire tree (M0) is compared to a selection model that allows the dN/dS ratio to vary along each branch (free ratio). Finally, the branch-site test was conducted by defining branches as foreground and background lineages; the foreground lineages are those on which an a priori hypothesis of adaptive evolution exists (Zhang et al. 2005)—in our case one based upon diet switch. The selection model allows dN/dS ratios to fall into one of three site classes: dN/dS between 0 and 1, dN/dS = 1, and dN/dS freely estimated for the foreground lineages only. The null model is one in which the foreground lineages freely estimated dN/dS ratio is set to 1. Selection is inferred if the freely estimated dN/dS ratio is >1 and the likelihood of the model is significantly greater than that of the null model. A Bayes empirical Bayes approach was used to calculate posterior probabilities that sites with dN/dS > 1 were subject to positive selection (Yang et al. 2005). The proportions of sites with the corresponding dN/dS (ω) values are labeled p1, p2, p3 or pfg (foreground), and pbg (background), depending on the test. The tests were conducted without removing sites with ambiguous data. We checked for convergence by repeating the analyses with various initial dN/dS values.
Sequence data from the human panel were automatically base called, assembled, and scanned for SNPs using Phred, Phrap, and polyPhred (Nickerson et al. 1997; Ewing and Green 1998; Ewing et al. 1998) and visually inspected using Consed (Gordon et al. 1998). The finished sequence was exported and haplotypes were inferred using PHASE (Stephens et al. 2001). Estimation of population genetic parameters and tests of neutrality were performed using DnaSP v. 4.0 (Rozas and Rozas 1999).
We used the statistical test Tajima's D (Tajima 1989) to quantify population genetic variation to identify deviations from expectations under the neutral theory of evolution (Kimura 1968). Tajima's D compares nucleotide polymorphism (θ) and nucleotide diversity (π), two estimates of 4Neμ (Ne is the effective population size, μ is the mutation rate), to identify deviations from neutrality. The test compares the relative abundance of low- and high-frequency polymorphisms. A selective sweep is predicted to eliminate nucleotide variation in the region, and as generations progress, mutations occur randomly throughout the swept region, leading to an excess of alleles that are found in very few individuals in a sample (rare alleles). A negative value of Tajima's D indicates an excess of rare alleles in the sample population, which can be caused by either recovery from a population bottleneck or a recent selective sweep. We used two methods to determine the significance of Tajima's D values. First, we used a standard coalescent with constant population size, as implemented in DnaSP (Rozas and Rozas 1999). We also generated simulations using the cosi simulation package, which has been calibrated to human sequence variation with populations similar to our samples (Schaffner et al. 2005). We generated 10,000 simulations using the parameters specified by Schaffner et al. (2005), except those specific to the enamelin region: recombination rate of 0.6 cm/Mb, length of 9500 bp, and 20 mutation sites. The recombination rate for the region is the sex-averaged recombination rate estimated by deCODE Genetics (Kong et al. 2002).
We used several programs to predict whether amino acid substitutions may have functional consequences: PolyPhen, SIFT, and PANTHER (Ramensky et al. 2002; Ng and Henikoff 2003; Brunham et al. 2005). The programs use sequence variability to predict how nucleotide substitutions affect protein function in a manner similar to a position-specific scoring matrix.
RESULTS
Human variation:
We examined nucleotide variation at the enamelin locus to confirm that direct sequence data support the signature of selection observed in genome scans and that evidence for positive selection at this locus is not a result of ascertainment and/or genotyping biases. Analysis of nucleotide variation reveals that a majority of the 32 SNPs identified at the enamelin locus are absent or at low frequency in all populations except the south Saharan Africans (Table 1 and Figure 2). Low levels of nucleotide variation in the populations outside of south Saharan Africa are consistent with a selective sweep at this locus. Tajima's D values calculated using the resequenced data from the European population are not consistent with neutrality, indicating a recent selective sweep (P < 0.01, Table 1). We used both a standard equilibrium model and simulations calibrated to human demographic history (Schaffner et al. 2005) to test for significance. The Asian and South American populations show evidence of a selective sweep; however, while the sample sizes are large (n = 40 and 20, respectively), lack of variation in these populations precludes statistical significance.
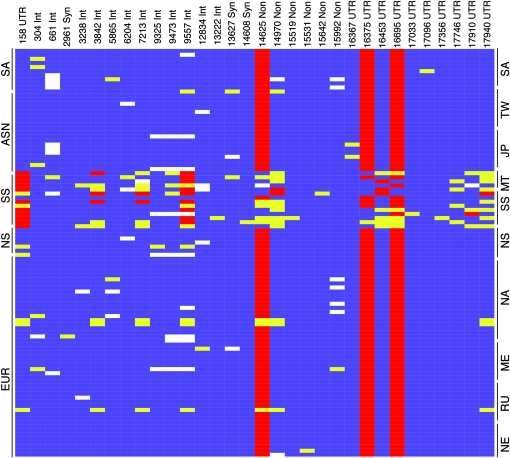
Figure 2.—
Visual genotype of enamelin locus from targeted resequencing. Individuals are represented in rows by population. For combined populations: EUR, European; NS, north Saharan African; SS, sub-Saharan African; ASN, Asian; SA, South American. For individual populations, NE, Northern European; RU, Russian; ME, Middle Eastern; NA, North American; NS, Africans north of Sahara; SS, Africans south of the Sahara; MT, Mbuti tribe; JP, Japanese; TW, Taiwanese; SA, South American. Each column represents a polymorphic site and is classified on the basis of location in the gene sequence, numbered from the first base 5′-UTR. Each rectangle indicates the genotypic state for the corresponding individual and SNP location; blue indicates homozygous for the ancestral allele, yellow indicates heterozygous, red indicates homozygous for the derived allele, and white indicates missing data. All populations, except the south Saharan Africans, have low nucleotide variation.
Nine of the 32 SNPs are located in enamelin exons; of these, 6 are nonsynonymous and cause an amino acid change. Only 2 of the 6 nonsynonymous SNPs were identified in more than one individual (Figure 2). For the SNP notation, the first letter is the ancestral allele, the number corresponds to the location from the first base in the 5′-UTR, and the second letter is the derived allele. The ancestral allele was determined by comparing the SNP to the orthologous position in the chimpanzee and rhesus genomes. The nonsynonymous SNP C14625T is at high frequency for the derived allele in the populations outside of south Saharan Africa (0.965). The derived allele is found at a frequency of 0.269 in the combined south Saharan African population (6/18 alleles in the sub-Saharan African panel and 1/8 in the Mbuti tribe). We evaluated the Fay and Wu's H (denoted Hasc) test statistic calculated by Voight et al. (2006) for the enamelin SNPs genotyped in the International HapMap Project (International HapMap Consortium 2005). Hasc is calculated for 50-marker windows and compared to a genomic empirical distribution. The enamelin SNPs in Northern and Western Europeans with calculated Hasc are all significantly negative (P = 0.05); additionally, all SNPs in the Asian combined population have a negative Hasc, and one meets the 5% significance cutoff. The two high-frequency nonsynonymous SNPs are located in prevalent enamelin cleavage products: SNP C14625T is located at amino acid 648, which is in the 25-kDa cleavage product, and SNP G14970A is at amino acid 763, which is found in the 34-kDa cleavage product.
The nonsynonymous high-frequency derived polymorphism C14625T results in a change from a polar residue to a nonpolar residue, specifically threonine to isoleucine. All primates sequenced in this study and species with enamelin sequence available online (including pig, cow, rat, and mouse) have threonine at the corresponding position. Evidence for lack of variation at the residue and the observed polarity-changing polymorphism suggest that the nonconservative change to an isoleucine may affect enamel formation, and therefore thickness. Methods used to predict the effects of nonsynonymous polymorphisms infer that the SNP in question may affect protein function; however, there is no three-dimensional structure for enamelin, which limits the parameters for prediction programs (Ramensky et al. 2002; Ng and Henikoff 2003; Brunham et al. 2005). The other nonsynonymous polymorphism G14970A results in a change from arginine to glutamine. While the change is not conservative, the amino acid change is predicted to have no functional consequence (Ramensky et al. 2002; Ng and Henikoff 2003; Brunham et al. 2005).
Variation between primates:
To understand the evolutionary history of enamelin, we sequenced enamelin exons in 12 primates and analyzed nucleotide changes for evidence of positive selection. Primates were chosen on the basis of their taxonomic relationships and dietary preferences (Figure 1). The phylogeny of these primates is well established (Purvis 1995; Boffelli et al. 2003). The dN/dS value for the data set, averaged over all sites, is 0.6799. We found that there are 18 nonsynonymous and 9 synonymous changes between human and chimpanzee. Averaging dN/dS across all sites is not a powerful method for detecting positive selection. While the average dN/dS for enamelin is high compared to other genes, we used more powerful methods to identify positive selection (see materials and methods). From the sites analysis, we conclude that enamelin has been subject to positive selection with 4% of the sites having dN/dS = 6.8 (P < 0.001) (Table 2).
TABLE 2.
Model comparisons for primate sequences
| Models compared | −2ΔlnL | Parameter estimates under selection model | Positively selected sites |
|---|---|---|---|
| Neutral (M1) vs. selection (M2) | 35.04** | p1 = 0.96, ω1 = 0.51 | 64, 68, 102, 110, 139, 146, 190, 257, 278, 337, 341, 354, 361, 426, 431, 525, 623, 644, 665, 672, 743, 846, 1056 |
| (d.f. = 2) | p2 = 0, ω2 = 1.00 | ||
| p3 = 0.04, ω3 = 6.76 | |||
| One-ratio (M0) vs. discrete (M3) | 63.22** | p1 = 0.33, ω1 = 0.51 | 64, 68, 102, 110, 139, 146, 190, 257, 278, 337, 341, 354, 361, 426, 431, 525, 587, 623, 639, 640, 644, 665, 672, 743, 760, 846, 1056 |
| (d.f. = 4) | p2 = 0.63, ω2 = 0.51 | ||
| p3 = 0.04, ω3 = 6.76 | |||
| β (M7) vs. β and ω (M8) | 35.06** | p0 = 0.96, p = 99.0, q = 93.2 | 64, 68, 102, 110, 139, 146, 190, 257, 278, 337, 341, 354, 361, 426, 431, 525, 587, 623, 639, 640, 644, 665, 672, 743, 760, 846, 1056 |
| (d.f. = 2) | (p1 = 0.044) | ||
| ω = 6.80 | |||
| β and ω = 1 (M8a) vs. β and ω (M8) | 17.52** | Same as above | Same as above |
| (d.f. = 1) | |||
| One-ratio (M0) vs. free-ratio | 25.55 | NA | NAa |
| (d.f. = 20) | |||
| Branch site neutral vs. selection | 43.17** | p0 = 0.395, ωbg = 0.030, ωfg = 0.030 | 102, 257, 672, 1056 |
| (d.f. =1) | p1 = 0.601 ωbg = 1.00, ωfg = 1.00 | ||
| p2a = 0.001, ωbg = 0.030, ωfg = 999 | |||
| p2b = 0.002, ωbg = 1.00, ωfg = 999 |
Boldface type indicates sites that have a prediction of P > 90% based on Bayes empirical Bayes. Italic type indicates sites that have a prediction of P > 80%. Underlining indicates sites that have a prediction of P > 70%. **Significant P < 0.001.
The test is not significant in our data set; therefore, we do not display the values from the free ratio.
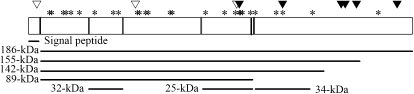
The locations of the amino acids under selection were predicted using a Bayes empirical Bayes approach (Yang et al. 2005) (Table 2). None of these sites correspond to the high-frequency nonsynonymous SNPs described above. The amino acids under selection are all located in the secreted protein; none are located in the 39-amino-acid signal sequence (Figure 3). The 32-kDa cleavage product is the most prevalent form of enamelin in the tooth enamel matrix; the 32-kDa products accumulate throughout the enamel matrix with higher abundance than other cleavage products. Three of the amino acids under selection are located in this 32-kDa cleavage product. In general, the majority of the sites under selection are concentrated in the N-terminal half of the protein. There is a concentration of charged changes between amino acids 354 and 639; only two of the eight changes in the region do not alter charge. One of the sites (665) corresponds to the C-terminal cleavage site for the 89-kDa cleavage product (which is later cleaved into the 32-kDa product) and the 25-kDa product. The locations of the amino acids under selection indicate regions that are potentially important in enamelin function. Positive selection on enamelin amino acids may be a result of shifting dietary pressures, leading to enamel thickness differences among primates.
Figure 3.—
Gene structure for enamelin. Predicted cleavage sites are denoted with vertical bars and the corresponding cleavage products are drawn below. Synonymous SNPs (▿) and nonsynonymous SNPs (▾) identified in the human polymorphism are indicated above the gene diagram. Additionally, we have indicated the sites predicted to be under positive selection by codeml with an asterisk.
Although primate diets are complex, diet categorization allows for phylogenetic and selection analyses. The primates used in our analysis can be divided into three major dietary groups: folivore, frugivore, and omnivore (see Figure 1 for classifications). Researchers using primate diet to understand species characteristics use similar classifications (Milton and May 1976; Richard 1985; Sussman 1987). The ancestral dietary states were inferred by parsimony (Swofford 2003). Two of the inferred diet shifts are from folivory to omnivory. The third change occurs on the lineage leading to New World monkeys, along which the diet changed from folivory to frugivory. Tooth enamel thickness and diet are correlated (Fleagle 1988; Schwartz 2000). Primate RNases and lysozymes have been shown to track with diet change at the molecular level (Messier and Stewart 1997; Zhang 2003, 2006). Messier and Stewart (1997) detected bursts of adaptive evolution corresponding to the evolution of foregut fermentation in colobine monkeys. Both lysozymes and RNases experienced episodes of adaptive evolution associated with diet specialization. Thus, we predicted that dietary shifts along the primate phylogeny might be correlated with bursts of adaptive evolution. Using the branch-site method in PAML, we tested the hypothesis that the three branches with inferred diet changes also experienced a burst of adaptive evolution. Branches with a diet shift will be referred to as foreground lineages. The null model, in which the foreground and background lineages have dN/dS between 0 and 1, was compared to the selection model that allows the foreground lineages to have dN/dS > 1 (see materials and methods). We concluded that positive selection acted on the branches, coinciding with changes in primate diet (−2ΔlnL = 44, d.f. = 1, P < 0.0001). The sites identified are a subset of those identified among primates in the sites model analysis. Our findings are consistent with previous studies documenting molecular change corresponding to dietary pressures.
DISCUSSION
Candidate loci identified by genome scans require detailed investigation to confirm their role in creating human phenotypic variability. Here we follow up on one such region, enamelin, a candidate identified by a previous genome scan (Kelley et al. 2006). Our analyses of enamelin sequence from 10 human populations and 12 primate species find evidence of positive selection on multiple timescales.
There is often a poor correspondence between the candidate regions identified by methods of detecting selection in the human genome. Imperfect overlap among the findings of these different methods is expected, because each method draws inference from different aspects of the data, performs best at different timescales, and has different susceptibility to demographic effects and other potential confounds (Biswas and Akey 2006). In European and Asian populations, enamelin has experienced a near-complete selective sweep. Little nucleotide polymorphism remains in these populations; therefore long-range haplotype tests, such as iHS (Voight et al. 2006), EHH (Sabeti et al. 2002), and LDD (Wang et al. 2006), cannot be used to detect selection on this region because these tests can be applied only in cases of incomplete selective sweeps or balancing selection (Kimura et al. 2007). A long-range approach recently designed specifically to evaluate evidence of selection after complete selective sweeps (MHH) finds evidence of a selective sweep at enamelin in the European (P < 0.036) and Asian (P < 0.035) HapMap Project populations (Kimura et al. 2007), confirming results from our population analysis. Additionally, while Tajima's D does not distinguish between population demographic history and positive selection, we simulated data using parameters that have been designed to replicate human data specifically adjusted to the multiple demographic events that have occurred in human population history (Schaffner et al. 2005), and enamelin remains an outlier.
enamelin has a high number of nonsynonymous changes between humans and chimpanzee. The average number of nonsynonymous changes per base pair between chimpanzee and humans is 0.002578 (Nielsen et al. 2005); for enamelin, the value is 0.007648, which is in the top 8% of the empirical distribution from Bustamante et al. (2005). In a scan for selection comparing divergence to polymorphism levels between humans and chimpanzees, enamelin is in the tail of the empirical distribution (P = 0.05166) (Bustamante et al. 2005), further evidence that selection is acting on enamelin in the human genome.
Identifying outliers using a genomewide polymorphism scan could have led to false positives due to demographics. For example, a population bottleneck can result in a loss of genetic variability similar to that observed after a selective sweep. However, dN/dS ratios, based on amino acid changes between species, look at selection on the species level and are not affected by population demographics. A previous genomewide scan using dN/dS ratios found that genes under positive selection also had an excess of high-frequency derived nonsynonymous SNPs, supporting their observation of positive selection(Nielsen et al. 2005); we see this phenomenon in enamelin. Population-specific selection considered in conjunction with the dN/dS analyses provides ample evidence of positive selection on the enamelin locus.
Diet shifts are associated with burst of adaptive evolution in enamelin. Evidence for adaptive evolution in enamelin in the human populations and population differences in enamel thickness suggest that enamelin may be evolving adaptively in response to diet changes. We have identified a nonsynonymous polymorphism in the enamelin locus that occurs at different frequencies in African and non-African populations. The presence of derived alleles at high frequency is consistent with positive selection (Fay and Wu 2000); there is significant evidence for selection in the European and Asian populations on the basis of Hasc (Voight et al. 2006). Additionally, it is expected that new mutations, especially amino-acid-altering ones, will be either neutral or slightly deleterious (Fay et al. 2001). Therefore, the presence of a nonsynonymous derived allele (SNP C14625T) at a high frequency in a population is uncommon, suggesting that positive selection has favored an increase in the allele frequency. The presence of the SNP in the 25-kDa cleavage product, as well as the high frequency of the derived allele in the European populations, suggests that C14625T may be functional. In addition to nonsynonymous allele frequency differences, people with African and non-African recent ancestry have significant differences in tooth enamel thickness (Harris et al. 2001). Specifically, African Americans have significantly thicker tooth enamel than European Americans. The observation that enamelin has been subject to positive selection in recent human history suggests that the identified polymorphism may be correlated to observed differences in tooth enamel thickness. These data suggest that tooth enamel in non-African populations may be adaptively thinning to account for changing diets in the out-of-Africa expansion. Our hypothesis is that the nonsynonymous polymorphism alters the molecular function of enamelin, resulting in a change in enamel thickness. enamelin provides an opportunity to look for an association between genotype and enamel thickness phenotype in human populations and to understand the role of adaptive evolution in creating human phenotypic diversity; the C14625T nonsynonymous SNP is one of the few molecular traits that has a good chance of showing a phenotypic effect on enamel thickness. Understanding the basic biology of tooth enamel formation gives us a basis for understanding the more complex interactions and processes occurring during enamel formation and could be important for the bioengineering of dental tissues.
Acknowledgments
We thank Josh Akey, Cindy Desmarais, David Hamm, Jeff Jensen, Al Kelley, Jeff Kidd, Sridhar Kudaravalli, Michael Nachman, Jonathan Pritchard, Stevan Springer, and Kayley Turkheimer. J.L.K. was supported by National Science Foundation (NSF) grant DIG 0709660 and a Sigma Xi Grant-in-Aid of Research and W.J.S. was supported by NSF grant DEB-0716761 and National Institutes of Health grants HD042563 and HD054631.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession nos. EU482096–EU482107.
References
- Akey, J. M., M. A. Eberle, M. J. Rieder, C. S. Carlson, M. D. Shriver et al., 2004. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova, M., J. P. Bielawski and Z. Yang, 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol. Biol. Evol. 18 1585–1592. [DOI] [PubMed] [Google Scholar]
- Biswas, S., and J. M. Akey, 2006. Genomic insights into positive selection. Trends Genet. 22 437–446. [DOI] [PubMed] [Google Scholar]
- Boffelli, D., J. McAuliffe, D. Ovcharenko, K. D. Lewis, I. Ovcharenko et al., 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299 1391–1394. [DOI] [PubMed] [Google Scholar]
- Brunham, L. R., R. R. Singaraja, T. D. Pape, A. Kejariwal, P. D. Thomas et al., 2005. Accurate prediction of the functional significance of single nucleotide polymorphisms and mutations in the ABCA1 gene. PLoS Genet. 1 e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante, C. D., A. Fledel-Alon, S. Williamson, R. Nielsen, M. T. Hubisz et al., 2005. Natural selection on protein-coding genes in the human genome. Nature 437 1153–1157. [DOI] [PubMed] [Google Scholar]
- Carlson, C. S., D. J. Thomas, M. A. Eberle, J. E. Swanson, R. J. Livingston et al., 2005. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 15 1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., S. Glanowski, R. Nielsen, P. Thomas, A. Kejariwal et al., 2003. Positive selection in the human genome inferred from human-chimp-mouse orthologous gene alignments. Cold Spring Harb. Symp. Quant. Biol. 68 471–477. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., M. J. Hubisz, C. D. Bustamante, S. H. Williamson and R. Nielsen, 2005. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 15 1496–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8 186–194. [PubMed] [Google Scholar]
- Ewing, B., L. Hillier, M. C. Wendl and P. Green, 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8 175–185. [DOI] [PubMed] [Google Scholar]
- Fay, J. C., and C. I. Wu, 2000. Hitchhiking under positive Darwinian selection. Genetics 155 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay, J. C., G. J. Wyckoff and C.-I. Wu, 2001. Positive and negative selection on the human genome. Genetics 158 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleagle, J. G., 1988. Primate Adaptation and Evolution. Academic Press, San Diego.
- Gantt, D. G., and J. A. Rafter, 1998. Evolutionary and functional significance of hominoid tooth enamel. Connect. Tissue Res. 39 195–206. [DOI] [PubMed] [Google Scholar]
- Goodall, J., 1986. The Chimpanzees of Gombe: Patterns of Behavior. Harvard University Press, Cambridge, MA.
- Gordon, D., C. Abajian and P. Green, 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8 195–202. [DOI] [PubMed] [Google Scholar]
- Gutierrez, S. J., M. Chaves, D. M. Torres and I. Briceno, 2007. Identification of a novel mutation in the enamalin gene in a family with autosomal-dominant amelogenesis imperfecta. Arch. Oral Biol. 52 503–506. [DOI] [PubMed] [Google Scholar]
- Harris, E. F., J. D. Hicks and B. D. Barcroft, 2001. Tissue contributions to sex and race: differences in tooth crown size of deciduous molars. Am. J. Phys. Anthropol. 115 223–237. [DOI] [PubMed] [Google Scholar]
- Hart, T. C., P. S. Hart, M. C. Gorry, M. D. Michalec, O. H. Ryu et al., 2003. Novel ENAM mutation responsible for autosomal recessive amelogenesis imperfecta and localised enamel defects. J. Med. Genet. 40 900–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, D. G., J. D. Thompson and T. J. Gibson, 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266 383–402. [DOI] [PubMed] [Google Scholar]
- Hinds, D. A., L. L. Stuve, G. B. Nilsen, E. Halperin, E. Eskin et al., 2005. Whole-genome patterns of common DNA variation in three human populations. Science 307 1072–1079. [DOI] [PubMed] [Google Scholar]
- Hlusko, L. J., G. Suwa, R. T. Kono and M. C. Mahaney, 2004. Genetics and the evolution of primate enamel thickness: a baboon model. Am. J. Phys. Anthropol. 124 223–233. [DOI] [PubMed] [Google Scholar]
- Hu, C. C., M. Fukae, T. Uchida, Q. Qian, C. H. Zhang et al., 1997. Cloning and characterization of porcine enamelin mRNAs. J. Dent. Res. 76 1720–1729. [DOI] [PubMed] [Google Scholar]
- Hu, C. C., T. C. Hart, B. R. Dupont, J. J. Chen, X. Sun et al., 2000. Cloning human enamelin cDNA, chromosomal localization, and analysis of expression during tooth development. J. Dent. Res. 79 912–919. [DOI] [PubMed] [Google Scholar]
- Hu, J. C., and Y. Yamakoshi, 2003. Enamelin and autosomal-dominant amelogenesis imperfecta. Crit. Rev. Oral Biol. Med. 14 387–398. [DOI] [PubMed] [Google Scholar]
- Hu, J. C., Y. Yamakoshi, F. Yamakoshi, P. H. Krebsbach and J. P. Simmer, 2005. Proteomics and genetics of dental enamel. Cells Tissues Organs 181 219–231. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium, 2005. A haplotype map of the human genome. Nature 437 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, J. L., J. Madeoy, J. C. Calhoun, W. Swanson and J. M. Akey, 2006. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 16 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida, M., T. Ariga, T. Shirakawa, H. Oguchi and Y. Sakiyama, 2002. Autosomal-dominant hypoplastic form of amelogenesis imperfecta caused by an enamelin gene mutation at the exon-intron boundary. J. Dent. Res. 81 738–742. [DOI] [PubMed] [Google Scholar]
- Kim, J. W., F. Seymen, B. P. Lin, B. Kiziltan, K. Gencay et al., 2005. ENAM mutations in autosomal-dominant amelogenesis imperfecta. J. Dent. Res. 84 278–282. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1968. Evolutionary rate at the molecular level. Nature 217 624–626. [DOI] [PubMed] [Google Scholar]
- Kimura, R., A. Fujimoto, K. Tokunaga and J. Ohashi, 2007. A practical genome scan for population-specific strong selective sweeps that have reached fixation. PLoS ONE 2 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, A., D. F. Gudbjartsson, J. Sainz, G. M. Jonsdottir, S. A. Gudjonsson et al., 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31 241–247. [DOI] [PubMed] [Google Scholar]
- Mardh, C. K., B. Backman, G. Holmgren, J. C. Hu, J. P. Simmer et al., 2002. A nonsense mutation in the enamelin gene causes local hypoplastic autosomal dominant amelogenesis imperfecta (AIH2). Hum. Mol. Genet. 11 1069–1074. [DOI] [PubMed] [Google Scholar]
- Messier, W., and C. B. Stewart, 1997. Episodic adaptive evolution of primate lysozymes. Nature 385 151–154. [DOI] [PubMed] [Google Scholar]
- Milton, K., and M. L. May, 1976. Body weight, diet and home range area in primates. Nature 259 459–462. [DOI] [PubMed] [Google Scholar]
- Ng, P. C., and S. Henikoff, 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, D. A., V. O. Tobe and S. L. Taylor, 1997. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 25 2745–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, R., C. Bustamante, A. G. Clark, S. Glanowski, T. B. Sackton et al., 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3 e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir, D., P. S. Hart, E. Firatli, G. Aren, O. H. Ryu et al., 2005. Phenotype of ENAM mutations is dosage-dependent. J. Dent. Res. 84 1036–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine, M. L., S. N. White, W. Luo, H. Fong, M. Sarikaya et al., 2001. Regulated gene expression dictates enamel structure and tooth function. Matrix Biol. 20 273–292. [DOI] [PubMed] [Google Scholar]
- Pavlic, A., M. Petelin and T. Battelino, 2007. Phenotype and enamel ultrastructure characteristics in patients with ENAM gene mutations g.13185-13186insAG and 8344delG. Arch. Oral Biol. 52 209–217. [DOI] [PubMed] [Google Scholar]
- Pickford, M., 2005. Incisor-molar relationships in chimpanzees and other hominoids: implications for diet and phylogeny. Primates 46 21–32. [DOI] [PubMed] [Google Scholar]
- Purvis, A., 1995. A composite estimate of primate phylogeny. Philos. Trans. R. Soc. Lond. B Biol. Sci. 348 405–421. [DOI] [PubMed] [Google Scholar]
- Rajpar, M. H., K. Harley, C. Laing, R. M. Davies and M. J. Dixon, 2001. Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta. Hum. Mol. Genet. 10 1673–1677. [DOI] [PubMed] [Google Scholar]
- Rambaut, A., 1996. Se-Al: sequence alignment editor. http://evolve.zoo.ox.ac.uk/.
- Ramensky, V., P. Bork and S. Sunyaev, 2002. Human nonsynonymous SNPs: server and survey. Nucleic Acids Res. 30 3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard, A. F., 1985. Primates in Nature. W. H. Freeman, New York.
- Rosenberg, N. A., J. K. Pritchard, J. L. Weber, H. M. Cann, K. K. Kidd et al., 2002. Genetic structure of human populations. Science 298 2381–2385. [DOI] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15 174–175. [DOI] [PubMed] [Google Scholar]
- Rozen, S., and H. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132 365–386. [DOI] [PubMed] [Google Scholar]
- Sabeti, P. C., D. E. Reich, J. M. Higgins, H. Z. Levine, D. J. Richter et al., 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419 832–837. [DOI] [PubMed] [Google Scholar]
- Schaffner, S. F., C. Foo, S. Gabriel, D. Reich, M. J. Daly et al., 2005. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 15 1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, G. T., 2000. Taxonomic and functional aspects of the patterning of enamel thickness distribution in extant large-bodied hominoids. Am. J. Phys. Anthropol. 111 221–244. [DOI] [PubMed] [Google Scholar]
- Shellis, R. P., A. D. Beynon, D. J. Reid and K. M. Hiiemae, 1998. Variations in molar enamel thickness among primates. J. Hum. Evol. 35 507–522. [DOI] [PubMed] [Google Scholar]
- Stanford, C. B., 1998. Chimpanzee and Red Colobus: The Ecology of Predator and Prey. Harvard University Press, Cambridge, MA.
- Stephens, M., N. J. Smith and P. Donnelly, 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman, R. W., 1987. Morpho-physiological analysis of diets: species-specific dietary patterns in primates and human dietary adaptations, Chapter 9 in The Evolution of Human Behavior: Primate Models, edited by W. G. Kinzey. State University of New York Press, Albany, NY.
- Swanson, W. J., R. Nielsen and Q. Yang, 2003. Pervasive adaptive evolution in mammalian fertilization proteins. Mol. Biol. Evol. 20 18–20. [DOI] [PubMed] [Google Scholar]
- Swofford, D. L., 2003. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. Sinauer Associates, Sunderland, MA.
- Tajima, F., 1989. The effect of change in population size on DNA polymorphism. Genetics 123 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teelen, S., 2007. Influence of chimpanzee predation on the red colobus population at Ngogo, Kibale National Park, Uganda. Primates 49 41–49. [DOI] [PubMed] [Google Scholar]
- Voight, B. F., S. Kudaravalli, X. Wen and J. K. Pritchard, 2006. A map of recent positive selection in the human genome. PLoS Biol. 4 e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, E. T., G. Kodama, P. Baldi and R. K. Moyzis, 2006. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc. Natl. Acad. Sci. USA 103 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, W. S., Z. Yang, N. Goldman and R. Nielsen, 2004. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15 568–573. [DOI] [PubMed] [Google Scholar]
- Yang, Z., R. Nielsen, N. Goldman and A. M. Pedersen, 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., W. S. Wong and R. Nielsen, 2005. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22 1107–1118. [DOI] [PubMed] [Google Scholar]
- Zhang, J., 2003. Parallel functional changes in the digestive RNases of ruminants and colobines by divergent amino acid substitutions. Mol. Biol. Evol. 20 1310–1317. [DOI] [PubMed] [Google Scholar]
- Zhang, J., 2006. Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat. Genet. 38 819–823. [DOI] [PubMed] [Google Scholar]
- Zhang, J., R. Nielsen and Z. Yang, 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22 2472–2479. [DOI] [PubMed] [Google Scholar]