Abstract
The functional divergence of duplicated genes is thought to play an important role in the evolution of new developmental and physiological pathways, but the role of positive selection in driving this process remains controversial. The objective of this study was to test whether amino acid differences among triplicated α-globin paralogs of the Norway rat (Rattus norvegicus) and the deer mouse (Peromyscus maniculatus) are attributable to a relaxation of purifying selection or to a history of positive selection that has adapted the gene products to new or modified physiological tasks. In each rodent species, the two paralogs at the 5′-end of the α-globin gene cluster (HBA-T1 and HBA-T2) are evolving in concert and are therefore identical or nearly identical in sequence. However, in each case, the HBA-T1 and HBA-T2 paralogs are distinguished from the third paralog at the 3′-end of the gene cluster (HBA-T3) by multiple amino acid substitutions. An analysis of genomic sequence data from several rodent species revealed that the HBA-T3 genes of Rattus and Peromyscus originated via independent, lineage-specific duplication events. In the independently derived HBA-T3 genes of both species, a likelihood analysis based on a codon-substitution model revealed that accelerated rates of amino acid substitution are attributable to positive directional selection, not to a relaxation of purifying selection. As a result of functional divergence among the triplicated α-globin genes in Rattus and Peromyscus, the red blood cells of both rodent species contain a mixture of functionally distinct α-chain hemoglobin isoforms that are predicted to have different oxygen-binding affinities. In P. maniculatus, a species that is able to sustain physiological function under conditions of chronic hypoxia at high altitude, the coexpression of distinct hemoglobin isoforms with graded oxygen affinities is expected to broaden the permissible range of arterial oxygen tensions for pulmonary/tissue oxygen transport.
THE duplication of protein-coding genes, followed by functional changes in one or both daughter copies, is thought to play a fundamental role in the evolution of new developmental and physiological pathways (Ohno et al. 1968; Ohno 1970; Kimura and Ohta 1974; Li 1983). However, the role of positive Darwinian selection in driving the functional divergence of duplicated genes remains controversial (Hughes 1994, 1999; Lynch and Force 2000; Lynch et al. 2001; Zhang 2003; Lynch and Katju 2004; Lynch 2007). One of the fundamental questions is whether the mutations that are responsible for the initial retention and subsequent divergence of newly created gene duplicates are typically advantageous or degenerative. According to the subfunctionalization model, the accumulation of degenerative mutations in one or both gene duplicates leads to a complementary loss of paralog-specific subfunctions (Force et al. 1999; Stoltzfus 1999; Lynch and Force 2000; Lynch 2007). The gene duplicates are then retained by purifying selection because inactivation of either paralog results in loss of the ancestral gene function.
According to the neofunctionalization model of Ohno (1970), the functional redundancy of duplicated genes entails a relaxation of purifying selection that results in the accumulation of mutations that are effectively neutral as long as the other paralog continues to perform the essential tasks of the ancestral, single-copy gene. In the vast majority of cases, the redundant gene duplicate will eventually be rendered functionless by inactivating mutations. In rare cases, the redundant gene copy will escape pseudogenization by fixing one or more mutations that fortuitously adapt the encoded protein to a new or modified function. An alternative model for the evolution of novel protein functions, suggested by Hughes (1994, 1999), Piatigorsky and Wistow (1991), and Piatigorsky (2007), invokes the presence of functional divergence prior to gene duplication and does not require an initial relaxation of functional constraint. This model, referred to as the “adaptive conflict” model by Lynch and Katju (2004), envisions an ancestral, single-copy gene that encodes a generalist protein that is capable of performing two or more distinct physiological functions. The product of this gene suffers from a “Jack of all trades, master of none” syndrome, as full optimization of the protein's multiple functions is constrained by antagonistic pleiotropy. Gene duplication then enables the two paralogous copies to become specialized on distinct subsets of the ancestral functions by eliminating pleiotropic constraints. The subfunctionalization model, Ohno's (1970) neofunctionalization model, and the adaptive-conflict model all invoke the accumulation of mutations that would have been off limits prior to duplication. The key distinction is that in the adaptive conflict model the partitioning of ancestral functions between the duplicated genes is brought about by positive selection for mutations that refine the restricted set of paralog-specific subfunctions.
A scenario similar to that envisioned by the adaptive conflict model was invoked by Goodman et al. (1975, 1987) to explain the evolution of heterotetrameric hemoglobin in gnathostome vertebrates from a homotetrameric precursor protein. Functional divergence of genes encoding the α- and β-globin subunits of the hemoglobin tetramer produced an allosteric mechanism of cooperative oxygen binding that opened up new pathways for the evolution of aerobic energy metabolism in vertebrates. Over the course of vertebrate evolution, subsequent rounds of gene duplication and divergence have given rise to families of α- and β-like globin genes that are ontogenetically regulated and biochemically optimized for oxygen transport under the vastly different physiological conditions that are encountered during the embryonic, fetal, and adult stages of development. The globin superfamily of genes therefore provides an excellent example of how physiological pathways can be elaborated and refined through functional and regulatory divergence of duplicated genes that encode different subunit polypeptides of the same multimeric protein.
In contrast to levels of divergence between globin genes that are expressed during different stages of development, paralogs that are coexpressed during the same stage of development are typically identical or nearly identical in sequence. For example, most mammals studied to date possess a tandemly duplicated pair of adult α-globin genes that have identical coding sequences and therefore encode identical polypeptides (Higgs et al. 1989; Hardison 2001). This pattern is generally thought to reflect a history of concerted evolution, mediated by gene conversion and/or unequal crossing over (Zimmer et al. 1980; Lam and Jeffreys 2006; Hoffmann et al. 2008).
Here we investigate two notable exceptions to this general pattern of concerted evolution in the α-globin gene family of mammals. The Norway rat (Rattus norvegicus) and the deer mouse (Peromyscus maniculatus) are unusual in that they each possess three transcriptionally active copies of adult α-globin. In each species, the two paralogs at the 5′-end of the α-globin gene cluster (HBA-T1 and HBA-T2) are evolving in concert and are therefore identical or nearly identical in sequence. However, in each case, the HBA-T1 and HBA-T2 paralogs are distinguished from the third paralog at the 3′-end of the gene cluster (HBA-T3) by multiple amino acid substitutions. Consequently, the red blood cells of these animals contain a heterogeneous mixture of α-chain hemoglobin isoforms that may have different biochemical properties. Depending on the nature of the amino acid differences among the different α-chain subunits, this type of hemoglobin isoform differentiation may have important physiological effects on blood-oxygen transport and red blood cell metabolism (Weber 2000, 2007).
The objective of this study was to determine whether the observed amino acid differences among the triplicated α-globin paralogs of Rattus and Peromyscus are attributable to a relaxation of purifying selection or to a history of positive selection that has adapted the gene products to new or modified physiological tasks. In principle, hypotheses about the role of positive selection in the initial retention and subsequent functional divergence of duplicated genes can be tested by using methods of comparative sequence analysis (Yang and Nielsen 2002; Bielawski and Yang 2003, 2004; Gu 2003; Raes and Van De Peer 2003; Aguileta et al. 2004; Zhang et al. 2005). We first conducted a comparative genomic analysis to reconstruct the history of gene duplication and divergence in the α-globin gene family of rodents. We then used a likelihood-based analysis of codon substitution and an in silico analysis of hemoglobin structure to assess whether positive selection has played a role in driving functional diversification of the adult α-globin genes in Rattus and Peromyscus.
MATERIALS AND METHODS
Nomenclature for globin genes:
Following the nomenclature of Aguileta et al. (2006), we refer to the embryonic ζ-globin gene, the adult α-globin gene, and the θ-globin gene, as HBZ, HBA, and HBQ, respectively. Since mammalian α-globin genes have undergone multiple rounds of duplication that have resulted in tandemly repeated sets of paralogous gene copies (Czelusniak et al. 1982; Proudfoot et al. 1982; Hardison and Gelinas 1986; Goodman et al. 1987; Flint et al. 1988, 2001; Hoffmann and Storz 2007; Hoffmann et al. 2008), we index each duplicated gene with the notation “-T” followed by a number that corresponds to the linkage order in the 5′-to-3′ orientation (Aguileta et al. 2006). For example, in Mus, HBA-T1 and HBA-T2 refer to the 5′ and 3′ α-globin paralogs, respectively. Following the convention of Dickerson and Geis (1983), each amino acid residue is indexed with an alphanumerical code that designates its position relative to seven α-helical domains of the α-globin polypeptide (A, B, C, E, F, G, and H).
Functional annotation of genomic sequences:
We annotated genomic contigs spanning the α-globin gene cluster in four rodent species: the brown Norway rat, R. norvegicus (family Muridae); the house mouse, Mus musculus (family Muridae); the deer mouse, P. maniculatus (family Cricetidae); and the guinea pig, Cavia porcellus (family Caviidae). All α-like globin genes were annotated by comparison with the human 16p13.3 genomic sequence (GenBank accession no. NT_037887). We analyzed publicly available genomic sequence data in the case of Rattus (NW_047334), Mus (AY016021), and Cavia (AC181986). In the case of Peromyscus, we isolated and characterized a genomic contig spanning the entire α-globin gene cluster by screening a bacterial artificial chromosome (BAC) library, as described below.
BAC isolation, sequencing, and analysis:
To isolate and characterize the α-globin gene cluster of P. maniculatus, we screened an EcoRI/EcoRI methylase BAC library using one labeled probe designed for the embryonic ζ-globin gene (HBZ-T1) and another labeled probe designed for the the 3′ adult α-globin gene (HBA-T3). The HBZ-T1 and HBA-T3 genes are located at the 5′- and 3′-ends of the α-globin gene cluster, respectively. The HBZ-T1 probe was designed using an alignment of orthologous sequences from Mus, Rattus, and humans, and the HBA-T3 probe was designed from a P. maniculatus sequence derived from a λ-bacteriophage genomic library (Storz et al. 2007b). After the hybridization screening, we end-sequenced probe-positive BAC clones to identify inserts that spanned the entire α-globin gene cluster. After selecting a single BAC clone for shotgun sequencing, a library with an average insert size of 4 kb was prepared using the PUC119 vector with BAC insert DNA sheared with a HydroShear device (Genemachines, San Carlos, CA). The library was sequenced by using M13 universal primers on an ABI3700 capillary sequencer with BigDye Terminator chemistry. Base calling and quality assessment were performed using Phred (Ewing and Green 1998; Ewing et al. 1998), assembly was performed using Phrap/Cross_match/Swat, and editing and finishing was performed using Consed/Autofinish (Gordon et al. 1998, 2001). The assembled sequence was masked for interspersed repeats by using the program RepeatMasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html) and was then annotated by an assessment of sequence homology with genomic sequence from Mus, Rattus, and humans. In comparisons between the Peromyscus BAC sequence and genomic sequences from the other species, we identified locally alignable chromosomal segments using PipMaker (Schwartz et al. 2000), MultiPipMaker (Schwede et al. 2003), and Mulan (Ovcharenko et al. 2005), and we used the BLAST suite of programs (Altschul et al. 1997) to identify sequence matches. In comparison to genomic sequence from Rattus and humans, the α-globin gene cluster in Mus exhibits an abrupt break in conserved synteny due to a translocation of the 3′-end of the gene cluster from chromosome 11 to chromosome 17 (Leder et al. 1981; Tufarelli et al. 2004). For this reason, we concentrated mainly on comparing the Peromyscus BAC sequence with genomic sequences from Rattus and humans.
Phylogenetic analysis:
For the comparative analysis of sequence variation of the four rodent species, we aligned a 4223-bp chromosomal fragment that extended 2 kb upstream of the start codon of each α-globin gene and 1 kb downstream of the stop codon. Sequences were aligned using MUSCLE (Edgar 2004), and coding regions were manually inspected to keep them in frame. Phylogenetic relationships among orthologous and paralogous α-globin sequences were inferred using a maximum-likelihood tree-building method. Maximum-likelihood tree searches were conducted using Treefinder (version February 2007; Jobb et al. 2004), and this program was also used to estimate parameters of a GTR + Γ model of nucleotide substitution. Measures of bootstrap support were based on 1000 pseudoreplicates.
In the case of multigene families, orthologous relationships among duplicated genes may often be obscured by a history of concerted evolution (Zimmer et al. 1980; Ohta 1990, 2000). As a result of gene conversion or unequal crossing over, paralogous genes within a species are often more similar to one another than either one is to its orthologous counterpart in closely related species. Gene conversion between tandemly duplicated globin genes of rodents has been especially well documented (Erhart et al. 1985, 1987; Storz et al. 2007a,b). However, since interparalog gene conversion is often largely restricted to coding sequence (Chen et al. 2007), orthologous relationships between duplicated genes can still be reliably inferred by examining flanking sequence that lies outside of identified gene conversion tracts (Hardison and Gelinas 1986; Hardison and Miller 1993; Storz et al. 2007a; Hoffmann et al. 2008). For this strategy to succeed, the length of the duplication block needs to exceed the length of the gene conversion tract so that there is a sufficient amount of “uncontaminated” sequence to preserve the historical record. To detect evidence of gene conversion among triplicated α-globin paralogs in Rattus and Peromyscus, we masked simple sequence repeats and used the program GENECONV (Sawyer 1989) with the G-scale parameter (mismatch penalty) set to 1.0.
To reconstruct the ancestral amino acid sequences of rodent α-globins, we used the maximum-likelihood approach of Yang (1995) and Koshi and Goldstein (1996).
Analysis of codon-substitution patterns:
We explored variation in the ratio of nonsynonymous-to-synonymous substitution rates (ω = dN/dS) among the rodent α-globin genes using a maximum-likelihood approach (Goldman and Yang 1994). We first applied lineage-specific likelihood models (Yang 1998; Yang and Nielsen 1998), which assume variation in selection pressure among lineages but no variation among sites in the alignment. To test for evidence of rate heterogeneity among lineages, we compared the one-ratio model, which assumes the same ω-ratio for all lineages, with the free-ratio model, which allows an independent ω-ratio for each branch of the phylogeny. We also implemented intermediate models to assess whether the HBA-T3 terminal branches of Rattus and Peromyscus were characterized by ω-ratios that differed from those of the remaining branches. The first model assumes two independent ω-ratios, one ratio for the HBA-T3 terminal branches of Rattus and Peromyscus and another ratio for the remaining branches in the phylogeny. The second model assumes three independent ω-ratios, one for the Rattus HBA-T3 terminal branch, one for the Peromyscus HBA-T3 terminal branch, and one for the remaining branches. In all cases we checked for the existence of multiple local optima by running all models three times with different starting values for ω (0.1, 1.0, and 2.0). Nested models were compared using likelihood-ratio tests (LRTs).
We then applied branch-site model A of Zhang et al. (2005), which assumes four classes of sites: site class 0 includes codons that are subject to stringent functional constraints with 0 < ω0 < 1 estimated from the data; site class 1 includes codons that are unconstrained with ω1 = 1 fixed; and site classes 2a and 2b include codons that are conserved or neutral on the background branches but can undergo a shift to positive directional selection on the foreground branches with ω2 estimated as a free parameter. We used model A to construct an LRT in which the null hypothesis was a simplified version of model A with ω2 = 1 fixed. This null model allows a shift from purifying selection on the background branches to relaxed constraint on the foreground branches. Consequently, the LRT involving the comparison of model A vs. the null model allows us to test whether accelerated rates of nonsynonymous substitution on the foreground branches are attributable to positive selection or to a relaxation of functional constraint. After obtaining maximum-likelihood estimates of parameters under the branch-site model, we used the Bayes empirical Bayes (BEB) method of Yang et al. (2005) to identify the specific sites that are inferred to be under positive selection. Specifically, the BEB approach was used to calculate the Bayesian posterior probabilities that a given site belongs to the site class with ω > 1. All model-based analyses of codon substitution were conducted using the codeml program in the PAML4 package (Yang 2007; http://abacus.gene.ucl.ac.uk/software/paml.html).
To quantify site-specific variation in functional constraint, we used the method of Valdar (2002). We used an alignment of 55 adult α-globin protein sequences from 39 mammalian species (representing 12 mammalian orders) to compute residue-specific conservation scores based on a modified PET91 distance.
Analysis of hemoglobin structure:
Structural modeling of the rodent hemoglobins was conducted using the Swiss model server (Schwede et al. 2003). Homology-based structural templates were selected from the Protein Data Bank (Berman et al. 2000). Structural validations and additional optimizations were conducted with Swiss-PdbViewer (Guex and Peitsch 1997) and the Procheck protein crystallography program from the CCP4 suite (Collaborative Computational Project 1994). Calculations of interatomic distances were performed using Swiss-Pdb Viewer (Guex and Peitsch 1997), and calculations of local binding energy minimization were conducted using AutoDock4 (Morris et al. 1998). All graphical representations of molecular structures were prepared using PyMOL (DeLano Scientific, San Carlos, CA).
RESULTS
Genomic structure of the α-globin gene cluster in rodents:
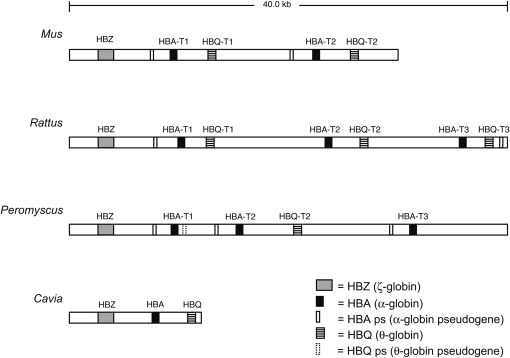
We isolated, subcloned, and sequenced a 184,952-bp BAC clone that contained the entire α-globin gene cluster of the deer mouse, P. maniculatus (GenBank accession no. EU053203). The Peromyscus contig contains five putatively functional α-like globin genes in the following order: 5′-HBZ, HBA-T1, HBA-T2, HBQ-T2, and HBA-T3-3′. The HBA-T2 and HBA-T3 genes correspond to the 5′ and 3′ α-globin genes, respectively, in Storz et al. (2007b). The five α-like globin genes in the cluster span ∼30 kb from the start codon of HBZ to the stop codon of HBA-T3, and this fragment aligns to nucleotide positions 15,592,379–15,556,379 of Rattus chromosome 10 (GenBank accession no. NW_047334). On the basis of patterns of conserved synteny with Rattus and Mus (Dawson et al. 1999; Ramsdell et al. 2006), we deduced that the α-globin gene cluster of P. maniculatus is located on chromosome 8. We found no evidence of additional genes immediately upstream of the HBZ gene. Downstream of HBA-T3, we identified the Peromyscus ortholog of Luc7L, which lies downstream of the α-globin gene cluster in Rattus, humans, and most other mammalian species studied to date (Flint et al. 2001; Tufarelli et al. 2004; Hughes et al. 2005). Farther downstream we found sequences matching most of the annotated genes found in comparable positions in the Rattus and human genomes (supplemental Table 1).
A comparison of genomic sequences spanning the α-globin gene cluster in Mus, Rattus, Peromyscus, and Cavia revealed variation in the number of tandemly duplicated HBA and HBQ genes (Figure 1). Whereas Cavia possesses a single copy of HBZ, HBA, and HBQ, copy number differences in Mus, Rattus, and Peromyscus suggest the hypothesis that a block triplication involving a linked pair of HBA and HBQ genes occurred in the ancestor of muroid rodents sometime after the divergence from Cavia. According to this block triplication hypothesis, the genomic structure of the α-globin gene cluster in Rattus closely approximates the ancestral state in the stem lineage of muroid rodents whereas the syntenic chromosomal region in Mus and Peromyscus must have experienced multiple lineage-specific deletions or translocations. Although Mus also possesses triplicated copies of a HBA–HBQ gene pair, the third (3′) block has been transposed from chromosome 11 to chromosome 17 (Leder et al. 1981; Tufarelli et al. 2004). The pair of HBA-T3 and HBQ-T3 genes on Mus chromosome 17 are pseudogenes, but it is not known whether they were already inactivated prior to the translocation. Consequently, Peromyscus and Rattus each have three adult α-globin genes (HBA-T1, HBA-T2, and HBA-T3) whereas Mus has only two functional copies. In Peromyscus, the block triplication hypothesis requires the secondary loss of HBQ-T1 from the first (5′) block and the loss of HBQ-T3 from the last (3′) block.
Figure 1.—
Genomic structure of the α-globin gene family in rodents.
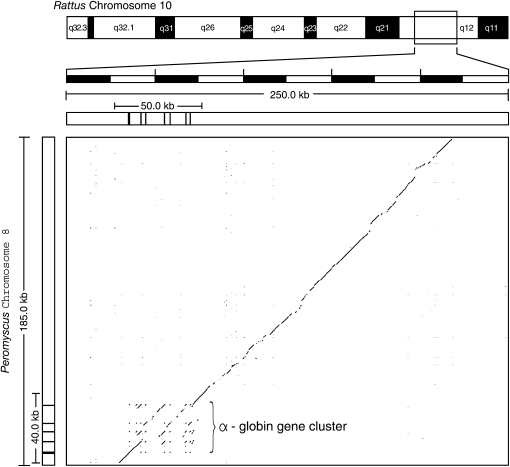
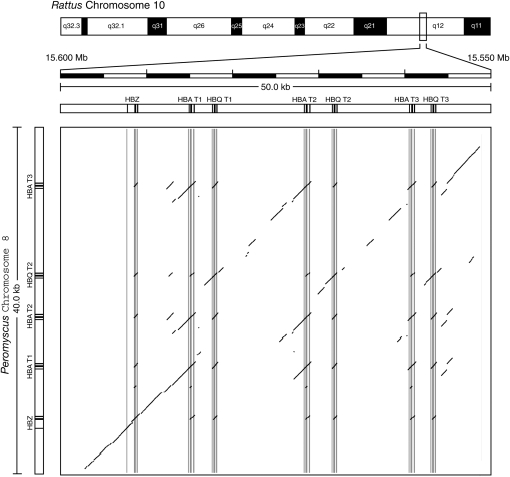
Comparison of the Peromyscus and Rattus sequences using PipMaker revealed extensive matches over the full ∼185 kb, indicating that synteny and gene order is conserved across the entire chromosomal region (Figure 2). In the bottom left of Figure 2, the clustering of off-diagonal elements shows multiple sequence matches among α-like globin genes between Rattus and Peromyscus. A more detailed dot plot of this region shows that the boundaries of duplication blocks are not clearly evident (Figure 3). If the triplicated HBA genes in Mus, Rattus, and Peromyscus are true 1:1 orthologs (i.e., if the block triplication containing the HBA–HBQ gene pair was inherited from the common ancestor of all three species), then the pattern shown in Figure 3 indicates that the intervening sequences between the duplication blocks have experienced multiple insertion and deletion events downstream of the T1 duplication block (and including the T1 duplication block in the case of Peromyscus). By contrast, the 5′-end of the gene cluster appears to have remained stable. This is indicated by the uniform intergenic distance between HBZ and HBA-T1 in all four rodent species (Figure 1) and the extensive colinearity of sequence matches in the region spanning HBZ and HBA-T1 in the Rattus and Peromyscus gene clusters (Figure 3).
Figure 2.—
Dot plot of the P. maniculatus BAC sequence (185 kb) against the rat chromosome 10 sequence (250 kb) with masked repeats.
Figure 3.—
Dot plot of a chromosomal fragment spanning the P. maniculatus α-globin gene cluster (40 kb) against the syntenic region of rat chromosome 10 sequence (50 kb) with masked repeats.
Inferring orthologous relationships:
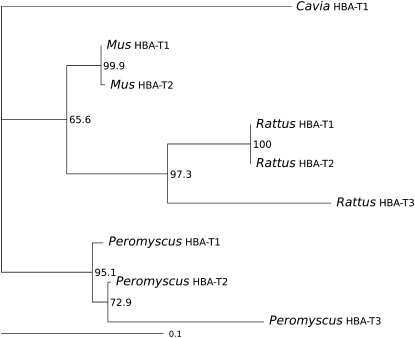
The block triplication hypothesis predicts that each paralogous clade of α-globin sequences should independently recover the true history of species divergence (((Mus, Rattus) Peromyscus) Cavia). Contrary to the predicted pattern, phylogenetic reconstructions based on the coding sequence and those based on upstream and downstream flanking sequence all yielded tree topologies in which α-globin sequences from the same species grouped together to the exclusion of sequences from other species (Figure 4). This pattern is partly attributable to a history of concerted evolution, as HBA-T2 has been partially converted by HBA-T1 in both Rattus and Peromyscus (Table 1). The same conversion tracts were identified when the analysis was restricted to noncoding sites and third codon positions (data not shown). A history of recurrent gene conversion between the HBA-T1 and HBA-T2 genes of Mus has been documented previously (Erhart et al. 1985).
Figure 4.—
Maximum-likelihood phylogeny of rodent α-globin genes based on coding sequence. Bootstrap support values are based on 1000 replicates.
TABLE 1.
Genomic positions of interparalog gene conversion tracts in the α-globin gene clusters of R. norvegicus and P. maniculatus
| Genes | P-valuea | Alignment position | Length (bp) | Affected gene region |
|---|---|---|---|---|
| R. norvegicus | ||||
| HBAT1; HBAT2 | <0.001 | 1172–2676 | 1505 | 5′-UTR: 35/35 bp |
| Exon 1: 95/95 bp | ||||
| Intron 1:115/115 bp | ||||
| Exon 2: 205/205 bp | ||||
| Intron 2: 32/185 bp | ||||
| HBAT2; HBAT3 | <0.010 | 3347–3618 | 272b | 3′ flanking sequence |
| P. maniculatus | ||||
| HBAT1; HBAT2 | <0.001 | 1551–2540 | 990 | 5′-UTR: 35/35 bp |
| Exon 1: 95/95 bp | ||||
| Intron 1: 124/124 bp | ||||
| Exon 2: 205/205 bp | ||||
| Intron 2: 8/173 bp | ||||
In each case the directionality of gene conversion is from the 5′ paralog to the neighboring 3′ paralog. No conversion tracts were identified in the HBA-T3 coding sequences of either species.
Calculated from the permutation test of Sawyer (1989).
The conversion tract starts 390 bp downstream of the stop codon.
Detailed analysis of sequence matches outside of identified gene conversion tracts in each pairwise combination of species confirmed evidence for 1:1 orthologous relationships among the HBA-T1 genes of Mus, Rattus, Peromyscus, and Cavia. These orthologous relationships are also corroborated by the sharing of SINE/B4 and SINE/Alu repetitive elements in the upstream flanking sequence of HBA-T1 in each species. By contrast, in Mus and Rattus, the T2 and T3 duplication blocks appear to have originated by a process of unequal crossing over that occurred after Peromyscus diverged from the common ancestor of these two species (supplemental Figure 1A), and the HBA-T3 gene in Peromyscus appears to be the product of a lineage-specific duplication event that involved at least one and possibly two successive rounds of unequal crossing over (supplemental Figure 1B). As a consequence of these lineage-specific duplication events, the HBA-T2 and HBA-T3 genes of Rattus are orthologous to the HBA-T1 and HBA-T2 genes of Peromyscus, respectively, but the newly created HBA-T3 gene of Peromyscus has no ortholog in Rattus. Evidence for lineage-specific duplications based on sequence matches is corroborated by (1) the sharing of derived SINE/B2 and SINE/Alu repetitive elements between the flanking regions of HBA-T2 and HBA-T3 in Rattus, but not in Peromyscus, and (2) the sharing of derived SINE/B2, SINE/Alu, and SINE/ID repetitive elements and a derived exon 3′ HBA pseudogene fragment between the flanking regions of HBA-T2 and HBA-T3 in Peromyscus, but not in Rattus.
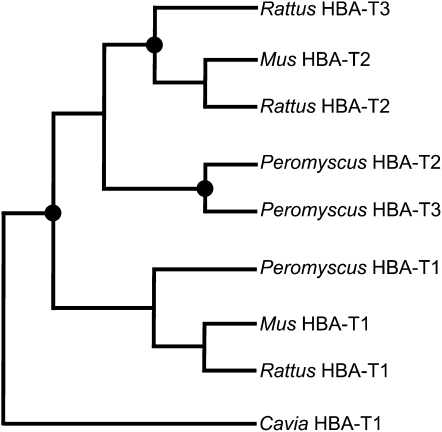
The phylogeny shown in Figure 5 provides an illustration of the inferred history of lineage-specific gene duplication based on (i) pairwise sequence matches in flanking regions and (ii) the distribution of shared, derived repetitive elements and pseudogene fragments in flanking regions. Since this tree depicts an inferred set of phylogenetic relationships that is not confounded by the effects of interparalog gene conversion, the branching order of HBA-T2 and HBA-T3 genes differs from that depicted in the phylogeny based on coding sequence shown in Figure 4.
Figure 5.—
Phylogenetic relationships among rodent α-globin genes inferred from sequence matches in flanking regions as well as shared, derived SINE elements and pseudogene fragments that lie outside of gene conversion tracts. The reconstructed history of gene duplication and species divergence indicates that the HBA-T3 genes of Rattus and Peromyscus originated via independent, lineage-specific duplication events. Nodes with solid circles denote duplication events.
Variation in selection pressure among branches and among sites:
To test for evidence of variation in ω-ratios across the inferred phylogeny of α-globin sequences, we first compared the one-ratio model with a free-ratio model that allowed an independent ω-ratio for each branch in the phylogeny. The one-ratio model yielded a log likelihood of −1564.37, with the estimate ω = 0.33, whereas the free-ratio model yielded a log likelihood of −1538.78. The LRT revealed that the free-ratio model provided a significantly better fit to the data than the one-ratio model (2Δℓ = 2 × 25.59 = 51.18, d.f. = 12, P < 0.001). This result suggests that variation in ω-ratios across the phylogeny of rodent α-globin sequences may be attributable to heterogeneous selection pressures. We then compared the one-ratio model to a two-ratio model that allowed different ω-ratios for the HBA-T3 terminal branches and the remaining branches of the α-globin phylogeny. The two-ratio model yielded a log likelihood of −1550.85, with estimates of ω = 1.19 for the HBA-T3 terminal branches and ω = 0.20 for the remaining branches (Table 2). The LRT revealed that the two-ratio model provided a significantly better fit to the data than the one-ratio model (2Δℓ = 2 × 13.52 = 27.04, d.f. = 1, P < 0.001). This result suggests that variation in ω-ratios across the phylogeny of rodent α-globin sequences may be attributable to positive selection in the separate HBA-T3 branches of Rattus and Peromyscus against a background of strong purifying selection in the remaining branches. Finally, to assess whether this result was attributable to just one of the HBA-T3 branches, we fit a three-ratio model to the data, which yielded a log likelihood of −1550.75, with estimates of ω = 1.38 for the branch leading to HBA-T3 of Rattus (HBA-T3Rat), ω = 1.04 for the branch leading to HBA-T3 of Peromyscus (HBA-T3Pero), and ω = 0.20 for the remaining branches, as before (Table 2). The LRT revealed that the three-ratio model did not provide a better fit to the data than the more simple two-ratio model (2Δℓ = 2 × 0.1 = 0.2, d.f. = 1, P > 0.05) due to the fact that ω-ratios for the HBA-T3Rat and HBA-T3Pero terminal branches were so closely similar that they collapsed into a single class. The fact that ω-ratios were >1 in both HBA-T3 terminal branches suggests a similar history of positive selection in each lineage.
TABLE 2.
Maximum-likelihood analysis of variation in the dN/dS ratio (= ω) across branches and across sites
| Model | Parameter estimates | ℓ | No. of parameters | Candidate sites for positive selection |
|---|---|---|---|---|
| Lineage models (Yang 1998) | ||||
| One ratio | ω = 0.33 | −1564.37 | 15 | NA |
| Two ratio | ωT3 = 1.19, ωBackground = 0.20 | −1550.85 | 16 | NA |
| Three ratio | ωT3 Rat = 1.38, ωT3 Pero = 1.04, ωBackground = 0.20 | −1550.75 | 17 | NA |
| Branch-site models (foreground branch: HBA-T3Rat) | ||||
| Null A | ω0-b = ω0-f = 0.03, (p0 = 0.54) | −1517.82 | 17 | NA |
| ω1-b = ω1-f = 1.00, (p1 = 0.25) | ||||
| ω2A-b = 0.03, ω2A-f = 1.00, (p2A = 0.14) | ||||
| ω2B-b = 1.00, ω2B-f = 1.00, (p2B = 0.07) | ||||
| A | ω0-b = ω0-f = 0.03, (p0 = 0.62) | −1511.54 | 18 | 19(AB1) Gly → |
| ω1-b = ω1-f = 1.00, (p1 = 0.29) | Asn** | |||
| ω2A-b = 0.03, ω2A-f = 19.62, (p2A = 0.06) | 24(B5) Tyr → Ile** | |||
| ω2B-b = 1.00, ω2B-f = 19.62, (p2B = 0.03) | 29(B10) Leu → Ile* | |||
| 38(C3) Thr → Ser* | ||||
| 39(C4) Thr → Ser* | ||||
| 48(CD6) Val → Thr** | ||||
| 50(CD8) Pro → Glu* | ||||
| Branch-site models (foreground branch: HBA-T3Pero) | ||||
| Null A | ω0-b = ω0-f = 0.03, (p0 = 0.56) | −1519.36 | 17 | NA |
| ω1-b = ω1-f = 1.00, (p1 = 0.29) | ||||
| ω2A-b = 0.03, ω2A-f = 1.00, (p2A = 0.09) | ||||
| ω2B-b = 1.00, ω2B-f = 1.00, (p2B = 0.05) | ||||
| A | ω0-b = ω0-f = 0.03, (p0 = 0.62) | −1512.79 | 18 | 32(B13) Met → Asp** |
| ω1-b = ω1-f = 1.00, (p1 = 0.32) | 36(C1) Phe → Pro** | |||
| ω2A-b = 0.03, ω2A-f = ∞, (p2A = 0.04) | 58(E7) His → Gln* | |||
| ω2B-b = 1.00, ω2B-f = ∞, (p2B = 0.02) |
Maximum-likelihood estimates of ω that are indicative of positive selection are in italics. **P > 0.95, *0.95 > P > 0.75.
Results of the lineage analyses were confirmed by estimates of pairwise ω-ratios, which do not depend on any particular tree topology. We calculated pairwise ω-ratios using the method of Yang and Nielsen (2000), as implemented in the yn00 program in PAML. In both Rattus and Peromyscus, pairwise comparisons between HBA-T3 and the other two HBA paralogs exhibited the highest ω-ratios (supplemental Table 2). In the case of Rattus, the HBA-T3 vs. HBA-T1 or HBA-T2 comparison yielded ω = 1.50 (dN = 0.140 ± 0.022, dS = 0.094 ± 0.033). In the case of Peromyscus, the HBA-T3 vs. HBA-T1 comparison yielded ω = 0.88 (dN = 0.095 ± 0.018, dS = 0.109 ± 0.036) and the HBA-T3 vs. HBA-T2 comparison yielded ω = 0.97 (dN = 0.084 ± 0.017, dS = 0.087 ± 0.032). The distribution of pairwise ω-ratios suggests that the rodent α-globin sequences are characterized by two contrasting modes of evolution. In principle, the accelerated rate of nonsynonymous substitutions in the HBA-T3 genes of Rattus and Peromyscus could be attributable to positive selection or to a relaxation of functional constraint. By contrast, the remaining comparisons between orthologous and paralogous gene pairs are characterized by relatively low ω-ratios (range = 0.14–0.39) that are indicative of strong purifying selection.
To perform a more definitive test to distinguish between the effects of positive selection and relaxed functional constraint as explanations for the accelerated rate of nonsynonymous substitution in the HBA-T3 terminal branches and to identify the specific substitutions in each lineage that may be responsible for adaptive modifications of hemoglobin function, we used the branch-site model A of Zhang et al. (2005; see also Yang and Nielsen 2002). In the first branch-site analysis, we treated HBA-T3Rat as the foreground branch and all other branches in the tree were treated as background branches. Branch-site model A provided a significantly better fit to the data than the null model A (2Δℓ = 2 × 6.28 = 12.56, d.f. = 1, P < 0.001) and suggested that 9% of sites are under positive selection in the HBA-T3Rat lineage with ω2 = 19.62 (Table 2). Similarly, when the analysis was repeated with HBA-T3Pero as the foreground branch, model A provided a significantly better fit to the data than the null model A (2Δℓ = 2 × 6.57 = 13.14, d.f. = 1, P < 0.001) and suggested that 6% of sites are under positive selection in the HBA-T3Pero lineage with ω2 = ∞ (Table 2). Since the LRT was highly significant, this result simply indicates that ω2 ≫ 1.0, but the precise value cannot be estimated (Z. Yang, personal communication). In each of the two branch-site tests, results were essentially identical when the HBA-T3 sequence from either Rattus or Peromyscus was treated as the foreground branch and the HBA-T3 sequence from the other species was removed from the data set. Since each of the α-globin paralogs of P. maniculatus are known to segregate functionally distinct protein alleles (Storz 2007; Storz et al. 2007b; J. F. Storz, unpublished results), we repeated all codeml analyses using HBA-T1, HBA-T2, and HBA-T3 sequences that were representative of different allele classes. Parameter estimates were highly similar in all cases (data not shown).
In summary, results of the codon-substitution analyses revealed an accelerated rate of nonsynonymous substitution in the HBA-T3 lineages of both Rattus and Peromyscus. Results of likelihood-ratio tests based on branch-site model A of Zhang et al. (2005) indicate that the accelerated rate of amino acid substitution in the HBA-T3Rat and HBA-T3Pero lineages is attributable to positive selection.
Effects of interparalog gene conversion on the LRTs:
Since the HBA-T2 coding sequences of Rattus and Peromyscus have experienced a history of interparalog gene conversion (Table 1), it is important to assess the effects of such conversion events on the LRTs. In general, gene conversion should not produce spurious evidence for positive selection because both synonymous and nonsynonymous sites are affected in a uniform manner (Aguileta et al. 2004). However, in phylogeny-based analyses, interparalog gene conversion can indirectly affect the LRTs by causing converted and unconverted segments of the same sequence to have different tree topologies (Anisimova et al. 2003). To assess the effects of different tree topologies on the LRTs, we conducted the same codeml analyses using the topology that depicts the inferred set of orthologous relationships among rodent α-globin sequences (Figure 5). As shown in supplemental Table 3, parameter estimates were similar when the codeml analyses were based on this alternative tree topology. Importantly, the branch-site tests of positive selection were still highly significant for both HBA-T3Rat and HBA-T3Pero and the same sets of Bayes-predicted sites were identified at the 0.95 probability threshhold. Given that the use of different tree topologies did not alter the results of the LRTs, and given that the accelerated rates of nonsynonymous substitution in the HBA-T3Rat and HBA-T3Pero lineages were readily apparent in the topology-independent pairwise analysis (supplemental Table 1), it appears that the results of our analysis were not unduly affected by the known history of gene conversion. Most importantly, there was no evidence of gene conversion in the HBA-T3 coding sequences of Rattus or Peromyscus (Table 1), as these are the two genes that were inferred to have experienced a history of positive selection.
Identifying sites involved in the adaptive modification of hemoglobin function:
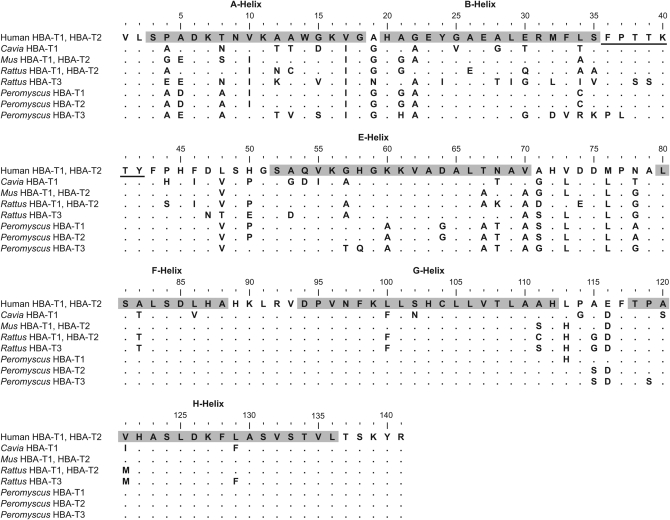
In Rattus, the HBA-T1 and HBA-T2 genes are identical in coding sequence, but each of these paralogs are distinguished from HBA-T3 by 32 amino acid substitutions (Figure 6). Maximum-likelihood reconstruction of the ancestral HBA sequence indicated that all 32 substitutions occurred in the branch leading to HBA-T3. Using a 95% posterior probability cutoff, the BEB method predicted that 3 of the 32 substitutions in the HBA-T3Rat lineage are attributable to positive selection. The three substitutions, 19(AB1)Gly → Asn, 24(B5)Tyr → Ile, and 48(CD13)Val → Thr, involved residue positions that were moderately to highly conserved (i.e., conservation scores were 0.69, 0.92, and 0.79, respectively). In Peromyscus, HBA-T1 and HBA-T2 are distinguished from one another by three amino acid substitutions, whereas each of these two paralogs are distinguished from HBA-T3 by 23 and 20 amino acid substitutions, respectively (Figure 6). Maximum-likelihood reconstruction of the ancestral HBA sequence indicated that 20 substitutions occurred in the branch leading to HBA-T3 and three substitutions occurred in the branch leading to HBA-T1. Using the same 95% posterior probability cutoff, the BEB method predicted that 2 of the 20 substitutions in the HBA-T3Pero lineage are attributable to positive selection. These two substitutions, 32(B13)Met → Asp and 36(CD1)Phe → Pro, involved residues with conservation scores of 0.75 and 0.93, respectively. None of the Bayes-predicted sites under positive selection represented parallel substitutions between the independently derived HBA-T3 genes of Rattus and Peromyscus. However, in the HBA-T3 genes of both species, a disproportionate number of amino acid substitutions were concentrated in the A, B, and C helices (Figure 6).
Figure 6.—
Structural alignment of mammalian α-globins. The A, B, E, F, G, and H α-helical domains are shaded, and the C helix is underlined.
Structural and functional variation in rodent hemoglobins:
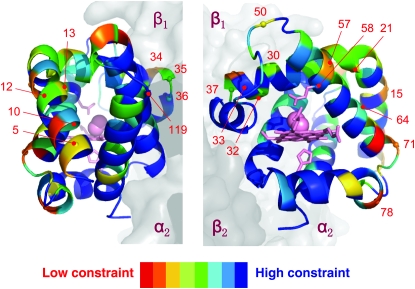
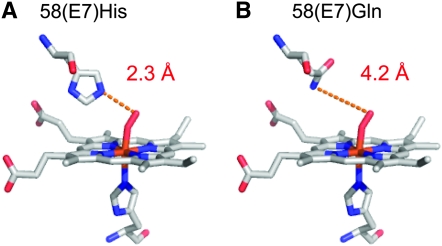
In the case of both Rattus and Peromyscus, hemoglobin tetramers that incorporate α-chain subunits encoded by the independently derived HBA-T3 genes are predicted to have higher oxygen-binding affinities than those that incorporate products of the HBA-T1 or HBA-T2 genes (Table 3). In the case of Rattus, it is not clear which of the 32 amino acid substitutions that distinguish HBA-T3 from the other two α-globin paralogs are responsible for the predicted isoform differences in hemoglobin–oxygen affinity. By contrast, in the case of Peromyscus, predicted differences in hemoglobin–oxygen affinity are primarily attributable to the 58(E7)His → Gln substitution in HBA-T3. This site had the third highest posterior probability in the BEB analysis, but it fell below the 0.95 threshhold (Table 2). The 58(E7)His → Gln substitution affects one of the more highly conserved residue positions in the α-chain polypeptide (Figure 7). In nearly all mammals studied to date, the E7 residue of the highly conserved E-helix domain is a histidine that plays a critical role in the reversible binding of oxygen to the heme iron. Specifically, the Nɛ atom of the imidazole side chain donates a hydrogen bond to the free atom of the bound dioxygen molecule, which helps stabilize the heme–ligand complex (Shaanan 1980; Phillips and Schoenborn 1981; Olson et al. 1988; Lukin et al. 2000; Perutz 2001). In the α-chain hemoglobin isoform that incorporates the product of HBA-T3, the substitution of glutamine for histidine at this E7 position is predicted to increase the Nɛ–O bond distance by 1.9 Å (Figure 8), thereby producing a minor shift in the three-dimensional coordinates of the heme–ligand complex. This shift is predicted to increase the intrinsic oxygen affinity of the E7(Gln)-containing isoform by reducing the free energy of oxygen binding by 0.45 kcal/mol relative to the E7(His)-containing isoform (Table 3). Functional experiments will be required to test this prediction of our model-based in silico analysis.
TABLE 3.
Energy calculation for dioxygen binding by different α-chain hemoglobins
| Hemoglobin α-chain | Binding energy (kcal/mol) |
|---|---|
| Cavia HBA-T1 | −1.80 |
| Mus HBA-T1 | −1.80 |
| Mus HBA-T2 | −1.80 |
| Peromyscus HBA-T1 | −1.80 |
| Peromyscus HBA-T2 | −1.80 |
| Peromyscus HBA-T3 | −1.35 |
| Rattus HBA-T1, -T2 | −1.80 |
| Rattus HBA-T3 | −1.74 |
Figure 7.—
Homology-based structural model of the P. maniculatus α-globin polypeptide, showing the location of 20 amino acid substitutions that are unique to the product of the HBA-T3 gene. Color coding of the ribbon diagram shows site-specific variation in conservation scores (see text for details). Residue positions in blue are invariant or nearly invariant across all mammals (and are therefore presumably subject to stringent functional constraints), whereas residue colors that are further toward the red end of the spectrum are more variable (and are therefore presumably subject to less stringent functional constraints).
Figure 8.—
Isoform differences in hydrogen-bond distance between the E7 residue side chain and the free atom of the bound dioxygen molecule. (A) Heme–ligand complex in the (E7)His-containing α-globin produced by the HBA-T1 and HBA-T2 genes of P. maniculatus. (B) Heme–ligand complex in the (E7)Gln-containing α-globin produced by the HBA-T3 gene of P. maniculatus.
In summary, in both Rattus and Peromyscus, amino acid substitutions between HBA-T3 and the other two α-globin paralogs are predicted to produce differences in oxygen-binding affinity among alternative α-chain hemoglobin isoforms. In Peromyscus, isoform differences in hemoglobin–oxygen affinity appear to be primarily attributable to a single amino acid substitution in a highly conserved heme–protein contact.
DISCUSSION
Our analysis of α-globin sequence variation in rodents has revealed that the functional differentiation of α-chain hemoglobin isoforms in both Rattus and Peromyscus has been brought about by two independent, lineage-specific rounds of gene duplication and divergence. In the independently derived HBA-T3 genes of both species, the application of codon-substitution models revealed that accelerated rates of amino acid substitution are attributable to positive directional selection, not to a relaxation of purifying selection. These results suggest that hemoglobin isoform differentiation has been driven by selection that favors some type of physiological division of labor between the functionally distinct α-globin paralogs. We first address the possible adaptive significance of this isoform differentiation with respect to blood-oxygen transport. We then discuss the possibility that functional divergence preceded the lineage-specific duplication events. Specifically, we consider whether the apparent specialization of function between duplicated α-globin genes evolved as a refinement of preexisting subfunctions in a pleiotropically constrained ancestral gene (as envisioned by the adaptive conflict model; Piatigorsky and Wistow 1991; Hughes 1994, 1999; Piatigorsky 2007) or whether the specialization of function between duplicated genes originated as a specialization of function between alternative alleles that were maintained as a single-locus balanced polymorphism (as envisioned by the balancing selection model; Spofford 1969; Proulx and Phillips 2006).
Functional significance of hemoglobin isoform differentiation:
Although there was very little overlap in the specific amino acid substitutions that accumulated in the HBA-T3 genes of Rattus and Peromyscus (Figure 6), in both species the product of the HBA-T3 gene is predicted to have a higher oxygen-binding affinity than the α-chain products of the paralogous HBA-T1 and HBA-T2 genes. In the case of Peromyscus, the predicted isoform differences in hemoglobin–oxygen affinity are primarily attributable to the 58(E7)His → Gln substitution in HBA-T3. Consistent with our modeling results (Table 3), protein engineering studies of human hemoglobin have revealed that the E7-Gln β-chain mutant results in an increased oxygen-binding affinity at low partial pressures of oxygen (pO2) relative to wild-type E7-His β-chain hemoglobin (Nagai et al. 1987). As a result of functional divergence among the triplicated α-globin genes in Rattus and Peromyscus, the red blood cells of both rodent species contain a mixture of functionally distinct hemoglobin isoforms that are predicted to have different oxygen-binding affinities. This hemoglobin isoform differentiation may have important effects on tissue oxygenation under conditions of anemia or high-altitude hypoxia (van Vliet and Huisman 1964; Weber 2000, 2007). The effects of high-altitude hypoxia are especially relevant in the case of P. maniculatus. This species has the broadest altitudinal distribution of any North American mammal and is able to survive and function under conditions of chronic hypoxia at elevations >4300 m (Storz 2007).
The expression of multiple hemoglobin isoforms with graded oxygen affinities is expected to broaden the permissible range of arterial oxygen tensions for pulmonary/tissue oxygen transport and may thus provide a regulatory reserve of oxygen transport capacity (Weber 2000, 2007). This cascade mechanism of blood-oxygen transport appears to have played an important role in the evolution of hypoxia tolerance in birds that are capable of flying at extremely high altitudes (Hiebl et al. 1987a,b,c, 1988, 1989; Weber et al. 1988a). The high-affinity hemoglobin isoforms are specialized on the task of pulmonary oxygen loading at low pO2, whereas the low-affinity isoforms are specialized on the task of oxygen unloading in the peripheral circulation (Weber et al. 1988a; Weber 2007).
In mammals, this form of hemoglobin isoform differentiation appears to be quite rare. Under conditions of high-altitude hypoxia, adult alpacas (Vicugna pacos) and yaks (Bos grunniens) are known to upregulate a fetal β-like globin gene, which results in the synthesis of a relatively high-affinity fetal hemoglobin (Reynafarje et al. 1975; Sarkar et al. 1999). This high-affinity fetal hemoglobin is adapted to placental/tissue oxygen transport in the hypoxic intrauterine environment and apparently can be co-opted for pulmonary/tissue oxygen transport under conditions of hypoxic stress during postnatal life. In addition to the coexpression of fetal and adult hemoglobins under hypoxic conditions, yaks also possess multiple adult hemoglobin isoforms due to functional divergence among tandemly duplicated α- and β-globin genes (Lalthantluanga et al. 1985; Weber et al. 1988b). Since yaks inhabit alpine environments at elevations of 3000–6000 m on the Tibetan Plateau, the cascaded oxygen affinities of the fetal and adult hemoglobin isoforms appear to play an important role in the hypoxia tolerance of these animals during both pre- and postnatal life. The differentiation in oxygen-binding affinity among α-chain hemoglobin isoforms of P. maniculatus may play a similar role in hypoxia tolerance and cold tolerance. For example, under conditions of cold stress in a hypoxic environment, regulatory adjustments in the stoichiometric ratio of high- and low-affinity hemoglobin isoforms may increase the circulatory conductance of oxygen in the blood stream, thereby providing an enhanced capacity for aerobic thermogenesis. Perhaps as importantly, the reduced autoxidation rate associated with the E7-Gln mutant would inhibit the formation of oxygen free radicals, which also occurs more readily at low pO2 (Dosek et al. 2007).
The possibility of functional divergence prior to duplication:
The evidence for positive selection on the independently derived HBA-T3 genes in Rattus and Peromyscus is consistent with predictions of the adaptive conflict model (Piatigorsky and Wistow 1991; Hughes 1994, 1999; Piatigorsky 2007) and the balancing selection model (Spofford 1969; Proulx and Phillips 2006). Both models invoke divergent selection on protein subfunctions prior to gene duplication, and neither model requires a post-duplication phase of relaxed functional constraint. In the case of the adaptive conflict model, gene duplication is favored because it alleviates pleiotropic constraints that hinder the full optimization of different protein subfunctions (Jensen 1976; Orgel 1977; Jensen and Byng 1981; Piatigorsky and Wistow 1991; Hughes 1994). In the case of the balancing selection model, gene duplication is favored because it alleviates the segregation load associated with the maintenance of alternative alleles by overdominant selection (Spofford 1969; Otto and Yong 2002; Walsh 2003; Proulx and Phillips 2006).
In addition to predicting a primary role for positive selection in driving the functional divergence of duplicated genes, the adaptive conflict model also postulates that the specialized subfunctions of duplicated genes were previously performed by a multifunctional, but pleiotropically constrained, ancestral gene. This unduplicated ancestral state may still be retained in extant species of murid or cricetid rodents, and in fact, M. musculus has essentially reverted back to this ancestral state as a result of the pseudogenization of HBA-T3.
In the case of species like P. maniculatus that inhabit high-altitude environments, there is a clear basis for pleiotropic constraints on blood-oxygen transport by hemoglobin. Under conditions of extreme hypoxia, a high hemoglobin–oxygen affinity is generally advantageous because there is a premium on pulmonary oxygen loading at low pO2 (Turek et al. 1973; Hsia 1998; Storz 2007). The physiological trade-off is that high hemoglobin–oxygen affinity hinders the release of oxygen in the tissue capillary beds. However, if the interrelated tasks of arterial oxygenation and tissue oxygenation are partitioned between the products of duplicated genes, then the coexpression of distinct hemoglobin isoforms with graded oxygen affinities may help to optimize blood-oxygen transport over a broad range of arterial oxygen tensions.
One possible alternative to the scenario envisioned by the adaptive conflict model is that the ancestral, single-copy gene was segregating two alternative alleles that performed distinct subfunctions. Under the balancing selection model for the initial fixation and subsequent retention of duplicated genes (Spofford 1969; Proulx and Phillips 2006), the segregation load associated with the maintenance of balanced polymorphism would create an immediate selective advantage for a duplication that converted allelic variation in protein function to nonallelic variation between the two nascent paralogs. In P. maniculatus, this model appears to be quite plausible because nonallelic differences in oxygen affinity between the product of HBA-T3 and those of the HBA-T1 and HBA-T2 genes are mirrored by differences in oxygen affinity between alternative protein alleles that are segregating at both HBA-T1 and HBA-T2 (Storz et al. 2007b; J. F. Storz, unpublished results). Moreover, the two alternative protein alleles are maintained as a long-term balanced polymorphism by spatially varying selection that favors different hemoglobin–oxygen affinities in different elevational zones (Storz et al. 2007b). Experiments involving wild-derived strains of P. maniculatus that carry different α-globin haplotypes in identical-by-descent condition have revealed that this allelic variation in blood–oxygen affinity contributes to fitness-related variation in aerobic capacity and thermogenic capacity under hypoxic conditions (Snyder 1981; Chappell and Snyder 1984; Chappell et al. 1988; Storz 2007). These findings demonstrate that levels of functional divergence between alleles segregating at the same locus may be similar in magnitude to levels of functional divergence between duplicated genes, although in this particular case, the amino acid changes that distinguish functionally distinct alleles at the HBA-T1 and HBA-T2 genes do not represent fixed differences relative to the HBA-T3 paralog. If there is variation in α-globin copy number among different species of Peromyscus, it should be possible to determine whether the products of derived gene duplicates in P. maniculatus perform functions that are performed by alternative alleles at the ancestral α-globin pro-ortholog in sibling species.
The role of duplicated globin genes in adaptive modifications of blood-oxygen transport:
In mammals, as in other gnathostome vertebrates, regulatory and functional divergence of α- and β-like globin genes has resulted in the production of distinct hemoglobin isoforms that are biochemically optimized for oxygen binding and oxygen transport during different stages of development (Hardison 1998, 2001; Nagel and Steinberg 2001). The hemoglobin isoform differentiation observed in Rattus and Peromyscus is unusual in that the functionally distinct α-globin paralogs are coexpressed in adult erythroid cells. In a number of hypoxia-tolerant animals, hemoglobin isoform differentiation appears to have played an important role in broadening the range of permissible arterial oxygen tensions for blood-oxygen transport (Weber 2000, 2007). In a similar fashion, divergence of duplicated genes that encode long-wavelength visual photopigments in butterflies has played an important role in broadening the range of spectral sensitivities (Frentiu et al. 2007a,b), and divergence of duplicated immunoglobulin VH genes (Tanaka and Nei 1989) and defensin genes (Hughes and Yeager 1997) has apparently helped to broaden the scope of host immune surveillance in mammals.
In summary, functional divergence among the triplicated α-globin paralogs of Rattus and Peromyscus stands in stark contrast to the pattern of concerted evolution that has been documented for the α-globin paralogs of most other mammals studied to date (Zimmer et al. 1980; Higgs et al. 1989; Hardison 2001). Additional work is required in both species to elucidate the adaptive significance of hemoglobin isoform differentiation in blood-oxygen transport. Due to the wealth of knowledge about structure–function relationships in vertebrate hemoglobin (Perutz 1983, 2001), the globin gene family provides an especially promising system for addressing questions about the evolution of duplicated genes because patterns of sequence divergence can be related to functional properties of the encoded proteins.
Acknowledgments
We thank D. Rand and two anonymous reviewers for comments on the manuscript. This work was funded by a National Science Foundation grant to J.F.S. (DEB-0614342), a Layman Award to J.F.S., and an Interdisciplinary Research Grant to J.F.S. and H.M. from the Nebraska Research Council.
Sequence data from this article have been deposited in the EMBL/GenBank Data Libraries under accession no. EU053203.
References
- Aguileta, G., J. P. Bielawski and Z. H. Yang, 2004. Gene conversion and functional divergence in the beta-globin gene family. J. Mol. Evol. 59 177–189. [DOI] [PubMed] [Google Scholar]
- Aguileta, G., J. P. Bielawski and Z. H. Yang, 2006. Proposed standard nomenclature for the alpha- and beta-globin gene families. Genes Genet. Syst. 81 367–371. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova, M., R. Nielsen and Z. Yang, 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat et al., 2000. The Protein Data Bank. Nucleic Acids Res. 28 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski, J. P., and Z. Yang, 2003. Maximum likelihood methods for detecting adaptive evolution after gene duplication. J. Struct. Funct. Genomics 3 201–212. [PubMed] [Google Scholar]
- Bielawski, J. P., and Z. H. Yang, 2004. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J. Mol. Evol. 59 121–132. [DOI] [PubMed] [Google Scholar]
- Chappell, M. A., and L. R. G. Snyder, 1984. Biochemical and physiological correlates of deer mouse α chain hemoglobin polymorphisms. Proc. Natl. Acad. Sci. USA 81 5484–5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell, M. A., J. P. Hayes and L. R. G. Snyder, 1988. Hemoglobin polymorphisms in deer mice (Peromyscus maniculatus): physiology of beta-globin variants and alpha-globin recombinants. Evolution 42 681–688. [DOI] [PubMed] [Google Scholar]
- Chen, J. M., D. N. Cooper, N. Chuzhanova, C. Ferec and G. P. Patrinos, 2007. Gene conversion: mechanisms, evolution and human disease. Nat. Rev, Genet. 8 726–775. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- Czelusniak, J., M. Goodman, D. Hewett-Emmett, M. L. Weiss, P. J. Venta et al., 1982. Phylogenetic origins and adaptive evolution of avian and mammalian haemoglobin genes. Nature 298 297–300. [DOI] [PubMed] [Google Scholar]
- Dawson, W. D., S. R. Young, Z. W. Wang, L. W. Liu, I. F. Greenbaum et al., 1999. Mus and Peromyscus homology established by FISH with three mouse paint probes. Mamm. Genome 10 730–733. [DOI] [PubMed] [Google Scholar]
- Dickerson, R. E., and I. Geis, 1983. Hemoglobin: Structure, Function, Evolution, and Pathology. Benjamin/Cummings, Menlo Park, CA.
- Dosek, A., H. Ohno, Z. Acs, A. W. Taylor and Z. Radak, 2007. High altitude and oxidative stress. Respir. Physiol. Neurobiol. 158 128–131. [DOI] [PubMed] [Google Scholar]
- Edgar, R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhart, M. A., K. S. Simons and S. Weaver, 1985. Evolution of mouse β-globin genes: a recent gene conversion in the Hbbs haplotype. Mol. Biol. Evol. 2 304–320. [DOI] [PubMed] [Google Scholar]
- Erhart, M. A., K. Piller and S. Weaver, 1987. Polymorphism and gene conversion in mouse α-globin haplotypes. Genetics 115 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., and P. Green, 1998. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8 186–194. [PubMed] [Google Scholar]
- Ewing, B., L. Hillier, M. Wendl and P. Green, 1998. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8 175–185. [DOI] [PubMed] [Google Scholar]
- Flint, J., A. M. Taylor and J. B. Clegg, 1988. Structure and evolution of the horse ζ-globin locus. J. Mol. Biol. 199 427–437. [DOI] [PubMed] [Google Scholar]
- Flint, J., C. Tufarelli, J. Peden, K. Clark, R. J. Daniels et al., 2001. Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the alpha globin cluster. Hum. Mol. Genet. 10 371–382. [DOI] [PubMed] [Google Scholar]
- Force, A., M. Lynch, F. B. Pickett, A. Amores, Y. L. Yan et al., 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu, F. D., G. D. Bernard, C. I. Cuevas, M. P. Sison-Mangus, K. L. Prudic et al., 2007. a Adaptive evolution of color vision as seen through the eyes of butterflies. Proc. Natl. Acad. Sci. USA 104 8634–8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu, F. D., G. D. Bernard, M. P. Sison-Mangus, A. Van Zant Brower and A. D. Briscoe, 2007. b Gene duplication is an evolutionary mechanism for expanding spectral diversity in the long wavelength photopigments of butterflies. Mol. Biol. Evol. 24 2016–2028. [DOI] [PubMed] [Google Scholar]
- Goldman, N., and Z. Yang, 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11 725–736. [DOI] [PubMed] [Google Scholar]
- Goodman, M., G. W. Moore and G. Matsuda, 1975. Darwinian evolution in the genealogy of haemoglobin. Nature 253 603–608. [DOI] [PubMed] [Google Scholar]
- Goodman, M., J. Czelusniak, B. F. Koop, D. A. Tagle and J. L. Slightom, 1987. Globins: a case study in molecular phylogeny. Cold Spring Harb. Symp. Quant. Biol. 52 875–890. [DOI] [PubMed] [Google Scholar]
- Gordon, D., C. Abajian and P. Green, 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8 195–202. [DOI] [PubMed] [Google Scholar]
- Gordon, D., C. Desmarais and P. Green, 2001. Automated finishing with Autofinish. Genome Res. 11 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X., 2003. Functional divergence in protein (family) sequence evolution. Genetica 118 133–141. [PubMed] [Google Scholar]
- Guex, N., and M. C. Peitsch, 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18 2714–2723. [DOI] [PubMed] [Google Scholar]
- Hardison, R., 1998. Hemoglobins from bacteria to man: evolution of different patterns of gene expression. J. Exp. Biol. 201 1099–1117. [DOI] [PubMed] [Google Scholar]
- Hardison, R., 2001. Organization, evolution and regulation of the globin genes, pp. 95–116 in Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management, edited by M. H. Steinberg, B. G. Forget, D. R. Higgs and R. L. Nagel. Cambridge University Press, Cambridge, UK.
- Hardison, R., and R. E. Gelinas, 1986. Assignment of orthologous relationships among mammalian α-globin genes by examining flanking regions reveals a rapid rate of evolution. Mol. Biol. Evol. 3 243–261. [DOI] [PubMed] [Google Scholar]
- Hardison, R., and W. Miller, 1993. Use of long sequence alignments to study the evolution and regulation of mammalian globin gene clusters. Mol. Biol. Evol. 10 73–102. [DOI] [PubMed] [Google Scholar]
- Hiebl, I., G. Braunitzer and D. Schneeganss, 1987. a The primary sequence of the major and minor hemoglobin components of adult Andean goose (Chloephaga melanoptera, Anatidae): the mutation Leu-Ser in position 55 of the β chains. Biol. Chem. Hoppe-Seyler 368 1385–1390. [DOI] [PubMed] [Google Scholar]
- Hiebl, I., J. Kosters and G. Braunitzer, 1987. b The primary structures of the major and minor hemoglobin component of adult goshawk (Accipiter gentilis, Accipitrinae). Biol. Chem. Hoppe-Seyler 368 333–342. [DOI] [PubMed] [Google Scholar]
- Hiebl, I., D. Schneeganss, F. Grimm, J. Kosters and G. Braunitzer, 1987. c The primary structures of the major and minor hemoglobin components of adult European black vulture (Aegypius monachus, Aegypiinae). Biol. Chem. Hoppe-Seyler 368 11–18. [DOI] [PubMed] [Google Scholar]
- Hiebl, I., R. E. Weber, D. Schneeganss, J. Kosters and G. Braunitzer, 1988. Structural adaptations in the major and minor hemoglobin components of adult Ruppell's griffon (Gyps ruepellii, Aegypiinae): a new molecular pattern for hypoxia tolerance. Biol. Chem. Hoppe-Seyler 369 217–232. [DOI] [PubMed] [Google Scholar]
- Hiebl, I., R. E. Weber, D. Schneeganss and G. Braunitzer, 1989. The primary structure and functional properties of the major and minor hemoglobin components of the adult white-headed vulture (Trigonoceps occipitalis, Aegypiinae). Biol. Chem. Hoppe-Seyler 370 699–706. [DOI] [PubMed] [Google Scholar]
- Higgs, D. R., M. A. Vickers, A. O. M. Wilkie, I.-M. Pretorius, A. P. Jarman et al., 1989. A review of the molecular genetics of the human α-globin gene cluster. Blood 73 1081–1104. [PubMed] [Google Scholar]
- Hoffmann, F. G., and J. F. Storz, 2007. The αD-globin gene originated via duplication of an embryonic α-like globin gene in the ancestor of tetrapod vertebrates. Mol. Biol. Evol. 24 1982–1990. [DOI] [PubMed] [Google Scholar]
- Hoffmann, F. G., J. C. Opazo and J. F. Storz, 2008. Rapid rates of lineage-specific gene duplication and deletion in the α-globin gene family. Mol. Biol. Evol. 25 591–602. [DOI] [PubMed] [Google Scholar]
- Hsia, C. C. W., 1998. Respiratory function of hemoglobin. N. Engl. J. Med. 338 239–247. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., 1994. The evolution of functionally novel proteins after gene duplication. Proc. R. Soc. Lond. Ser. B Biol. Sci. 256 119–124. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., 1999. Adaptive Evolution of Genes and Genomes. Oxford University Press, New York.
- Hughes, A. L., and M. Yeager, 1997. Coordinated amino acid changes in the evolution of mammalian defensins. J. Mol. Evol. 44 675–682. [DOI] [PubMed] [Google Scholar]
- Hughes, J. R., J.-F. Cheng, N. Ventress, S. Prabhakar, K. Clark et al., 2005. Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc. Natl. Acad. Sci. USA 102 9830–9835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, R. A., 1976. Enzyme recruitment in the evolution of new function. Annu. Rev. Microbiol. 30 409–425. [DOI] [PubMed] [Google Scholar]
- Jensen, R. A., and G. S. Byng, 1981. The partitioning of biochemical pathways with isozyme systems. Isozymes 5 143–174. [PubMed] [Google Scholar]
- Jobb, G., A. von Haeseler and K. Strimmer, 2004. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 4 18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kimura, M., and T. Ohta, 1974. On some principles governing molecular evolution. Proc. Natl. Acad. Sci. USA 71 2848–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshi, J. M., and R. A. Goldstein, 1996. Probabilistic reconstruction of ancestral protein sequences. J. Mol. Evol. 42 313–320. [DOI] [PubMed] [Google Scholar]
- Lalthantluanga, R., H. Wiesner and G. Braunitzer, 1985. Studies on yak hemoglobin (Bos grunniens, Bovidae): structural basis for high intrinsic oxygen affinity. Biol. Chem. Hoppe-Seyler 366 63–68. [DOI] [PubMed] [Google Scholar]
- Lam, K.-W. L., and A. Jeffreys, 2006. Processes of copy-number change in human DNA: the dynamics of α-globin gene deletion. Proc. Natl. Acad. Sci. USA 103 8921–8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder, A., D. Swan, F. Ruddle, P. D'Eustachio and P. Leder, 1981. Dispersion of α-like globin genes of the mouse to three different chromosomes. Nature 293 196–200. [DOI] [PubMed] [Google Scholar]
- Li, W.-H., 1983. Evolution of duplicate genes and pseudogenes, pp. 14–37 in Evolution of Genes and Proteins, edited by M. Nei and R. K. Koehn. Sinauer Associates, Sunderland, MA.
- Lukin, J. A., V. Simplaceanu, M. Zou, N. T. Ho and C. Ho, 2000. NMR reveals hydrogen bonds between oxygen and distal histidines in oxyhemoglobin. Proc. Natl. Acad. Sci. USA 97 10354–10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., 2007. The Origins of Genome Architecture. Sinauer Associates, Sunderland, MA.
- Lynch, M., and A. Force, 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and V. Katju, 2004. The altered evolutionary trajectories of gene duplicates. Trends Genet. 20 544–549. [DOI] [PubMed] [Google Scholar]
- Lynch, M., M. O'Hely, B. Walsh and A. Force, 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart et al., 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19 1639–1662. [Google Scholar]
- Nagai, K., B. Luisi, D. Shih, G. Miyazaki, K. Imai et al., 1987. Distal residues in the oxygen binding site of haemoglobin studied by protein engineering. Nature 329 858–860. [DOI] [PubMed] [Google Scholar]
- Nagel, R. L., and M. H. Steinberg, 2001. Hemoglobins of the embryo and fetus and minor hemoglobins of adults, pp. 197–230 in Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management, edited by M. H. Steinberg, B. G. Forget, D. R. Higgs and R. L. Nagel. Cambridge University Press, New York.
- Ohno, S., 1970. Evolution by Gene Duplication. Springer-Verlag, New York.
- Ohno, S., U. Wolf and N. B. Atkin, 1968. Evolution from fish to mammals by gene duplication. Hereditas 59 169–187. [DOI] [PubMed] [Google Scholar]
- Ohta, T., 1990. How gene families evolve. Theor. Popul. Biol. 37 213–219. [DOI] [PubMed] [Google Scholar]
- Ohta, T., 2000. Evolution of gene families. Gene 259 45–52. [DOI] [PubMed] [Google Scholar]
- Olson, J. S., A. J. Mathews, R. J. Rohlfs, B. A. Springer, K. D. Egeberg et al., 1988. The role of the distal histidine in myoglobin and hemoglobin. Nature 336 265–266. [DOI] [PubMed] [Google Scholar]
- Orgel, L. E., 1977. Gene-duplication and the origin of proteins with novel functions. J. Theor. Biol. 67: 773. [DOI] [PubMed]
- Otto, S. P., and P. Yong, 2002. The evolution of gene duplicates, pp. 451–483 in Homology Effects, edited by J. C. Dunlap and C.-T. Wu. Academic Press, San Diego. [DOI] [PubMed]
- Ovcharenko, I., G. G. Loots, B. M. Giardine, M. M. Hou, J. Ma et al., 2005. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 15 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz, M. F., 1983. Species adaptation in a protein molecule. Mol. Biol. Evol. 1 1–28. [DOI] [PubMed] [Google Scholar]
- Perutz, M. F., 2001. Molecular anatomy and physiology of hemoglobin, pp. 174–196 in Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management, edited by M. H. Steinberg, B. G. Forget, D. R. Higgs and R. L. Nagel. Cambridge University Press, Cambridge, UK.
- Phillips, S. E. V., and B. Schoenborn, 1981. Neutron diffraction reveals oxygen-histidine hydrogen bond in oxymyoglobin. Nature 292 81. [DOI] [PubMed] [Google Scholar]
- Piatigorsky, J., 2007. Gene Sharing and Evolution: The Diversity of Protein Functions. Harvard University Press, Cambridge, MA.
- Piatigorsky, J., and G. J. Wistow, 1991. The recruitment of crystallins: new functions precede gene duplication. Science 252 1078–1079. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N. J., A. Gil and T. Maniatis, 1982. The structure of the human ζ-globin gene and a closely linked, nearly identical pseudogene. Cell 31 553–563. [DOI] [PubMed] [Google Scholar]
- Proulx, S. R., and P. C. Phillips, 2006. Allelic divergence precedes and promotes gene duplication. Evolution 60 881–892. [PubMed] [Google Scholar]
- Raes, J., and Y. Van de Peer, 2003. Gene duplication, the evolution of novel gene functions, and detecting functional divergence of duplicates in silico. Appl. Bioinformatics 2 91–101. [PubMed] [Google Scholar]
- Ramsdell, C. M., E. L. Thames, J. L. Weston and M. J. Dewey, 2006. Development of a deer mouse whole-genome radiation hybrid panel and comparative mapping of Mus chromosome 11 loci. Mamm. Genome 17 37–48. [DOI] [PubMed] [Google Scholar]
- Reynafarje, C., J. Faura, D. Villavicencio, A. Curaca, B. Reynafarje et al., 1975. Oxygen transport of hemoglobin in high-altitude animals (Camelidae). J. Appl. Physiol. 38 806–810. [DOI] [PubMed] [Google Scholar]
- Sarkar, M., R. N. Pal, D. N. Das, D. B. Mondal, T. K. Mohanty et al., 1999. Postnatal persistence of foetal haemoglobin in yaks. Aust. Vet. J. 77 190. [DOI] [PubMed] [Google Scholar]
- Sawyer, S. A., 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6 526–536. [DOI] [PubMed] [Google Scholar]
- Schwartz, S., Z. Zhang, K. A. Frazer, A. Smit, C. Riemer et al., 2000. PipMaker: aWeb server for aligning two genomic DNA sequences. Genome Res. 10 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede, T., J. Kopp, N. Guex and M. C. Peitsch, 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaanan, B., 1980. The crystal structure of oxyhaemoglobin at 2.1 A resolution. J. Mol. Biol. 142: 531. [DOI] [PubMed]
- Snyder, L. R. G., 1981. Deer mouse hemoglobins: Is there genetic adaptation to high altitude? Bioscience 31 299–304. [Google Scholar]
- Spofford, J. B., 1969. Heterosis and the evolution of duplications. Am. Nat. 103 407–432. [Google Scholar]
- Stoltzfus, A., 1999. On the possibility of constructive neutral evolution. J. Mol. Evol. 49 169–181. [DOI] [PubMed] [Google Scholar]
- Storz, J. F., 2007. Hemoglobin function and physiological adaptation to hypoxia in high-altitude mammals. J. Mammal. 88 24–31. [Google Scholar]
- Storz, J. F., M. Baze, J. L. Waite, F. G. Hoffmann, J. C. Opazo et al., 2007. a Complex signatures of selection and gene conversion in the duplicated globin genes of house mice. Genetics 177 481–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz, J. F., S. J. Sabatino, F. G. Hoffmann, E. J. Gering, H. Moriyama et al., 2007. b The molecular basis of high-altitude adaptation in deer mice. PloS Genet. 3(e45): 448–459. [DOI] [PMC free article] [PubMed]
- Tanaka, T., and M. Nei, 1989. Positive Darwinian selection observed at the variable-region genes of immunoglobulins. Mol. Biol. Evol. 6 447–459. [DOI] [PubMed] [Google Scholar]
- Tufarelli, C., R. Hardison, W. Miller, J. Hughes, K. Clark et al., 2004. Comparative analysis of the alpha-like globin clusters in mouse, rat, and human chromosomes indicates a mechanism underlying breaks in conserved synteny. Genome Res. 14 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek, Z., F. Kreuzer and L. Hoofd, 1973. Advantage or disadvantage of a decrease of blood oxygen affinity for tissue oxygen supply at hypoxia. A theoretical study comparing man and rat. Pflügers Arch. 342: 185–187. [DOI] [PubMed]
- Valdar, W. S., 2002. Scoring residue conservation. Proteins 48 227–241. [DOI] [PubMed] [Google Scholar]
- van Vliet, G., and T. H. Huisman, 1964. Changes in the haemoglobin types of sheep as a response to anaemia. Biochem. J. 93 401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, B., 2003. Population-genetic models of the fates of duplicate genes. Genetica 118 279–294. [PubMed] [Google Scholar]
- Weber, R. E., 2000. Adaptations for oxygen transport: lessons from fish hemoglobins, pp. 23–37 in Hemoglobin Function in Vertebrates: Molecular Adaptation in Extreme and Temperate Environments, edited by G. Di Prisco, B. Giardina and R. E. Weber. Springer-Verlag, New York.
- Weber, R. E., 2007. High-altitude adaptations in vertebrate hemoglobins. Respir. Physiol. Neurobiol. 158 132–142. [DOI] [PubMed] [Google Scholar]
- Weber, R. E., I. Hiebl and G. Braunitzer, 1988. a High altitude and hemoglobin function in the vultures Gyps rueppellii and Aegypius monachus. Biol. Chem. Hoppe-Seyler 369 233–240. [PubMed] [Google Scholar]
- Weber, R. E., R. Lalthantluanga and G. Braunitzer, 1988. b Functional characterization of fetal and adult yak hemoglobins: an oxygen binding cascade and its molecular basis. Arch. Biochem. Biophys. 263 199–203. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1995. A space-time process model for the evolution of DNA sequences. Genetics 139 993–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15 568–573. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 2007. PAML4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and R. Nielsen, 1998. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J. Mol. Evol. 46 409–418. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and R. Nielsen, 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17 32–43. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and R. Nielsen, 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19 908–917. [DOI] [PubMed] [Google Scholar]
- Yang, Z. H., W. S. W. Wong and R. Nielsen, 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22 1107–1118. [DOI] [PubMed] [Google Scholar]
- Zhang, J., 2003. Evolution by gene duplication. Trends Ecol. Evol. 18 292–298. [Google Scholar]
- Zhang, J. Z., R. Nielsen and Z. H. Yang, 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22 2472–2479. [DOI] [PubMed] [Google Scholar]
- Zimmer, E. A., S. L. Martin, S. M. Beverley, Y. W. Kan and A. C. Wilson, 1980. Rapid duplication and loss of genes coding for the α-chains of hemoglobin. Proc. Natl. Acad. Sci. USA 77 2158–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]