Abstract
Nonhomologous end joining (NHEJ) is an important DNA double-strand-break (DSB) repair pathway that requires three protein complexes in Saccharomyces cerevisiae: the Ku heterodimer (Yku70-Yku80), MRX (Mre11-Rad50-Xrs2), and DNA ligase IV (Dnl4-Lif1), as well as the ligase-associated protein Nej1. Here we use chromatin immunoprecipitation from yeast to dissect the recruitment and release of these protein complexes at HO-endonuclease-induced DSBs undergoing productive NHEJ. Results revealed that Ku and MRX assembled at a DSB independently and rapidly after DSB formation. Ligase IV appeared at the DSB later than Ku and MRX and in a strongly Ku-dependent manner. Ligase binding was extensive but slightly delayed in rad50 yeast. Ligase IV binding occurred independently of Nej1, but instead promoted loading of Nej1. Interestingly, dissociation of Ku and ligase from unrepaired DSBs depended on the presence of an intact MRX complex and ATP binding by Rad50, suggesting a possible role of MRX in terminating a NHEJ repair phase. This activity correlated with extended DSB resection, but limited degradation of DSB ends occurred even in MRX mutants with persistently bound Ku. These findings reveal the in vivo assembly of the NHEJ repair complex and shed light on the mechanisms controlling DSB repair pathway utilization.
NONHOMOLOGOUS end joining (NHEJ) is a principal mechanism for repairing DNA double-strand breaks (DSBs) in which the two DSB ends are directly rejoined (Wilson 2007). As such, NHEJ is critical for maintaining genome stability. Many NHEJ proteins are known, but how they cooperate to execute repair in a living cell is poorly understood. In the model organism Saccharomyces cerevisiae, three major preformed protein complexes are required for all NHEJ reactions: Ku, the MRX complex, and DNA ligase IV (Dudasova et al. 2004; Daley et al. 2005). Yku70 and Yku80 form the yeast Ku heterodimer, which by homology with human Ku is inferred to form a ring that binds DNA by sliding a DSB end through its opening (Walker et al. 2001). This binding is a principal means of DSB recognition that is critical for NHEJ, but, interestingly, unimportant for the competing homologous recombination (HR) pathway.
Mre11, Rad50, and Xrs2 (Nbs1 in mammalian cells) form the MRX complex (Usui et al. 1998), which also binds DNA but without a requirement for DSB ends as with Ku (Trujillo et al. 2003). Also unlike Ku, MRX plays a role in HR, possibly regulating repair pathway utilization through actions in 5′ resection of DSB ends (Connelly and Leach 2002; Symington 2002). Rad50 is composed of two globular ATPase domains separated by a long coiled-coil region that self-associates at its end (Anderson et al. 2001; Wiltzius et al. 2005). Mre11 has an N-terminal nuclease domain, which binds near the Rad50 ATPase to create a DNA-binding head (Usui et al. 1998; Hopfner et al. 2001). Mre11 is also required for interaction between Rad50 and Xrs2 (Usui et al. 1998; Chen et al. 2001). Xrs2 harbors N-terminal FHA and BRCT domains and a C-terminal Mre11-binding domain (Shima et al. 2005) and is also required for efficient DNA binding by MRX (Trujillo et al. 2003).
Yeast DNA ligase IV is composed of Dnl4 (homologous to human Lig4) (Wilson et al. 1997) and Lif1 (XRCC4 in humans) (Herrmann et al. 1998). Dnl4 is a typical ATP-dependent DNA ligase with tandem C-terminal BRCT domains that interact with a coiled-coil region of Lif1 (Dore et al. 2006). This interaction is strong and physically stabilizes Dnl4 (Herrmann et al. 1998), but further actions of Lif1 are enigmatic. A third ligase-associated protein is Nej1 (XLF/Cernunnos in humans) (Revy et al. 2006), which plays a further poorly defined supporting role through less stable interactions with the globular head of Lif1 (Frank-Vaillant and Marcand 2001). Unlike Ku and MRX, which function in telomere maintenance (Boulton and Jackson 1998; Tsukamoto et al. 2001), and, in the case of MRX, HR and DNA damage checkpoints (Sung et al. 2000; D'Amours and Jackson 2002), the only known function of DNA ligase IV is NHEJ. Indeed, facilitation of ligase IV action is the unifying objective of NHEJ.
Several interactions between these protein complexes have been identified and characterized, specifically between Yku80-Dnl4, Xrs2-Lif1, Mre11-Yku80, and Lif1-Nej1 (Palmbos et al. 2005; Deshpande and Wilson 2007). However, the order of their assembly onto DSB ends, their relationship and interdependency during binding, and their dissociation after repair are poorly described. Here, we used the HO endonuclease to generate a single DSB in gene promoters, guided by known nucleosome positions in these regions. Chromatin immunoprecipitation (ChIP) and physical analysis of DSB status were combined to correlate protein recruitment with the time course of DSB formation and subsequent repair. The data support a model in which Ku and MRX bind independently and rapidly to a DSB and then coordinately recruit DNA ligase IV, which itself helps load Nej1, with Ku having the greater role in ligase binding. Interestingly, NHEJ protein dissociation also appears to be an active process that depends on an intact MRX complex and ATP binding by Rad50, implying a role of MRX in repair pathway switching. The collected findings define the time course of many early events in the life of DSB.
MATERIALS AND METHODS
Yeast strains:
All yeast strains were isogenic derivatives of BY4741 (Brachmann et al. 1998). Genotypes are listed in supplemental Table S1. Gene disruptions and modified alleles were made using a PCR-mediated technique (Brachmann et al. 1998) or a URA3 pop-in/pop-out method (McCormick et al. 1995; Peterson et al. 1995). Disruptions were confirmed by PCR and cut site and competitive quantitative PCR (CQ–PCR) control alleles by sequencing (see the supplemental Methods for details of allele construction). All strains were grown at 30°. Media were as described (Karathanasis and Wilson 2002).
Survival assay:
Overnight cultures in rich dextrose liquid medium (YPAD) were inoculated into YPA medium with 3% glycerol as the carbon source and grown overnight to a final OD600 of 0.3–0.6. Galactose was then added to 2% final concentration to induce HO expression. At varying times after induction aliquots were serially diluted, plated to YPAD medium, and incubated at 30° for 3 days. Survival rate was measured as the ratio of corrected colony counts at each time point to corrected counts before galactose addition.
Epitope tagging:
13Myc and 3HA tagging of target proteins was performed with a PCR-based recombination technique using plasmids pFA6a-13Myc-His3MX6 and pFA6a-3HA-kanMX6 (Longtine et al. 1998). 3FLAG tagging involved an overlapping PCR and homologous recombination method. 3FLAG sequence with a stop codon was designed into three overlapping primers and fused to LEU2 by three sequential PCR reactions. The final PCR product contained 45-bp tails to drive its homologous integration upon transformation after the last YKU80 codon with a downstream LEU2 marker. All tagged alleles were confirmed with PCR, sequencing, and Western blot.
DSB induction:
For the GAL1-cs, overnight cultures in YPAD liquid medium were inoculated into YPA medium with 3% glycerol as the carbon source and grown overnight to a final OD600 of 0.6–0.8. Galactose was then added to 2% final concentration to induce HO expression. After a 60-min incubation at 30°, 200 ml of cells were spun down and resuspended into the same volume of YPAD. After various times of further incubation, 30-ml samples (∼3 × 108 cells) were withdrawn for ChIP and DSB analyses. The time 0 sample was taken just before galactose addition. The 60-min sample was taken when galactose medium was exchanged with YPAD. The ILV1-cs experiment was similar except that no glucose medium exchange was used so that HO expression was maintained throughout. In the G1 arrest experiment, bar1 GAL1-cs strains (supplemental Table S1) were arrested by adding α-factor to glycerol cultures at 50 ng/ml final concentration, followed by continued incubation for 4 hr. Galactose was then added and the experiment continued as above. α-Factor was maintained in the culture after glucose addition at 60 min. Cultures were verified by microscopy to have at least 98% unbudded/shmoo cells throughout the experiment.
Chromatin immunoprecipitation:
Cell lysis and chromatin immunoprecipitation were performed as described (Aparicio et al. 2005). Input DNAs, i.e., after chromatin preparation but before immunoprecipitation, were split and used for ChIP as well as parallel CQ–PCR analyses with the primer pairs for both ChIP and DSB monitoring. Immunoprecipitation antibodies were anti-FLAG (M2) (Sigma-Aldrich), c-Myc (9E10), and HA (F-7) (Santa Cruz Biotechnology).
CQ–PCR:
In CQ–PCR, the same set of primers is used for amplification of both the target and an internal control template (supplemental Figure S4). Specifically, the internal control, a piece of AmpR flanked by 19- to 23-bp sequences common to promoter containing the DSB, was integrated into a different chromosome of the assay strains as described in the supplemental Methods. Allele pairings were designed so that the target and internal control PCR products were the same size (±5%) and had the same G/C content (±1%) to ensure similar amplification efficiency. To distinguish the two PCR products upon electrophoresis, they were digested after PCR with BanI restriction endonuclease, which cuts only the control product into smaller fragments. For DSB monitoring, the two primers flanked the HO cut site so that DSB presence led to a decrease in the target-to-control ratio. For ChIP, the primers were on the same side of the cut site to reveal the binding of proteins to the DSB end by enrichment of target PCR product over control. CQ–PCR primers are shown in supplemental Figures S1 and S2. See the supplemental Methods for details of the PCR reactions, BanI digestion, polyacrylamide gel electrophoresis, and product quantification.
Southern blotting:
Genomic DNA extracted at different time points after HO induction in the GAL1 cut-site system was digested with HindIII, separated on a 0.8% agarose gel, and transferred to Zeta-Probe membrane (Bio-Rad, Hercules, CA). The blot was simultaneously probed with two 32P-labeled DNA fragments, one located on one side of the HO cut site on chromosome II and one within the APN1 gene on chromosome XI. The relative amount of DNA in each band was determined using a Typhoon phosphorimager. Disappearance and reappearance of the intact HO cut-site-containing fragment (2.6 kb), expressed as a ratio relative to the APN1 control (3.5 kb), reveals DSB formation and subsequent NHEJ. Disappearance of the HO-cut fragment (1.1 kb) reveals the combined effects of NHEJ repair and DSB resection.
Ligation-mediated PCR:
Five microliters of input DNA from the ChIP assay or genomic DNA at 5 ng/μl was mixed in a 20-μl reaction with 10 pmol preannealed adaptor (OW2797 + OW2798) and 400 units T4 DNA ligase (NEB) in the supplied buffer and incubated at 16° overnight. OW2797 is 5′-TTCCGGCTGGCTGGTTTATTGTGTT and OW2798 is 5′-CAATAAACCAGCCAGCCGGAA, which upon annealing form a TGTT 3′ overhang complementary to the HO-generated overhang. Ligation products were then diluted 10-fold and 1 μl was used for PCR with primers OW2502 (see supplemental Figure S2) and OW2796 (complement of OW2798 and also a reverse primer in the ChIP AmpR control allele; see supplemental Figure S2). The PCR reaction gives rise to two products, the ligation-mediated PCR (LM–PCR) product (275 bp) and an internal control product amplified from the ChIP AmpR control allele (197 bp). The PCR products were separated on an acrylamide gel and quantified (see supplemental Methods). The level of HO-induced DSBs capable of being ligated to the adapter was expressed as a ratio of LM–PCR product to the internal control.
RESULTS
Generating DSBs in gene promoters:
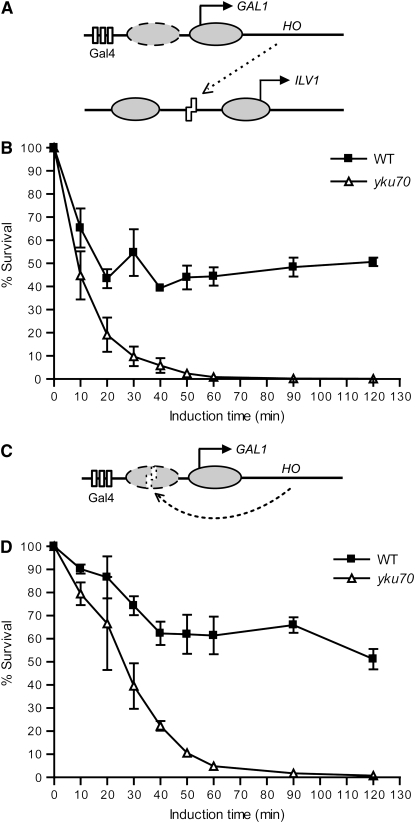
We sought to correlate DSB formation and subsequent repair with NHEJ protein binding at the same site, which required DSBs informative for the pre- to post-NHEJ phases. Mega-endonucleases have been extensively used to generate DSBs in chromosomes (Haber 1995) and are clearly influenced by chromatin status, since the HO endonuclease will not cleave at HML and HMR (Weiss and Simpson 1998; Ravindra et al. 1999). We reasoned that placing cut sites in other chromosomal positions that were either nucleosome-bound or nucleosome-free could similarly regulate mega-endonuclease cutting. Consensus cut sites were inserted into the well-studied promoters of GAL1 and ILV1 (supplemental Figures S1 and S2). A GAL1 cut site (GAL1-cs) was placed within a positioned nucleosome that is bound when the promoter is inactive. This nucleosome becomes undetectable when GAL1 is activated by galactose (Reagan and Majors 1998; Li and Smerdon 2002), suggesting a means of regulating cut-site accessibility. The sequence and length of the replaced region are otherwise unimportant for GAL1 function (Reagan and Majors 1998). In contrast, an ILV1 cut site (ILV1-cs) was placed in a constitutively nucleosome-free promoter region, where again precise sequence and length are unimportant (Moreira et al. 2002). Consistent with our hypothesis, constitutive expression of HO or I-SceI mega-endonucleases killed yku70 yeast bearing an ILV1-cs, but only killed yku70 yeast bearing a GAL1-cs when the promoter was active and open, and even then less robustly (supplemental Figure S3 and data not shown).
The above results suggested the DSB systems used in further experiments. Specifically, the HO coding sequence was placed under control of the native GAL1 promoter in chromosome II in strains with either ILV1 or GAL1 cut sites and an uncleavable MATa-inc allele (Figure 1, A and C). In the GAL1-cs system, the cut site was in the same promoter that controlled HO expression. This GAL1-cs is protected by a nucleosome until the promoter is activated by galactose, expression of the endonuclease terminates upon DSB formation, and addition of glucose has the dual shut-off effect of ending endonuclease expression and making the cut site inaccessible to recleavage. To estimate the efficiency of DSB formation and repair in the two systems, we first performed survival assays after transient HO induction in wild-type and yku70 yeast. Essentially, any DSB will be lethal in this NHEJ-deficient mutant, thus revealing the extent of cutting, while the strain difference indicates the extent of repair in wild type. HO-generated DSBs correlated with galactose exposure time in each system, but DSB induction and repair were not equally efficient (Figure 1, B and D). More than 90% of cells were killed in the ILV1-cs yku70 strain after 30 min of HO induction, while only ∼60% were killed in the GAL1-cs yku70 strain in the same time. In wild type, the ILV1-cs system plateaued at ∼40% survival over a 2-hr induction, while the GAL1-cs system showed higher (∼60%) survival. Further evidence that the extensive survival after glucose addition in the GAL1-cs system was due to NHEJ is that, in parallel experiments, the plating efficiency to raffinose–galactose medium was only 0.0016%, because in such systems multiple rounds of cleavage greatly reduce survival. The GAL1-cs system was thus adopted for dissecting NHEJ protein binding and dissociation at a DSB, due to its better HO shut off and associated repair capacity. For detecting the assembly order of NHEJ complexes, we used the ILV1-cs system for its faster cutting.
Figure 1.—
DSB systems used in this study. (A) DSB induction paradigm for ILV1-cs strains. Ovals represent nucleosomes, arrows represent transcription start sites, rectangles represent Gal4-binding sites, and zigzags represent the HO recognition site. The dashed nucleosome leaves the GAL1 promoter upon activation by galactose (Boeger et al. 2003; Reinke and Horz 2003). See text for details. (B) Survival assay of wild-type and yku70 mutant ILV1-cs strains. Yeast cells grown in galactose liquid medium for the indicated times were plated on glucose medium and incubated at 30° for 3 days. Colony counts at each time point are expressed as a percentage of the value at time 0. Data points are the mean ± SEM of three independent experiments. (C) DSB induction paradigm for GAL1-cs strains, similar to A. (D) Survival assay of GAL1-cs strains, similar to B. Time-dependent killing and repair in wild type are evident in both systems, but with slower cutting and more extensive repair with the GAL1-cs.
Ku binding is independent of MRX and DNA ligase IV, but Ku dissociation requires MRX:
We used a CQ–PCR approach for analyzing both DSB formation and DNA recovered by ChIP (see materials and methods and supplemental Figure S4). Briefly, DSB formation is revealed as loss of a PCR product using primers that flank the HO cut site, while specific protein binding in ChIP is detected by enrichment of a PCR product adjacent to the cut site, each in comparison to an internal control amplified with the same primers. For ChIP, 13Myc epitope tags were added to the C termini of Yku80, Xrs2, and Dnl4 to represent the various NHEJ complexes. The resulting proteins were fully functional for NHEJ, as strains containing these alleles displayed the same survival curve as wild type (data not shown). To dissect the interdependency of binding of these complexes, we further deleted individual representative genes (yku70, rad50, or dnl4).
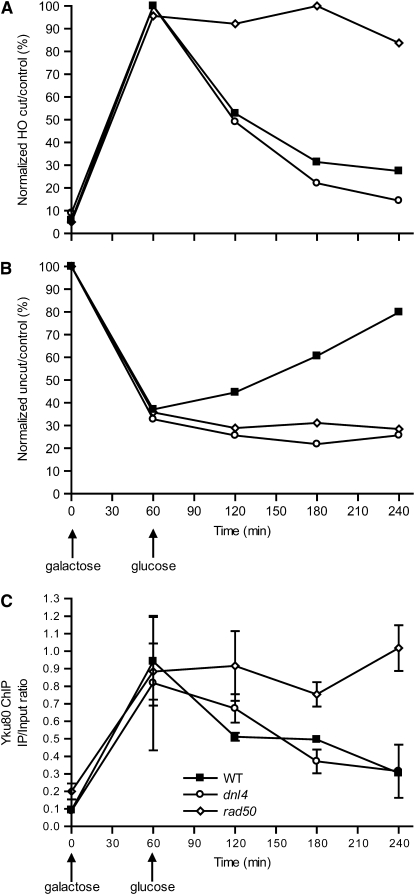
For our first GAL1-cs experiments, HO expression was induced with galactose for 60 min and then repressed with glucose to follow repair and associated protein binding over time. Yku80 ChIP was performed in wild-type, rad50, and dnl4 null mutant strains. For correlation, we first determined the kinetics of DSB formation and repair (Figure 2A). After 60 min of HO induction, ∼70% of NHEJ-deficient rad50 and dnl4 cells had broken chromosomes, whereas only 50% of chromosomes were broken in wild type. This difference likely reflects the competition of cutting and early repair in wild type. After HO shut off, wild-type yeast repaired 50% of the broken chromosomes over the next 60 min and an additional 10% during the next 120 min. The rest of the DSBs were not repaired. As expected, rad50 and dnl4 strains were grossly repair deficient. The slow increase of intact cut sites in these strains could be due to either alternative repair pathways or, more likely, growth of cells that did not suffer a DSB. Indeed, such growth could also explain the late increase of intact cut sites in wild type, so that most NHEJ occurred in the hour following HO shut off, with a restoration half-time of ∼25 min.
Figure 2.—
Appearance and disappearance of Yku80, Xrs2, and Dnl4 at a DSB. A DSB was induced in the GAL1 promoter (see Figure 1C) in various yeast strains by a 60-min galactose exposure followed by transfer to glucose to terminate HO expression. (A, C, and E) Chromatin fractions were prepared at different time points and portions used in the CQ–PCR assay to follow DSB formation and subsequent repair. (B, D, and F) Separate portions of the chromatin preparations were subjected to ChIP analysis using antibodies directed against tagged Yku80 (B), Xrs2 (D), and Dnl4 (F). Specific binding of each protein to the DSB is expressed as the immunoprecipitation-to-input PCR ratio as described in materials and methods. The complete experiment was performed at least two times for each strain with similar results; one representative experiment is shown. The mean ± SEM of two separate CQ–PCR reactions from the input DNA (A, C, and E) or ChIP samples (B, D, and F) of that experiment are shown.
The NHEJ repair period correlated precisely with the time of peak Yku80 ChIP signal in the wild-type and dnl4 strains, importantly including the appearance of Yku80 at the DSB, its persistence during repair, and its disappearance after repair (Figure 2B). Ku binding was not dependent on either MRX or DNA ligase IV, as demonstrated by similar Yku80 ChIP during the early time points in rad50 and dnl4 yeast. However, dissociation of Ku from the DSB was markedly dependent on MRX, but not DNA ligase IV, since only rad50 mutation caused persistent Yku80 ChIP signal at later time points, a phenomenon examined in more detail below.
Recruitment and dissociation of MRX are largely independent of Ku and DNA ligase IV:
Similar ChIP analysis of Xrs2 showed that this protein, and presumably MRX, was recruited to the GAL1-cs DSB at early time points, with a peak corresponding to peak NHEJ repair (Figure 2, C and D). It was again dissociated at later time points, similar to Ku. Examining yku70 and dnl4 mutant strains revealed a moderate decrease in the maximal intensity of Xrs2 binding at the earliest time points, but binding nonetheless occurred and was indistinguishable by 30 min after HO shut off. This suggests a possible early role of Ku and DNA ligase IV in promoting MRX binding in the early NHEJ phase, a role that becomes less important at later times. Accordingly, Xrs2 dissociation was equivalent in wild-type, yku70, and dnl4 strains. Thus, neither recruitment nor dissociation of MRX at a DSB strictly requires the other NHEJ complexes, but early binding appears to be enhanced by these factors.
Recruitment of Dnl4 requires Ku, but its dissociation requires MRX:
Similar to Yku80 and Xrs2, Dnl4 was recruited to the GAL1-cs DSB at early time points and left the site after repair in wild-type cells (Figure 2, E and F). In the yku70 strain, Dnl4 binding was markedly reduced throughout the time course. Thus, Ku is required for the binding of Dnl4 to a DSB during NHEJ, consistent with prior observations and known interactions between Ku and DNA ligase IV (Hsu et al. 2002; Palmbos et al. 2005; Zhang et al. 2007). In contrast, Dnl4 binding was excessive and persistent at later times in the rad50 strain, demonstrating that MRX is essential for the dissociation of DNA ligase IV from the DSB just as it was for Ku. There was a slight reduction in Dnl4 binding at the earliest time point in the rad50 mutant but this was not statistically significant and, importantly, possibly masked by the dissociation defect (note the very different slope of Dnl4 binding between 60 and 90 min in wild-type and rad50 yeasts). In total, the behaviors of Ku and DNA ligase IV were very similar, consistent with a close functional association between them, but with the important distinction that DNA ligase IV recruitment depended on Ku but not vice versa. As a result, we cannot distinguish whether the MRX-dependent dissociation activity is directed at both Ku and DNA ligase IV or just at Ku with DNA ligase IV following as a consequence of its dependence on Ku.
Timing of Ku, MRX, and DNA ligase IV assembly at a DSB:
The above results imply an order to NHEJ complex assembly, which we tested by constructing yeast strains in which Yku80, Xrs2, and Dnl4 were tagged with different C-terminal epitopes in the same strain (Yku80-3FLAG, Xrs2-3HA, and Dnl4-13Myc). This allowed measurement of relative binding kinetics by parallel ChIP analysis of a single chromatin fraction. Survival assays again showed these strains to have normal NHEJ efficiency and thus that the tagged proteins were functional even when combined (data not shown; see also supplemental Figure S5A). We first performed the experiment with the GAL1-cs system and observed that all three complexes assembled and dissociated at the DSB with the same apparent kinetics (supplemental Figure S5B). No recruitment order could be resolved, likely because DSB formation across the population was slower than protein binding to the DSB. We therefore repeated the experiment with the ILV1-cs system, in this case maintaining HO induction throughout the experiment. As expected from the survival curves (Figure 1B), CQ–PCR documented very rapid cutting that was essentially complete by 25 min, markedly improving the time resolution of the experiment (Figure 3A). Indeed, we now observed differences in the binding kinetics of Yku80, Xrs2, and Dnl4, with binding of Yku80 and Xrs2 preceding Dnl4 by ∼10 min throughout the time course (Figure 3B). Yku80 and Xrs2 clearly began to appear 15 min after HO induction while Dnl4 binding was not evident at this time and took 25 min to reach similar levels. Plotting normalized DSB formation and protein binding on the same graph (Figure 3C) emphasizes that Yku80 and Xrs2 binding could not be easily distinguished from each other, or from DSB formation, at the earliest time points. Thus both MRX and Ku binding are rapid events following DSB formation.
Figure 3.—
Time course of initial binding of Yku80, Xrs2, and Dnl4 at a DSB. A DSB was induced in the ILV1 promoter (see Figure 1A) in a single yeast strain in which each of Yku80, Xrs2, and Lif1 were tagged for ChIP analysis with different antibodies. HO induction was initiated with galactose at time 0 and maintained throughout the experiment. (A) DSB monitoring similar to Figure 2, A, C, and E. (B) ChIP assay of Yku80, Xrs2, and Dnl4 similar to Figure 2, B, D, and F, performed on fractions of the same chromatin preparation as in A. (C) The same data in A and B normalized to allow plotting on the same graph. DSB formation is now expressed as the percentage of alleles broken, where 100% is the maximum level of breakage observed during the experiment. ChIP data were also normalized to span from the smallest to the largest values. Dnl4 binding was delayed ∼10 min relative to Yku80 and Xrs2 binding, which themselves could not be distinguished and occurred coincidentally with DSB formation at the earliest time points.
Recruitment of Nej1 requires DNA ligase IV, but not vice versa:
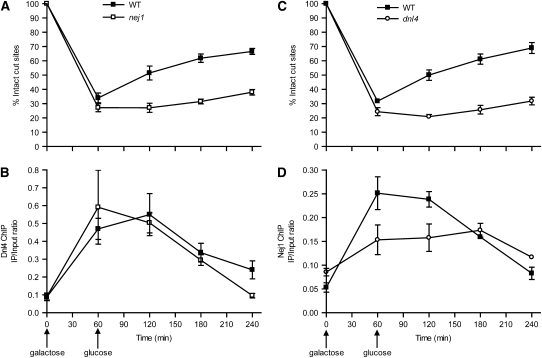
As expected, nej1 mutation also abolished NHEJ of the GAL1-cs DSB (Figure 4A). One possible explanation for this is that Nej1 might assist in the binding of Dnl4 to the DSB. However, Dnl4 binding was unchanged in the nej1 mutant (Figure 4B). Ku binding is inferred to also be normal from this result, since failure of Ku recruitment would be inconsistent with normal ligase recruitment (see above). It is thus likely that the remainder of the NHEJ complex assembles but is nonfunctional in a nej1 mutant, indicating that Nej1 must fulfill some function downstream of ligase binding. To study Nej1 binding, we C-terminally tagged it with a FLAG epitope and performed ChIP using the GAL1-cs system. Once again, Nej1 bound to the DSB at early times and dissociated later (Figure 4D). Here, Nej1 binding proved to be strongly decreased in a dnl4 mutant at the times corresponding to peak NHEJ in wild type (Figure 4D). Thus, Dnl4 helps recruit Nej1 to the DSB, consistent with the known Dnl4-Lif1-Nej1 interactions (Deshpande and Wilson 2007) and with an inferred downstream function of Nej1.
Figure 4.—
Dnl4 helps recruit Nej1 to a DSB, but not vice versa. (A and C) DSB monitoring in wild-type, nej1, and dnl4 strains similar to Figure 2, A, C, and E. (B and D) Dnl4 and Nej1 ChIP analysis, respectively, similar to Figure 2, B, D, and F, performed on fractions of the same chromatin preparations as in A and C.
Ku dissociation correlates with bulk resection:
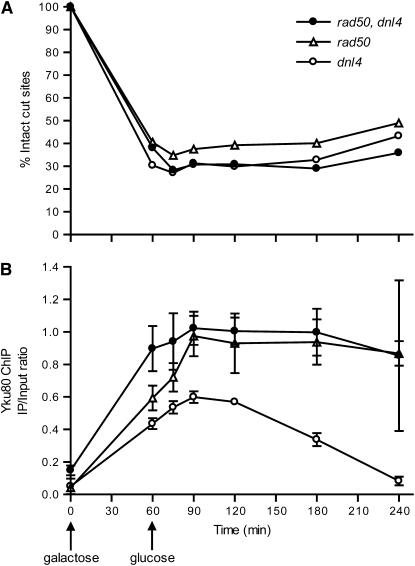
An unanticipated result in the above experiments was the persistent binding of Ku and DNA ligase IV in yeast lacking MRX. Importantly, the persistent Yku80–DSB association in rad50 cells was not simply a result of failed NHEJ since the dnl4 mutant was as repair deficient as rad50 (Figure 2A) but showed Ku binding and dissociation equivalent to wild type (Figure 2B). Moreover, Ku persistence was induced in the dnl4 background by further rad50 mutation (Figure 5B), demonstrating the specificity of the effect to MRX distinct from NHEJ and suggesting an active MRX-dependent dissociation of NHEJ proteins, at least at ends that failed NHEJ. The most obvious mechanism for this dissociation would be MRX-stimulated resection of DSB ends for HR, which seems incompatible with continued NHEJ protein binding. However, a distinct mode of NHEJ protein dissociation independent of resection was also possible, which we next attempted to distinguish.
Figure 5.—
Yku80 ChIP in rad50 dnl4 double-mutant yeast. (A) DSB monitoring in dnl4, rad50, and double-mutant strains, similar to Figure 2, A, C, and E. As expected, all strains are repair deficient. (B) Yku80 ChIP was performed in these same strains, similar to Figure 2B. rad50 mutation caused persistent binding of Yku80 even in the dnl4 background, demonstrating that this effect is specific to MRX function and not NHEJ efficiency.
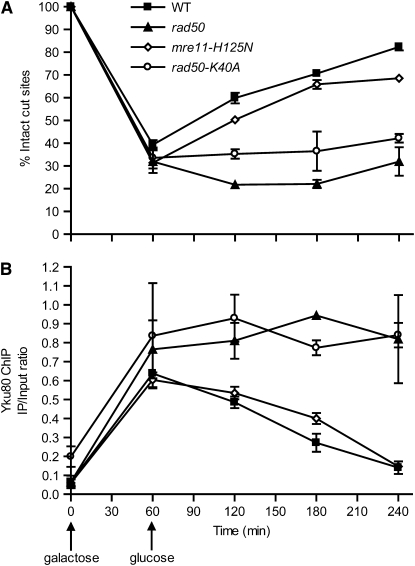
We first constructed two chromosomal MRX point mutants in the Yku80-13Myc background. rad50-K40A alters the Rad50 Walker type A ATP-binding motif. MRX containing this mutation shows defects in ATPase activity, ATP-dependent DNA unwinding, and ATP-stimulated endonuclease activities (Chen et al. 2005). mre11-H125N disrupts Mre11 nuclease activity (Moreau et al. 1999). Neither mutation impairs MRX complex formation according to in vitro biochemical assays (Chen et al. 2005; Krogh et al. 2005). The DSB repair curves of these mutants (Figure 6A) were consistent with previous reports that rad50-K40A mutation results in NHEJ deficiency while mre11-H125N cells are proficient in NHEJ (Moreau et al. 1999; Chen et al. 2005). ChIP of Yku80 demonstrated that Rad50-K40A mutation led to persistent Ku binding at the induced GAL1-cs DSB, similar to rad50 (Figure 6B), whereas mre11-H125N mutation showed similar kinetics as wild type, even though not all DSBs undergo NHEJ. These data suggest that the MRX ATPase is required for dissociation of Ku from a DSB but that the nuclease activity is not.
Figure 6.—
MRX ATP-binding activity, but not nuclease activity, is required for Ku dissociation from a DSB. (A) DSB monitoring in wild-type, mre11-H125N, and rad50-K40A yeast, similar to Figure 2, A, C, and E. (B) ChIP assay of Yku80 in parallel with A, similar to Figure 2, B, D, and F.
We next monitored resection in the GAL1-cs system, including HO shut off at 60 min to allow direct correlation of results. Resection was first monitored as disappearance of an HO-cut HindIII restriction fragment relative to a control fragment from a different chromosome. In this system, DSB repair contributes to the disappearance of the cut band in NHEJ-proficient cells (Figure 7C). This can explain the more rapid disappearance in wild type as compared to dnl4 and yku70 (Figure 7, A and B). However, rad50 mutation caused a marked further slowing of band disappearance that cannot be attributed to failed NHEJ (Figure 7, A and B). Therefore, retention of Ku at the DSB at later times appears to correlate with decreased end resection in the rad50 mutant, which is consistent either with resection causing Ku dissociation or with Ku dissociation being required for the onset of resection.
Figure 7.—
Persistent Ku binding is correlated with reduced end resection. (A) DNAs prepared from wild type, rad50, dnl4, and yku70 yeast using the same time course as in Figure 2 were digested with HindIII and subjected to Southern blot analysis with probes specific to the GAL-cs and APN1 control. Uncut and HO-cut GAL-cs bands are indicated. Bands were subsequently quantified with a phosphorimager. (B) The ratio of the HO-cut band to the APN1 control was normalized to the ratio at 60 min, the time of HO shut off when DSB formation was maximal. Disappearance of the HO-cut band at subsequent times results from NHEJ (wild-type strain only) and/or resection (all strains). (C) The ratio of uncut HO band to the APN1 control was normalized to the ratio at time 0 to allow monitoring of DSB formation and repair in a manner analogous to the CQ–PCR method in previous figures.
We next repeated the resection and Yku80 ChIP experiments in yeast arrested in G1 by α-factor, which was previously demonstrated to prevent 5′ resection (Aylon et al. 2004; Ira et al. 2004) and therefore was expected to allow study of MRX status independent of resection. rad50 cells did show a complete lack of resection in G1 (Figure 8A), more dramatic than in asynchronous cells, which correlated with persistent Ku–DSB association (Figure 8C). However, we did in fact observe resection in G1 in other strains (Figure 8A). As above, disappearance of the HO-cut band could be attributed to NHEJ in wild-type cells (Figure 8B), but band loss, presumably by resection, was as extensive in NHEJ-deficient dnl4 cells (Figure 8A). Indeed, Aylon et al. (2004) observed the same phenomenon under constitutive HO induction. Nonetheless, when comparing G1 and asynchronous wild-type and dnl4 cells, there was a modest decrease in the rate and extent of resection at the latest time points (180 and 240 min; compare Figures 7B and 8A) that corresponded to a similar relative decrease in the rate and extent of Ku loss at these times (compare Figures 2B and 8C). This tends to support the correlation between resection and Ku loss, but the extent of MRX-dependent resection observed even in G1 prevents a clear distinction of resection from other possible MRX functions in Ku dissociation.
Figure 8.—
Yku80 ChIP and Southern analysis of resection in G1 cells. (A and B) DSB analysis of resection and repair, similar to Figure 7, B and C, of wild-type, dnl4, and rad50 strains arrested in G1 by α-factor. (C) Yku80 ChIP, performed as in Figure 2, B, D, and F, except using cells maintained in G1 by α-factor arrest. MRX-dependent resection and Ku dissociation occurred in G1, but to a lesser final extent than in asynchronous cells.
We also used LM–PCR to monitor resection. This method depends on ligation of an oligonucleotide linker to the HO-cut end, similar to Frank-Vaillant and Marcand (2002), where even minimal resection would lead to impaired ligation and loss of signal. We observed, similar to published results (Frank-Vaillant and Marcand 2002), an ∼30-min period after HO shut off in all strains during which cut DSB ends remained intact, as evidenced by a stable induced LM–PCR signal (Figure 9A). The signal subsequently decreased to baseline, which again presumably reflects resection in NHEJ-deficient mutants. Most strikingly, rad50 mutants showed a pattern indistinguishable from wild-type, yku70, or dnl4 in both asynchronous (Figure 9A) and G1-arrested (Figure 9B) cultures. It thus appears that some level of initial end degradation occurs that is distinct from the more extensive resection measured by Southern blot, since the former, unlike the latter, neither requires MRX nor is correlated with the status of Ku binding at later times (compare Figures 7–9).
Figure 9.—
LM–PCR analysis of resection reveals MRX-independent nucleotide loss. Wild-type, rad50, dnl4, and yku70 cells growing either exponentially (A) or arrested by α-factor (B) were induced to express HO with galactose and repressed with glucose 60 min later. LM–PCR was performed at various times as described in materials and methods using an oligonucleotide adapter capable of being ligated to an unresected HO-cut DSB. The signal from the LM–PCR product was expressed as a ratio relative to the AmpR control product and then normalized to the maximum value for each strain. LM–PCR signal persisted for a time and then disappeared in a manner independent of cell cycle stage, NHEJ, and MRX status.
DISCUSSION
The goal of NHEJ is to direct DNA ligase IV action to at least one of the two strand breaks in a DSB, which requires coordination between the various protein components. We systematically studied NHEJ protein assembly at DSBs in vivo to establish a framework for understanding this coordination. Working in order of time, Ku and MRX each appeared at a DSB very soon after its formation (Figure 3), consistent with various observations in different species. In Chinese hamster cells, enhanced green fluorescence protein-labeled Ku80 started to accumulate at laser-damaged DNA within a few seconds (Mari et al. 2006). In yeast, YFP-Mre11 focus formation peaked within 6 min following γ-irradiation, the earliest detectable event (Lisby et al. 2004). Importantly, Ku foci at single DSBs have not been observed microscopically, indicating the importance of ChIP for exploring this question. The fact that Ku and MRX binding were nearly coincident with break formation, even though these proteins are not known to exist in a preformed complex, suggests that the binding of each might be driven by the DSB substrate. Indeed, both have inherent DNA-binding affinity, but by different mechanisms that do (Ku) (Griffith et al. 1992) or do not (MRX) (Furuse et al. 1998) require a free DSB end. Independent Ku binding was confirmed by its normal recruitment in a rad50 mutant (Figure 2B). MRX also bound to a DSB in a yku70 mutant, ultimately achieving levels equivalent to wild type (Figure 2D). However, the observed delay in maximal MRX binding in the absence of Ku suggests a role of Ku in stabilizing MRX for NHEJ, as also concluded in a recent report (Zhang et al. 2007). A difference is that Zhang et al. (2007) observed decreased MRX binding at all time points in a Ku mutant, perhaps because they used constitutive HO induction for 2 hr while we monitored a DSB for 3 hr after HO shut off. Indeed, a key feature of our study was the use of cells executing productive NHEJ in a time frame suitable for examining all repair phases, including NHEJ protein dissociation and transition to HR.
Very rapid Ku and MRX binding is somewhat difficult to reconcile with a recent suggestion that RSC-mediated chromatin remodeling around the MAT locus is required to prepare a DSB for Ku and MRX binding (Shim et al. 2007). Activation of RSC and movement of nucleosomes would take time and create a delay in dependent downstream events, a delay that we would expect to have observed in the ILV1-cs system. Indeed, it was noted that RSC, Ku, and Mre11 arrive at a DSB at “almost the same time” (Shim et al. 2007), and RSC recruitment is in fact impaired in Ku- and MRX-deficient strains (Shim et al. 2005), making a simple linear pathway difficult to reconcile. Given the special status of MAT with regard to DSB formation, it is further possible that chromatin regulation there represents a special case not common to all DSBs. Alternatively, the relatively large nucleosome-free region in the ILV1 promoter (Moreira et al. 2002) might minimize the need for chromatin remodeling, although rapid Ku binding has been observed in other systems as summarized above. It is of considerable interest to continue to explore the influence of DSB location and chromatin status on NHEJ protein recruitment. Our non-MAT systems with well-described chromatin status should prove useful for this purpose.
In contrast to Ku and MRX, DNA ligase IV appearance at a DSB was measurably delayed relative to break formation (Figure 3C). Notably, this was observed only with the ILV1-cs, emphasizing the importance of rapid DSB kinetics when following events that occur rapidly, since slow DSB formation limits time resolution. It is noteworthy that the ∼10-min delay in initial ligase binding was slower than the observed ∼25-min DSB resolution half-time (Figure 2A). This difference might suggest that events other than ligase binding, such as synapsis of ends, are rate limiting in NHEJ. However, recleavage by HO would confound this interpretation. We minimized this problem by the glucose-dependent switch to HO-cut-site inaccessibility in the GAL1-cs system (Figure 1; supplemental Figure 3), as judged by the fact that no new DSBs were formed after glucose addition and by the robust early NHEJ repair that could be detected in the GAL1-cs (Figure 2A) but not in the ILV1-cs system (not shown). Nonetheless, we do not know the speed of rechromatinization at repaired DSBs, and thus the extent of GAL1-cs recleavage.
The delay in ligase binding suggests that it might be dependent on the Ku and/or MRX proteins that arrive first, despite the fact that Lif1 has inherent DNA-binding affinity (Modesti et al. 1999). We recently described Yku80-Dnl4 and Xrs2-Lif1 protein contacts (Palmbos et al. 2005), one or both of which could account for active ligase recruitment. However, we and others have consistently observed that ligase binding at a DSB depends strongly on Ku (Figure 2F) (Nick McElhinny et al. 2000; Calsou et al. 2003; Zhang et al. 2007), while MRX loss caused at most a delay in ligase binding (Figure 2F). This suggests that Ku is the dominant player in ligase recruitment.
Although ligase binds later than Ku and MRX, it might have reciprocal influences on their status and/or stability. Regarding Ku, Zhang et al. (2007), using both in vitro and noncrosslink ChIP approaches, demonstrated that the stability of Ku at a DSB is reduced in the absence of Dnl4 or Lif1. We did not observe any reduction in Ku binding in dnl4 strains, and binding was robust and persistent in the absence of both MRX and Dnl4 (Figure 5). However, crosslink ChIP may be insensitive to a possible rapid equilibrium of Ku entry and exit, whether by dissociation or translocation along the DNA. Regarding MRX, we did observe a paradoxical reduced intensity of early MRX binding in dnl4 yeast (Figure 2D) even though ligase bound later (Figure 3). These data need not be conflicting, given that our experiments are population measurements, so that in the slower-cutting GAL1-cs system cells in many different states are being simultaneously assessed. Thus, ligase promotion of early MRX stability, or perhaps of a distinct mode of MRX binding productive for NHEJ, might be evident as a delay in maximal MRX signal. Normal MRX binding at later times when ligase is excluded from DSB ends (Figure 2, D and F) would reflect a different mode of binding relevant to events such as resection.
Continuing the NHEJ reaction sequence, ligase presence at the DSB is required for productive engagement of Nej1 (Figure 4D). Although we did not explicitly measure the timing of Nej1 binding, and anticipate that it would be similar to Dnl4, our results nonetheless conceptually place Nej1 downstream of Ku and ligase. Ku and ligase bind regardless of Nej1, but for unknown reasons cannot complete the reaction without Nej1. These results agree with Nej1-independent binding of Lif1 to chromatin (Ahnesorg and Jackson 2007), corroborating them for Dnl4 binding to a fixed DSB, and refute the suggestion that Dnl4-Lif1 does not enter the nucleus in the absence of Nej1 (Valencia et al. 2001). They also agree with findings that mammalian XLF relies on DNA ligase IV to mobilize to damaged chromatin, but is not required for the recruitment of other NHEJ proteins (Wu et al. 2007).
As expected, NHEJ proteins leave DSB sites soon after repair is completed (Figures 2 and 4). In some cases, this might represent simple product dissociation, but Ku in particular would appear to require active removal, given the “ring a string” manner in which it binds DNA (Walker et al. 2001). Nonetheless, Ku left in a time course similar to that of the ligase (Figure 2). Unfortunately, ChIP cannot distinguish dissociation and linear translocation away from the DSB site. Active Ku dissociation from repaired DSBs could conceivably depend on MRX similar to unrepaired sites, but assessment of this phenomenon was not possible because MRX mutants are themselves NHEJ deficient.
Strikingly, disappearance of Ku and DNA ligase IV from unrepaired DSBs strongly depended on MRX (Figure 2, B and F). In the absence of MRX, the assembled Ku–ligase complex was stable for up to 3 hr, reminiscent of the stability with which Ku binds DSB ends in vitro (Milne et al. 1996; Zhang et al. 2007). This presumably accounts for the hyper-recruitment of Ku and Lif1 observed at constitutively induced DSBs in mre11 and rad50 strains (Zhang et al. 2007); our method demonstrates that this is caused by decreased dissociation, not increased association. Ku dissociation in particular required MRX regardless of whether Dnl4 was present (Figure 5), with peak binding in dnl4 yeast restricted to the period during which unrepaired DSB ends are stable and unresected (Frank-Vaillant and Marcand 2002). These data demonstrate that at least Ku is actively dissociated from DSBs in an MRX-dependent manner, consistent with the surprisingly rapid turnover of Ku molecules at laser damage in vivo (Mari et al. 2006).
The mechanism by which MRX influences Ku dissociation is enigmatic. Protein contacts between MRX and Ku (Palmbos et al. 2005) might play a role, but our results do not demand this. Indeed, MRX mutants also show persistence of Sae2 (Lisby et al. 2004) and histones (Tsukuda et al. 2005) at DSBs, suggesting a general mechanism by which MRX turns proteins over (Ivanov et al. 1994). Data here correlate Ku dissociation with bulk resection in preparation for HR, but it is not clear to what extent Ku dissociation is a de facto result of resection or whether its dissociation is required to allow resection to occur. Ku loss leads to more rapid resection (Lee et al. 1998; Zhang et al. 2007), increases HR efficiency in species with low HR (Ninomiya et al. 2004; Nayak et al. 2006), and partially rescues radiosensitivity of mre11 yeast (Bressan et al. 1999), all suggesting that Ku removal is necessary to promote HR. At the same time, resection clearly requires more than just Ku dissociation, since mre11 yku70 double mutants resect as slowly as mre11 (Lee et al. 1998). Delineating the MRX role in protein dissociation will require separating MRX action from resection. We tried to do this in G1 where resection has been reported to be inactive (Ira et al. 2004), but observed substantial MRX-dependent resection (Figure 8) consistent with a different report (Aylon et al. 2004). Intriguingly, however, we could separate MRX action from an apparent limited degradation of DSB ends, which does not require MRX and proceeds at DSB ends bound by Ku (Figure 9). It might be that going past this initial limited resection requires dissociation of Ku and other proteins, perhaps placing MRX in a group of ATPases executing disassembly of various protein intermediates along the DSB repair pathway (Symington and Heyer 2006).
Acknowledgments
We thank members of the Wilson laboratory for many helpful discussions and critical reading of the manuscript and Mark Johnston for helpful early discussion regarding the use of gene promoters. This work was supported by Public Health Service grant CA102563.
References
- Ahnesorg, P., and S. P. Jackson, 2007. The non-homologous end-joining protein Nej1p is a target of the DNA damage checkpoint. DNA Repair 6 190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, D. E., K. M. Trujillo, P. Sung and H. P. Erickson, 2001. Structure of the Rad50 × Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J. Biol. Chem. 276 37027–37033. [DOI] [PubMed] [Google Scholar]
- Aparicio, O., J. V. Geisberg, E. Sekinger, A. Yang, Z. Moqtaderi et al., 2005. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo, pp. 21.3.1–21.3.33 in Current Protocols in Molecular Biology, edited by F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith and K. Struhl. John Wiley & Sons, New York. [DOI] [PubMed]
- Aylon, Y., B. Liefshitz and M. Kupiec, 2004. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23 4868–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeger, H., J. Griesenbeck, J. S. Strattan and R. D. Kornberg, 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11 1587–1598. [DOI] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 115–132. [DOI] [PubMed] [Google Scholar]
- Bressan, D. A., B. K. Baxter and J. H. Petrini, 1999. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 7681–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calsou, P., C. Delteil, P. Frit, J. Drouet and B. Salles, 2003. Coordinated assembly of Ku and p460 subunits of the DNA-dependent protein kinase on DNA ends is necessary for XRCC4-ligase IV recruitment. J. Mol. Biol. 326 93–103. [DOI] [PubMed] [Google Scholar]
- Chen, L., K. Trujillo, W. Ramos, P. Sung and A. E. Tomkinson, 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8 1105–1115. [DOI] [PubMed] [Google Scholar]
- Chen, L., K. M. Trujillo, S. Van Komen, D. H. Roh, L. Krejci et al., 2005. Effect of amino acid substitutions in the Rad50 ATP binding domain on DNA double strand break repair in yeast. J. Biol. Chem. 280 2620–2627. [DOI] [PubMed] [Google Scholar]
- Connelly, J. C., and D. R. Leach, 2002. Tethering on the brink: the evolutionarily conserved Mre11-Rad50 complex. Trends Biochem. Sci. 27 410–418. [DOI] [PubMed] [Google Scholar]
- Daley, J. M., P. L. Palmbos, D. Wu and T. E. Wilson, 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39 431–451. [DOI] [PubMed] [Google Scholar]
- D'Amours, D., and S. P. Jackson, 2002. The mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell Biol. 3 317–327. [DOI] [PubMed] [Google Scholar]
- Deshpande, R. A., and T. E. Wilson, 2007. Modes of interaction among yeast Nej1, Lif1 and Dnl4 proteins and comparison to human XLF, XRCC4 and Lig4. DNA Repair 6 1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore, A. S., N. Furnham, O. R. Davies, B. L. Sibanda, D. Y. Chirgadze et al., 2006. Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode. DNA Repair 5 362–368. [DOI] [PubMed] [Google Scholar]
- Dudasova, Z., A. Dudas and M. Chovanec, 2004. Non-homologous end-joining factors of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 28 581–601. [DOI] [PubMed] [Google Scholar]
- Frank-Vaillant, M., and S. Marcand, 2001. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 15 3005–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Vaillant, M., and S. Marcand, 2002. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell 10 1189–1199. [DOI] [PubMed] [Google Scholar]
- Furuse, M., Y. Nagase, H. Tsubouchi, K. Murakami-Murofushi, T. Shibata et al., 1998. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 17 6412–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith, A. J., P. R. Blier, T. Mimori and J. A. Hardin, 1992. Ku polypeptides synthesized in vitro assemble into complexes which recognize ends of double-stranded DNA. J. Biol. Chem. 267 331–338. [PubMed] [Google Scholar]
- Haber, J. E., 1995. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. BioEssays 17 609–620. [DOI] [PubMed] [Google Scholar]
- Herrmann, G., T. Lindahl and P. Schar, 1998. Saccharomyces cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 17 4188–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner, K. P., A. Karcher, L. Craig, T. T. Woo, J. P. Carney et al., 2001. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105 473–485. [DOI] [PubMed] [Google Scholar]
- Hsu, H. L., S. M. Yannone and D. J. Chen, 2002. Defining interactions between DNA-PK and ligase IV/XRCC4. DNA Repair 1 225–235. [DOI] [PubMed] [Google Scholar]
- Ira, G., A. Pellicioli, A. Balijja, X. Wang, S. Fiorani et al., 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, E. L., N. Sugawara, C. I. White, F. Fabre and J. E. Haber, 1994. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 14 3414–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karathanasis, E., and T. E. Wilson, 2002. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics 161 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, B. O., B. Llorente, A. Lam and L. S. Symington, 2005. Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics 171 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. E., J. K. Moore, A. Holmes, K. Umezu, R. D. Kolodner et al., 1998. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94 399–409. [DOI] [PubMed] [Google Scholar]
- Li, S., and M. J. Smerdon, 2002. Nucleosome structure and repair of N-methylpurines in the GAL1-10 genes of Saccharomyces cerevisiae. J. Biol. Chem. 277 44651–44659. [DOI] [PubMed] [Google Scholar]
- Lisby, M., J. H. Barlow, R. C. Burgess and R. Rothstein, 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118 699–713. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Mari, P. O., B. I. Florea, S. P. Persengiev, N. S. Verkaik, H. T. Bruggenwirth et al., 2006. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. USA 103 18597–18602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, S. P., J. K. Ng, S. Taylor, L. M. Flynn, R. E. Hammer et al., 1995. Mutagenesis of the human apolipoprotein B gene in a yeast artificial chromosome reveals the site of attachment for apolipoprotein(a). Proc. Natl. Acad. Sci. USA 92 10147–10151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne, G. T., S. Jin, K. B. Shannon and D. T. Weaver, 1996. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 16 4189–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesti, M., J. E. Hesse and M. Gellert, 1999. DNA binding of XRCC4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO J. 18 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau, S., J. R. Ferguson and L. S. Symington, 1999. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 19 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira, J. M., W. Horz and S. Holmberg, 2002. Neither Reb1p nor poly(dA*T) elements are responsible for the highly specific chromatin organization at the ILV1 promoter. J. Biol. Chem. 277 3202–3209. [DOI] [PubMed] [Google Scholar]
- Nayak, T., E. Szewczyk, C. E. Oakley, A. Osmani, L. Ukil et al., 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny, S. A., C. M. Snowden, J. McCarville and D. A. Ramsden, 2000. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 20 2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya, Y., K. Suzuki, C. Ishii and H. Inoue, 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 101 12248–12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmbos, P. L., J. M. Daley and T. E. Wilson, 2005. Mutations of the Yku80 C terminus and Xrs2 FHA domain specifically block yeast nonhomologous end joining. Mol. Cell. Biol. 25 10782–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, K. R., Q. L. Li, C. H. Clegg, T. Furukawa, P. A. Navas et al., 1995. Use of yeast artificial chromosomes (YACs) in studies of mammalian development: production of beta-globin locus YAC mice carrying human globin developmental mutants. Proc. Natl. Acad. Sci. USA 92 5655–5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindra, A., K. Weiss and R. T. Simpson, 1999. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol. Cell. Biol. 19 7944–7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagan, M. S., and J. E. Majors, 1998. The chromatin structure of the GAL1 promoter forms independently of Reb1p in Saccharomyces cerevisiae. Mol. Gen. Genet. 259 142–149. [DOI] [PubMed] [Google Scholar]
- Reinke, H., and W. Horz, 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11 1599–1607. [DOI] [PubMed] [Google Scholar]
- Revy, P., L. Malivert and J. P. de Villartay, 2006. Cernunnos-XLF, a recently identified non-homologous end-joining factor required for the development of the immune system. Curr. Opin. Allergy Clin. Immunol. 6 416–420. [DOI] [PubMed] [Google Scholar]
- Shim, E. Y., J. L. Ma, J. H. Oum, Y. Yanez and S. E. Lee, 2005. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell. Biol. 25 3934–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim, E. Y., S. J. Hong, J. H. Oum, Y. Yanez, Y. Zhang et al., 2007. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 27 1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima, H., M. Suzuki and M. Shinohara, 2005. Isolation and characterization of novel xrs2 mutations in Saccharomyces cerevisiae. Genetics 170 71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, P., K. M. Trujillo and S. Van Komen, 2000. Recombination factors of Saccharomyces cerevisiae. Mutat. Res. 451 257–275. [DOI] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington, L. S., and W. D. Heyer, 2006. Some disassembly required: role of DNA translocases in the disruption of recombination intermediates and dead-end complexes. Genes Dev. 20 2479–2486. [DOI] [PubMed] [Google Scholar]
- Trujillo, K. M., D. H. Roh, L. Chen, S. Van Komen, A. Tomkinson et al., 2003. Yeast xrs2 binds DNA and helps target Rad50 and Mre11 to DNA ends. J. Biol. Chem. 278 48957–48964. [DOI] [PubMed] [Google Scholar]
- Tsukamoto, Y., A. K. Taggart and V. A. Zakian, 2001. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 11 1328–1335. [DOI] [PubMed] [Google Scholar]
- Tsukuda, T., A. B. Fleming, J. A. Nickoloff and M. A. Osley, 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui, T., T. Ohta, H. Oshiumi, J. Tomizawa, H. Ogawa et al., 1998. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell 95 705–716. [DOI] [PubMed] [Google Scholar]
- Valencia, M., M. Bentele, M. B. Vaze, G. Herrmann, E. Kraus et al., 2001. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 414 666–669. [DOI] [PubMed] [Google Scholar]
- Walker, J. R., R. A. Corpina and J. Goldberg, 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412 607–614. [DOI] [PubMed] [Google Scholar]
- Weiss, K., and R. T. Simpson, 1998. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLalpha. Mol. Cell. Biol. 18 5392–5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, T. E., 2007. Nonhomologous end-joining: mechanisms, conservation and relationship to illegitimate recombination, pp. 487–513 in Molecular Genetics of Recombination (Topics in Current Genetics), edited by A. Aguilera and R. Rothstein. Springer-Verlag, Berlin/Heidelberg, Germany/New York.
- Wilson, T. E., U. Grawunder and M. R. Lieber, 1997. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388 495–498. [DOI] [PubMed] [Google Scholar]
- Wiltzius, J. J., M. Hohl, J. C. Fleming and J. H. Petrini, 2005. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 12 403–407. [DOI] [PubMed] [Google Scholar]
- Wu, P. Y., P. Frit, L. Malivert, P. Revy, D. Biard et al., 2007. Interplay between cernunnos-XLF and NHEJ proteins at DNA ends in the cell. J. Biol. Chem. 282 31937–31943. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., M. L. Hefferin, L. Chen, E. Y. Shim, H. M. Tseng et al., 2007. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 14 639–646. [DOI] [PubMed] [Google Scholar]