Abstract
Strain LH530, a mutant of Escherichia coli K-12, was reported by others to show increased outer membrane permeability, temperature-sensitive growth, and reduced synthesis of lipid A. The unmapped mutant gene was found to be suppressed by high-copy-number plasmids carrying the wild-type acpT gene, which encodes a protein that catalyzes a post-translational protein modification, the attachment of 4′-phosphopantetheine. We mapped the strain LH530 mutation to a gene of unknown function, yejM, known to encode an inner membrane protein. The mutation is a yejM nonsense mutation that produces a truncated protein lacking the predicted periplasmic domain. Reconstruction of the mutation gave a strain having the same phenotypes as LH530. In contrast to the nonsense mutants, deletion of the entire yejM gene was lethal. Suppression by AcpT overexpression of the yejM nonsense mutants encoding the truncated proteins was specific to AcpT. Moreover, AcpT overexpression also suppressed the lethality due to deletion of the entire yejM gene and this suppression also did not require that AcpT be enzymatically active. The mechanism whereby overexpression of a specific cytosolic protein bypasses the essentiality of an inner membrane protein is unknown.
THE cell envelopes of gram-negative bacteria consist of an inner membrane, an outer membrane, and a rigid layer of peptidoglycan located between the two membranes. The outer membrane provides a protective barrier that prevents the diffusion of hydrophobic and large hydrophilic antibiotics into the cell and consists of an inner leaflet composed of phospholipids and an outer leaflet composed of lipopolysaccharides. The structure of lipopolysaccharide can be divided into three components: lipid A, the core oligosaccharide, and the O-antigen. The lipid A portion of the lipopolysaccharide anchors the lipopolysaccharide to the outer leaflet of the outer membrane. The synthesis of lipid A in Escherichia coli has been well studied and serves as a model system for understanding this process (Raetz et al. 2007). The lipid A molecules synthesized by E. coli and other gram-negative bacteria consist of a β-1′-6-linked disaccharide of N-acetylglucosamine that is modified by the attachment of fatty acids, phosphates, and additional saccharides (Raetz et al. 2007). Additional covalent modifications to the core lipid A molecule, such as the addition of a palmitate or modification of the 1′- or 4′-phosphates with l-4-aminoarabinose or phosphoethanolamine, occur when the outer membrane is stressed (Raetz et al. 2007).
Mutations have been identified in several lipid A biosynthetic genes that result in reduction of lipid A synthesis and a hypersensitivity to hydrophobic and large hydrophilic antibiotics. These mutations occur in the lpxA, lpxC, and lpxD genes (Normark 1970; Tsuruoka et al. 1988; Galloway and Raetz 1990; Kloser et al. 1996; Vaara and Nurminen 1999). Both lpxA and lpxD encode enzymes that transfer a fatty acid, β-hydroxymyristate, to nascent lipid A species (Crowell et al. 1986; Kelly et al. 1993) whereas lpxC encodes an enzyme that deacetylates a precursor of lipid A (Young et al. 1995).
Hirvas et al. (1997) isolated another E. coli mutant strain, strain LH530, that was hypersensitive to hydrophobic and large hydrophilic antibiotics and showed temperature-sensitive growth. Strain LH530 was reported to be defective in lipid A biosynthesis but the mutant gene was neither mapped nor isolated. However, in their attempts to isolate the responsible gene, these workers identified a gene called ORF195 (or o195), but now called acpT, that encodes a phosphopantetheinyl transferase (Lambalot et al. 1996; Flugel et al. 2000; De Lay and Cronan 2006), which suppressed the temperature-sensitive phenotype of mutant strain LH530. However, strain LH530 carried a wild-type copy of acpT and growth at 42° occurred only upon high levels of overexpression (Hirvas et al. 1997). Therefore, growth was due to suppression rather than complementation. Strain LH530 had other phenotypes consistent with a defect in lipid A synthesis; i.e., the strain leaked a periplasmic enzyme, but not a cytoplasmic enzyme, and its outer membrane contained abnormally low levels of the OmpF porin (Nurminen et al. 1997).
We studied this mutation due to our interest in phosphopantetheinyl transferases (Flugel et al. 2000; De Lay and Cronan 2006). We report the mapping of the mutation responsible for the temperature-sensitive phenotype and other phenotypes of strain LH530. The mutant strain and the yejM gene have also been further characterized, including the demonstration that yejM is an essential gene. Moreover, we demonstrate that suppression of the temperature-sensitive phenotype of strain LH530 upon overexpression of AcpT is not due to its phosphopantetheinyl transferase activity and that AcpT overexpression also suppresses the lethality of yejM null mutants.
MATERIALS AND METHODS
Bacterial strains and plasmids:
The bacterial strains used in this study were derivatives of strain E. coli K-12. All of these E. coli strains are listed in Table 1 except for the Tn10 insertion strains used for gene mapping, which were previously described. (Singer et al. 1989; Nichols et al. 1998). All plasmids used in this study are listed in Table 2. The primers used in this study (supplemental Table 1S) were purchased from Integrated DNA Technologies. Fosmid pCC1Fos was obtained from Epicentre. Site-directed mutagenesis was done by the Quickchange PCR method (Stratagene, La Jolla, CA).
TABLE 1.
Strains used in this study
| Strain | Relevant features | Source or reference |
|---|---|---|
| CAG12177 | MG1655 atoS∷Tn10 | Singer et al. (1989) |
| DH5α | recA1 endA1 | Strain collection |
| HT253 | W3110 pdxJ8∷ΔTn10 | Strain collection |
| JM105 | F′ lacIq proAB/endA Δ(lac-proAB) | Yanisch-Perron et al. (1985) |
| LH530 | F′ lacIq proAB/endA Δ(lac-proAB) Ts | Hirvas et al. (1997) |
| MC1061 | araD139 Δ(ara-leu)7696 | Strain collection |
| Top10F′ | F′ lacIq Tn10 (TetR)/φ80∷lacZ ΔM15 ΔlacX74 recA1 endA1 | Invitrogen (San Diego) |
| JW0757-1 | ΔaraBAD ΔlacZ ΔbioA746∷kan | CGSC; Baba et al. (2006) |
| JW0014-1 | ΔaraBAD ΔlacZ ΔdnaJ735∷kan | CGSC; Baba et al. (2006) |
| CY1783 | ΔaraBAD ΔlacZ bioA746∷lacZY | This study |
| CY1817 | ΔaraBAD ΔlacZ ΔdnaJ735∷lacZY | This study |
| NRD142 | LH530 atoS∷Tn10 | This study |
| NRD148 | JM105 atoS∷Tn10 Ts | This study |
| NRD150 | MC1061 ΔompC∷kan | This study |
| NRD151 | MC1061 ΔyojI∷kan | This study |
| NRD153 | MC1061 ΔrcsC∷kan | This study |
| NRD159 | MC1061 ΔatoB∷kan | This study |
| NRD161 | MC1061 ΔnapB∷kan | This study |
| NRD163 | MC1061 ΔccmH∷kan | This study |
| NRD169 | MC1061 ΔyejK∷kan | This study |
| NRD178a | MC1061 ΔyejM∷kan | This study |
| NRD183 | JM105 ΔyejM∷kan carrying pNRD217 | This study |
| NRD184 | MC1061 yejM569∷kan | This study |
| NRD187 | JM105 yejM569∷kan | This study |
CGSC, Coli Genetic Stock Center (Yale University).
This strain also carries a chromosomal duplication of the yejM region (see text).
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant features | Source or reference |
|---|---|---|
| pBAD24 | Ampr, araBAD promoter-based expression vector, pUC-type ori | Guzman et al. (1995) |
| pBAD30 | Ampr, araBAD promoter-based expression vector, p15a ori | Guzman et al. (1995) |
| pBAD322 | AmpR, araBAD promoter-based expression vector, complete pBR322 origin | Cronan (2006) |
| pCR2.1 | Ampr, Kanr, cloning vector, pUC ori | Invitrogen |
| pKD4 | oriRγ, Kanr cassette flanked by FRT sites | Datsenko and Wanner (2000) |
| pKD46 | AmpR, RepA1019(Ts), λ, γ, β, and exo expressed from an araBAD promotor | Datsenko and Wanner (2000) |
| pQE70-Sfp | Ampr, pQE70-derived plasmid encoding Sfp with a C-terminal His6 tag | Mofid et al. (1999) |
| pYon113 | Ampr, acpT under the control of an araBAD promoter | Flugel et al. (2000) |
| pNRD28 | acpS amplified from JM105 using primers AcpSBAD For and Rev and TOPO cloned into pCR2.1 | This study |
| pNRD83 | EcoRI–HindIII acpS fragment of pNRD28 inserted into the same sites of pBAD322 | This study |
| pNRD192 | EcoRI–XbaI acpS fragment of pQE70-Sfp inserted into the same sites of pBAD30 | This study |
| pNRD203 | EcoRI–HindIII acpS acpS fragment of pNRD83 inserted into the same sites of pBAD24 | This study |
| pNRD204 | EcoRI–XbaI sfp acpS fragment of pNRD192 inserted into the same sites of pBAD18 | This study |
| pNRD205 | Encodes AcpT D91E. Site-directed mutation in acpT in pYon113 was generated using primers AcpTD91E For and Rev. | This study |
| pNRD206 | Encodes AcpT E137S. Site-directed mutation in acpT in pYon113 was generated using primers AcpTE137S For and Rev. | This study |
| pNRD207 | Encodes AcpT K141A. Site-directed mutation of acpT in pYon113 using primers AcpTK141A For and Rev. | This study |
| pNRD216 | yejM amplified from JM105 using primers YejM For and Rev primers and TOPO cloned into pCR2.1 | This study |
| pNRD217 | NcoI–XbaI yejM containing fragment from pNRD216 inserted into the same sites of pBAD322 | This study |
Strain NRD142 was constructed by phage P1vir transduction of the Tn10 of strain CAG12177 into strain LH530 (Miller 1972) followed by screening of the tetracycline-resistant transductants for maintenance of the temperature-sensitive phenotype. Strain NRD148 was generated by transducing the Tn10 from strain NRD142 into strain JM105 and screening the resulting tetracycline transductants for a strain that had acquired a temperature-sensitive phenotype. The ΔompC strain NRD150 was constructed by λ-Red recombinase-mediated gene replacement (Datsenko and Wanner 2000) using the PCR product generated from the template pKD4 (Datsenko and Wanner 2000) with the OmpCKO For and Rev primers. The strains carrying deletions of yojI, rcsC, atoB, napB, ccmH, yejM, and yejK were generated via λ-Red recombinase-mediated gene replacement using the PCR product generated from the template pKD4, using the pair of primers named for the gene (supplemental Table 1S). Strain NRD183 was constructed by transduction of the kanamycin resistance construct from strain NRD178 into strain JM105 carrying plasmid pNRD217. This transduction was performed in the presence of arabinose, and transductants were selected on LB plates containing kanamycin and arabinose. The wild-type yejM gene was modified by introducing a UGA termination codon in place of the codon for Y190 by an UGA (opal) codon and by substituting a kanamycin resistance cassette for the yejM sequences downstream of the mutated codon by λ-Red-mediated recombination using a PCR product generated from the template plasmid pKD4 using primers YejMKO4 For and YejMKO Rev. Kanamycin-resistant recombinants were selected on LB plates containing kanamycin at 28°. Strain NRD187 was generated by transduction of the kanamycin resistance determinant from strain NRD184 into strain JM105. The lacZY transcriptional fusions to bioA and dnaJ were constructed as described by Ellermeier et al. (2002) except that vectors with improved ribosome-binding sites (J. Slauch, personal communication) were used and pCY580 (Cronan 2003) was used as the source of Flp recombinase in the latter construction. β-Galactosidase activities were determined according to Miller (1972).
Culture media and growth conditions:
Strains were grown in LB liquid medium or on LB agar plates. LB medium was supplemented with final concentrations of glucose (0.2%) or arabinose (0.08%). Antibiotics were used at the following concentrations: 100 mg liter−1 ampicillin, 50 mg liter−1 kanamycin, and 5 mg liter−1 tetracycline.
Gene mapping:
The mutation conferring temperature sensitivity to strain LH530 was initially localized on the E. coli K-12 chromosome by transduction of strain LH530 with P1vir lysates made from a collection of strains harboring Tn10 insertions spaced around the E. coli chromosome (Singer et al. 1989; Nichols et al. 1998). P1vir lysates were made from nearly all of the strains in this collection that have a Tn10 insertion within the first 56 min of the E. coli chromosome. These P1vir lysates were used to transduce the Tn10 insertion from each of these strains into strain LH530. The transductants that acquired the Tn10 were selected for the ability of these strains to grow at 28° on LB plates containing tetracycline. Between 48 and 102 of tetracycline-resistant transductants obtained from each transduction were screened for the loss of temperature sensitivity by streaking each transductant on two LB plates containing tetracycline followed by incubation of one plate at 28° and the other at 42°. Two Tn10 insertions cotransduced with the temperature-sensitive mutation of strain LH530. One Tn10 was inserted into napA whereas the other was inserted into atoS. To further define the location of the mutation, the cotransduction frequencies of the mutation with a kanamycin marker replacing yejK, ccmH, napB, yojI, ompC, rcsC, or atoB were determined. More than 200 kanamycin-resistant transductants resulting from transduction of strain LH530 with phage P1vir lysates grown on strains NRD169, NRD163, NRD161, NRD151, NRD150, NRD153, or NRD159 were screened for temperature sensitivity as described above. Once a very closely linked marker was identified, the gene replaced by that marker, together with ∼2500 bp of adjacent genomic DNA from both strain LH530 and the parental strain JM105, was sequenced after amplification by PCR using the YejK, YejL, and YejM primers. Each PCR product was directly sequenced with both primers used for amplification by the Core Sequencing Unit at the Keck Center for Comparative and Functional Genomics at the University of Illinois.
RESULTS
Mapping of the mutation responsible for the temperature-sensitive phenotype of the E. coli K-12 strain LH530:
Hirvas et al. (1997) previously identified a temperature-sensitive mutant named strain LH530 that had a phenotype similar to that of mutants defective in lipid A synthesis; i.e., the mutant was sensitive to hydrophobic and large hydrophilic antibiotics at permissive temperatures and was reported to have subnormal rates of lipid A synthesis at all temperatures. Hirvas et al. (1997) found that plasmids carrying a gene named acpT (then called ORF195) suppressed the temperature-sensitive phenotype of this mutant strain, but only when the AcpT-encoding plasmids were of high copy number. However, the acpT gene of strain LH530 had the same sequence as the parental wild-type strain, indicating suppression rather than complementation. The mutation responsible for the temperature-sensitive phenotype of strain LH530 was not identified or mapped.
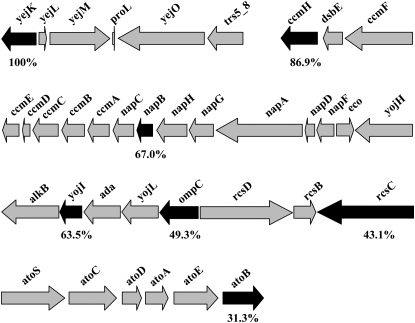
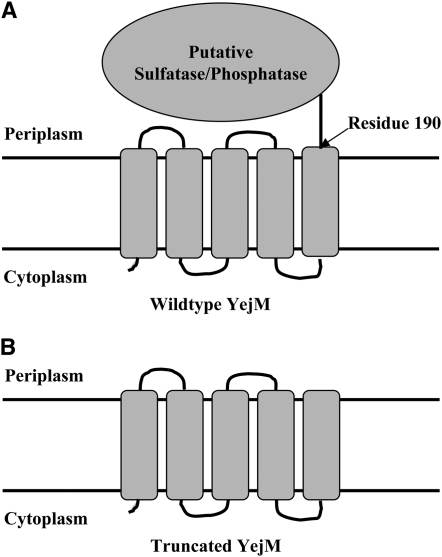
We mapped the mutation responsible for the temperature-sensitive phenotype of strain LH530 by transducing strain LH530 with phage P1vir lysates of a collection of strains, each of which carried a transposon Tn10 antibiotic resistance element inserted at a known chromosomal location (Singer et al. 1989; Nichols et al. 1998). Upon screening the first 56 min of the E. coli chromosome, the temperature-sensitive mutation of strain LH530 was found to be ∼53 and 29% linked to Tn10 insertions in napA and atoS, respectively (Figure 1). To further resolve the location of the mutation, a set of nonessential genes located in this region of the E. coli chromosome (yejK, ccmH, napB, yojI, ompC, rcsC, and atoB) were replaced with kanamycin resistance cassettes via λ-Red recombinase-mediated gene replacement (Datsenko and Wanner 2000) and the linkage of the mutation with these markers was determined by measuring cotransduction frequencies (Figure 1). The temperature-sensitive mutation of strain LH530 was found to very tightly linked (100%) to the kanamycin resistance cassette inserted in place of yejK. We then amplified the genes yejK, yejL, and yejM and the intergenic regions between these genes from strain LH530 and its parental strain JM105 and sequenced the PCR products. The sequencing data showed a single difference between the two genomic sequences: a G570A mutation in the yejM gene of strain LH530. The G570A mutation resulted in replacement of the UGG codon of tryptophan 190 with a UGA termination codon. YejM has previously been shown to be an inner membrane protein predicted to be composed of a transmembrane domain consisting of five putative membrane-spanning helices and a large periplasmic domain (Daley et al. 2005) (Figure 2). The nonsense mutation would truncate the expressed protein to 190 residues from 586 residues and would cleanly remove the predicted periplasmic domain while leaving the predicted transmembrane domain intact (Figure 2).
Figure 1.—
The segment of the E. coli chromosome that contains yejM. Transductions using strain LH530 as the recipient were performed using P1vir lysates made from derivatives of strain MC1061, which had one of the genes denoted by a solid arrow replaced with a kanamycin resistance cassette. Transductants were selected on LB–kanamycin plates incubated at 28° for 2 days. From each cross at least 200 kanamycin-resistant transductants were then screened for temperature-sensitive growth. Below each of the solid arrows is the frequency at which the kanamycin marker replacing the gene of interest cotransduced with the gene that relieved the temperature-sensitive phenotype of strain LH530.
Figure 2.—
An illustration of the putative structure of the full-length YejM (A) and the truncated form of YejM produced by strain LH530 (B) based on the data and predictions of Daley et al. (2005). YejM is an inner membrane protein that contains five putative membrane-spanning helices and a periplasmic C-terminal domain. The periplasmic domain of YejM is predicted to have sulfatase/phosphatase activity in the UniProtKB/Swiss-Prot database.
We linked the yejM mutation of strain LH530 to the Tn10 insertions within napA and atoS by transduction and then used these strains as donors to transduce strains JM105 and MG1655 to tetracycline resistance. The resulting transductants were then screened for temperature-sensitive growth. The temperature-sensitivity profile observed in strain NRD187, the derivative of strain JM105 that had acquired the yejM(Ts) mutation, was identical to that observed for strain LH530 (data not shown). However, although the yejM(Ts) derivative of strain MG1655 was clearly temperature sensitive, this strain showed greater growth at 42° than did the strain JM105 derivatives (data not shown). This difference in behavior can be attributed to the fact that strain MG1655 grew significantly faster than strain JM105 and this faster growth accentuated the leakiness (formation of small colonies) shown by strain LH530 upon prolonged incubation at 42°.
Characterization of the yejM(Ts) mutation:
To confirm that the premature termination codon in the yejM gene of strain LH530 was responsible for the temperature-sensitive growth of strain LH530, we used λ-Red recombinase-mediated gene replacement to generate the nonsense (G570A) mutation of strain LH530 in the wild-type gene and to concomitantly replace the yejM sequence downstream of the mutation with a kanamycin resistance cassette. This construct was then transduced into strain JM105 to generate strain NRD187. We then compared the abilities of strains JM105, LH530, and NRD187 to grow on LB plates incubated at 28° or 42° overnight. Although all three strains grew well at 28°, only strain JM105 grew at 42° following overnight incubation (Figure 3). However, upon prolonged incubation, strain LH530 formed small colonies at 42° whereas strain NRD187 did not. This difference suggested that in strain LH530 basal readthrough of the UGA nonsense codon (Bjornsson and Isaksson 1993) resulted in low levels of full-length YejM. Strains LH530 and NRD187 had essentially identical lipid A phenotypes (see below) and, as expected from the previous report (Hirvas et al. 1997), both strains were much more sensitive to rifampicin and erythromycin than strain JM105.
Figure 3.—
The growth of strains JM105, NRD187, and LH530 on LB plates incubated overnight at either 28° or 42°.
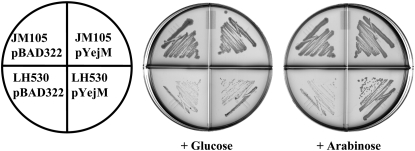
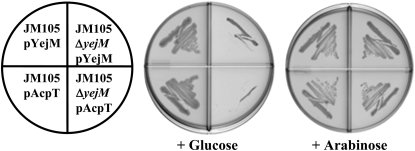
We performed complementation experiments to assess whether or not expression of a wild-type copy of yejM either from its native promoter on a cosmid vector or from an araBAD promoter on a multicopy plasmid allowed growth of strain LH530 at 42°. A fragment of DNA that contained yejM, together with the upstream and downstream genes yejL and proL, respectively, was amplified from strain JM105. This genome segment was then inserted into the single-copy F-factor-based cosmid pCC1Fos. The presence of this construct in strain LH530 allowed growth at the nonpermissive temperature (data not shown). We then placed the yejM gene under the araBAD promoter of pBAD322 and tested the ability of the resulting plasmid, pNRD217, to complement growth of strain LH530 at 42° on plates containing either glucose or arabinose. Strain LH530 containing pNRD217 grew on the LB–arabinose-containing plates, but did not grow on the plates containing glucose (which represses expression from the araBAD promoter) (Figure 4). The introduction of plasmid pNRD217 into strain NRD187 similarly allowed growth at 42° on LB plates containing arabinose but not on those containing glucose (data not shown). The inability to recover a plasmid-encoding yejM by complementation of the mutation of LH530 indicates that the clone bank screened by Hirvas et al. (1997) lacked a yejM plasmid presumably due to a bias in construction of the bank.
Figure 4.—
Complementation of the temperature-sensitive strain LH530 by a plasmid-encoded copy of yejM. Strain JM105 or the derived temperature-sensitive mutant, strain LH530, carrying either the vector pBAD322 or a plasmid containing a copy of yejM under the control of the araBAD promoter (pNRD217 called pYejM in the figure) were tested for the ability to grow at 42° on LB plates containing either glucose or arabinose.
The yejM(Ts) mutation is suppressed by overexpression of active and inactive forms of AcpT, but not by high-level expression of two other phosphopantetheinyl transferases, AcpS and Sfp:
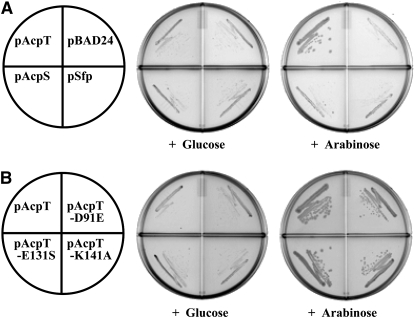
Hirvas et al. (1997) have previously shown that the temperature-sensitive phenotype of strain LH530 is suppressed by high-copy-number plasmids carrying acpT, which encodes a phosphopantetheinyl transferase that plays a minor role in E. coli K-12 lipid metabolism (De Lay and Cronan 2006). Phosphopantetheinyl transferases are enzymes that transfer the 4′-phosphopantetheinyl moiety of CoA to a conserved serine residue of a carrier protein such as the acyl carrier proteins of lipid synthesis (Lambalot et al. 1996). Attachment of this prosthetic group is mandatory because the thiol group of the phosphopantetheinyl moiety carries the intermediates of fatty acid biosynthesis. We tested the ability of three phosphopantetheinyl transferases, AcpT, AcpS, and Sfp, to suppress the temperature-sensitive phenotype of LH530 upon overexpression. E. coli AcpS is a trimeric phosphopantetheinyl transferase that catalyzes the modification of acyl carrier proteins of fatty acid biosynthesis (Flugel et al. 2000) whereas Sfp is a monomeric Bacillus subtilis enzyme that catalyzes phosphopantetheinylation of carrier proteins involved in secondary metabolic processes such as nonribosomal peptide and polyketide syntheses (Lambalot et al. 1996; Mootz et al. 2001). As previously reported by Hirvas et al. (1997), high-level overexpression of AcpT suppressed the temperature-sensitive phenotype of strain LH530 (Figure 5) whereas low levels of overexpression did not (data not shown). However, neither overexpression of AcpS nor high-level expression of Sfp suppressed the temperature-sensitive phenotype of strain LH530 (Figure 5). Similar results were obtained for strain NRD187, which carries the in vitro-constructed yejM nonsense allele (data not shown).
Figure 5.—
Suppression analyses of strain LH530 by plasmids encoding either diverse phosphopantetheinyl transferases or site-directed mutants of the AcpT phosphopantetheinyl transferase. (A) Derivatives of strain LH530 carrying the vector pBAD24 or a plasmid that expressed sfp, acpS, or acpT under the control of an araBAD promoter (pNRD204, pNRD83, and pYon113, respectively) were tested for the ability to grow at 42° on LB plates containing glucose or the inducer, arabinose. (B) The ability of wild-type AcpT or of several site-directed mutants of AcpT that eliminate phosphopantetheinyl transferase activity in vivo (Table 2) was tested for the ability to suppress the defective growth of strain LH530 at 42° on LB plates containing either glucose or arabinose.
The lack of suppression of the temperature-sensitive phenotype of strain LH530 by high-level expression of AcpS or Sfp suggested that the phosphopantetheinyl transferase activity of AcpT was not required for suppression. Thus, we made amino acid substitutions for three key AcpT residues (D91E, E137S, and K141A) conserved among phosphopantetheinyl transferases (Lambalot et al. 1996) since they are involved in binding of the CoA substrate by Sfp and AcpS (Reuter et al. 1999; Parris et al. 2000; Mofid et al. 2004). The AcpT amino acid substitutions were substitutions previously shown to eliminate Sfp phosphopantetheinyl transferase activity (Mofid et al. 2004) and were constructed in the pBAD24 vector. Since, in agreement with a prior report (Lambalot et al. 1996), we found that, upon overexpression, AcpT was inactive and largely insoluble, we assayed the activities of these proteins by the abilities of the plasmids to support growth of a strain carrying a conditional mutation in acpS (Flugel et al. 2000; De Lay and Cronan 2006). Upon high-level expression, all of the mutant acpT alleles lacked the ability to functionally replace AcpS whereas the wild-type allele showed the expected strong complementation (Flugel et al. 2000; De Lay and Cronan 2006) (data not shown). However, the plasmids encoding the wild-type and mutant AcpT proteins all suppressed the temperature-sensitive phenotype of strain LH530 (Figure 5). Therefore, suppression of yejM nonsense mutations did not require that AcpT be enzymatically active.
yejM is an essential gene, but its essentiality is suppressed by AcpT overexpression:
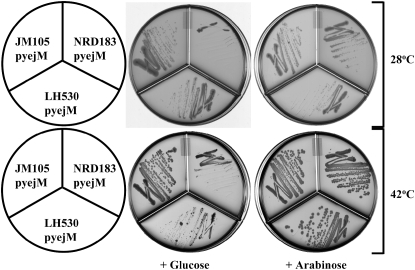
The temperature-sensitive phenotype of strain LH530, which was due to the production of a truncated form of YejM lacking the C-terminal periplasmic domain, raised the question of the effects of a deletion of the entire yejM gene. Repeated attempts to delete yejM from strain MC1061 by λ-Red recombinase-mediated gene replacement were unsuccessful despite our ready success in deleting other genes by this method. We eventually isolated a single yejM deletion candidate, strain NRD178. We confirmed the presence of the yejM deletion in this strain using PCR primers that yielded a product only if the yejM deletion was present. However, the difficulty that we experienced in construction of this strain suggested that this yejM deletion strain might have a second characteristic that allowed viability of the deletion strain. We therefore transduced the yejM deletion into a derivative of strain JM105 that carried a second plasmid-borne copy of yejM under control of the araBAD promoter. Transduction was performed in the presence of arabinose and the transduction mixtures were plated on LB plates containing arabinose and kanamycin. Six transductants were tested for the ability to grow on LB plates containing glucose (where expression of plasmid-borne yejM was repressed) or arabinose (where expression of plasmid-borne yejM was induced). All six transductants grew on LB–arabinose plates but not on LB–glucose plates (Figure 6). Similar results were obtained when this experiment was performed with an MG1655 ΔyejM strain carrying pNRD217, the yejM plasmid (data not shown). Therefore, transduction seemed to have resolved the ΔyejM allele from another character present in the original strain, NRD178. Further analysis of the genome of the original yejM deletion strain NRD178, using PCR primers that generated products of different lengths depending upon the presence or absence of a wild-type copy of yejM, demonstrated that the original yejM deletion strain, strain NRD178, contained a duplication of the region of the genome that includes yejM. The duplicated region contained a copy of wild-type yejM plus a copy of the ΔyejM allele (data not shown). We attribute the chromosomal arrangement of strain NRD178 to λ-Red-mediated recombination into one copy of a preexisting chromosomal duplication of the yejM region present in the cell undergoing the recombination event. Such spontaneous duplications of bacterial chromosomes are well documented (Roth et al. 1996). Moreover, an F-factor-based cosmid generated from strain NRD178 genomic DNA contained a genome fragment that spanned the region between fruA and ccmF (including a wild-type copy of yejM) but lacked the kanamycin-marked ΔyejM deletion. This cosmid also complemented the ΔyejM deletion strain NRD186 (data not shown). This suggests that this duplicated region could include all of the genes between fruA and ccmF, consistent with the sizes of many documented duplications (Roth et al. 1996).
Figure 6.—
Determination of the essentiality of yejM. Strains JM105, LH530, and NRD183, a derivative of strain JM105 containing a deletion of yejM that carries plasmid pNRD217 encoding yejM (called pyejM above) under the control of the araBAD promoter, were tested for the ability to grow overnight at 28° or 42° on LB plates containing either arabinose or glucose.
To further test whether or not yejM is essential, a ΔyejM deletion was constructed by a λ-Red recombinase-mediated gene replacement in strain DY330 (Yu et al. 2000) carrying the yejM plasmid pNRD217. Two of the recombinants obtained were shown to contain the yejM deletion by PCR and both recombinants (one of which is shown in Figure 6) grew only upon induction of the yejM expression from the plasmid.
Given that yejM is essential for growth, whereas the protein lacking the periplasmic domain allows growth at low-growth temperatures, the question was raised of whether or not AcpT overexpression would suppress the lethal phenotype of strains carrying a total deletion of the yejM gene. We found that the ΔyejM deletion mutation was readily transduced into a derivative of JM105 that carried a multicopy plasmid with acpT expression under an inducible (araBAD) promoter. The strain grew under inducing conditions but not under conditions that repressed expression of the plasmid-encoded acpT (Figure 7). The observed growth was very similar to that given by induction of a plasmid-borne copy of yejM and thus AcpT overproduction fully suppressed the effects of this otherwise lethal mutation. As reported above for suppression of the temperature sensitivity of the strain expressing the truncated YejM, suppression of the yejM null mutation did not require AcpT enzymatic activity. When the yejM null mutation was transduced into a derivative of JM105 expressing the inactive E137S AcpT encoded by plasmid pNRD206, growth proceeded as seen with wild-type AcpT (Figure 7). The stain expressing the E137S AcpT grew well in the presence of arabinose, but failed to grow when expression was blocked by the absence of arabinose (in either the presence or the absence of glucose) (data not shown).
Figure 7.—
Suppression of the lethality of a yejM deletion strain by overexpression of AcpT. The yejM deletion of strain NRD178 was transduced via phage P1vir into strain JM105 containing a plasmid that expressed either yejM (pNRD217) or acpT (pYon113) from the arabinose-inducible araBAD promoter. Transductants that acquired the yejM deletion were selected on LB–kanamycin–arabinose plates. A transductant from each cross, together with the parental strains, was streaked onto plates of LB–glucose or LB–arabinose that were incubated overnight at 30°.
Does AcpT overexpression induce the heat-shock response?
One possibility for suppression of yejM mutations by AcpT overexpression is that overexpression triggers a stress response that induces synthesis of a protein that replaces (or bypasses) YejM function. One stress response known to be induced by high-level expression of insoluble proteins such as AcpT is the heat-shock response. Hence, we have asked if the heat-shock regulon is induced upon AcpT overexpression by assaying expression of the classical heat-shock protein, DnaJ, by use of a chromosomal dnaJ-lacZY fusion construct. Construction of the fusion eliminated DnaJ function, but since DnaJ is functionally replaced by either of two other cochaperone proteins, DjlA and CbpA (Gur et al. 2004), DnaJ mutants behave normally in regards to heat shock. Use of this fusion provides an assay for DnaK expression since it is encoded by the promoter-proximal gene in the dnaK-dnaJ operon (Cowing et al. 1985; Bardwell et al. 1986).
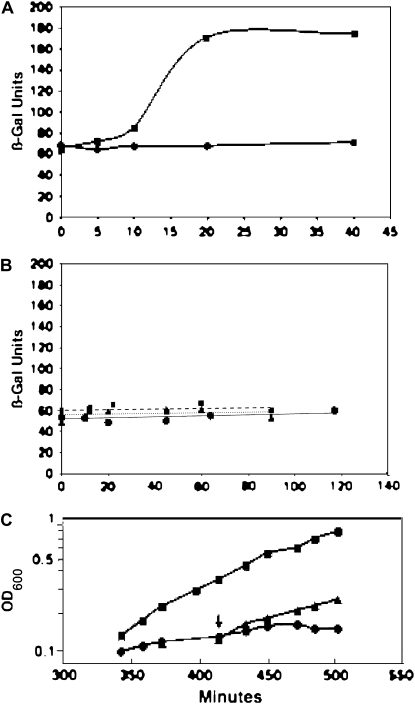
Following construction of the dnaJ-lacZY fusion strain CY1817, the behavior of the strain was tested in a classical 30°–42° temperature shift. The strain showed the expected rapid increase in dnaK-dnaJ expression (assayed by β-galactosidase activity) soon after a temperature shift that reached a plateau about threefold greater than the 30° expression level ∼30 min after shift (Figure 8A) as expected from the increases in DnaK levels seen in the same temperature-shift regimen (Gross 1996). As a control, a lacZY fusion to a housekeeping biosynthetic gene, bioA, was assayed in parallel. In contrast to the dnaJ fusion, the bioA∷lacZY fusion strain showed no effect of temperature; the β-galactosidase activity was constant (Figure 8A).
Figure 8.—
Effects of heat shock and AcpT overproduction on dnaJ induction. (A) Behavior of the dnaJ∷lacZY fusion in the heat-shock response. Cultures of the dnaJ∷lacZY fusion strain CY1817 (squares) or the bioA∷lacZY fusion strain CY1783 (circles) growing exponentially in early log phase at 30° were shifted to 42° at time zero and samples were taken for β-galactosidase assays at the times given. β-Galactosidase activities are given in Miller units (Miller 1972). (B) Derivatives of strain CY1817 carrying plasmids encoding either the wild-type (pYon13) or mutant (pNRD207) AcpT proteins or the empty vector (pBAD24) were grown at 30° in LB–ampicillin. Exponentially growing cultures then received arabinose to a final concentration of 0.2% to induce AcpT overexpression and samples were taken for β-galactosidase assays at the times given. Triangles, vector control; squares, pYon13; and circles, pNRD207. β-Galactosidase activities are given in Miller units and the ordinate scale is identical to that of A. (C) The cells of a culture of strain NRD183 grown in LB–ampicillin medium containing 0.2% arabinose at 30° were harvested by centrifugation, washed, and resuspended in LB medium. The culture was then diluted to an OD600 of 0.05 and split in half. One culture received 0.2% arabinose whereas the other half remained unsupplemented. The cultures were then grown overnight at 30°. The washed cells of both cultures were recovered as before and resuspended at an OD600 of 0.1 in the same medium in which they had been grown. The turbidities of the cultures during growth at 30° were monitored with periodic dilution to keep the cultures in exponential growth until the growth of the culture lacking arabinose began to slow (at ∼5 hr after dilution of the overnight cultures). Half of this culture was then added to a prewarmed flask containing sufficient arabinose to give a final concentration of 0.2%. The growth of the three cultures was then monitored. Squares denote the culture grown with arabinose throughout the experiment; circles denote the culture lacking arabinose throughout the experiment; triangles denote the culture supplemented with arabinose following growth in the absence of arabinose.
The dnaJ-lacZY fusion strain CY1817 was then transformed with plasmids encoding the wild type, the E137S mutant AcpT, or the empty vector. Cultures of these strains were grown 30°, protein expression was induced with 0.2% arabinose, and β-galactosidase activities were measured. The low-growth temperature was chosen to decrease heat-shock protein expression levels and thereby increase the sensitivity of the assay. All three cultures showed a constant level of expression over a 90-min time course (Figure 8B), indicating no induction of the heat-shock response. A possible caveat to this experiment is that the time course may be too brief to allow induction of the heat-shock regulon, although induction of the heat-shock response upon expression of other insoluble proteins was seen during roughly comparable time intervals (Lesley et al. 2002; Smith 2007). However, each protein seems to be a different case and thus we asked how long induction of AcpT overexpression takes to bypass loss of YejM. Strain NRD183, which contains a complete deletion of yejM and a plasmid encoding AcpT under arabinose control, was grown in the presence of arabinose at 30° and the cells of early log phase cultures were recovered by centrifugation. The cells were then washed with and suspended in medium lacking arabinose. This culture was then divided into halves. One-half received 0.2% arabinose, and the second received water. These cultures were then incubated at 30° and their growth was followed by turbidity. When growth of the culture lacking arabinose began to slow due to depletion of AcpT, the culture was split and one-half was supplemented with arabinose. The time required before growth of the arabinose-supplemented culture, accelerated relative to the culture lacking arabinose, was taken as a measure of the time required for accumulation of AcpT levels sufficient to bypass the yejM deletion. Acceleration of growth by the addition of arabinose was seen within 36 min of inducer addition (Figure 8C) and thus the time course chosen to test induction of the heat-shock response was deemed appropriate.
DISCUSSION
YejM is an inner membrane protein predicted to have five membrane-spanning helices and a large C-terminal periplasmic domain (Figure 2) (Daley et al. 2005) that shares sequence similarities with sulfatases and phosphatases (Swiss Protein database). If the topological prediction is accurate, then the G205A yejM mutant allele of strain LH530 would encode a YejM protein containing the complete transmembrane domain, but lacking the entire C-terminal periplasmic domain. We constructed the yejM mutation in vitro and the resulting strain was temperature sensitive. Moreover, the original and constructed yejM G205A mutant strains had similar phenotypes with respect to lipid A biosynthesis and hypersensitivity to antibiotics. Chromatographic and mass spectral analyses (C. Michael Reynolds, personal communication) showed that both strains (LH530 and NRD187) have essentially normal lipid A compositions in that the bulk of the lipid A produced by these strains and their parent strain JM105 was the hexa-acylated lipid A 1,4′-bis-phosphate species normally found in E. coli (Raetz et al. 2007). However, the two mutant strains both synthesized small but discernible levels of a hepta-acylated lipid A species that contains an additional palmitoyl group (C. Michael Reynolds, personal communication). We believe that the presence of this lipid A species in the mutant strains is a secondary response to problems in maintaining outer membrane integrity (Raetz et al. 2007). Previous studies have shown that addition of compounds that affect outer membrane integrity, such as cationic peptides, result in the synthesis of palmitoylated lipid A in Salmonella enterica serovar Typhimurium (Zhou et al. 1999). An LpxM-deficient mutant strain of E. coli O157:H7 has also been reported to accumulate this lipid A species (Raetz et al. 2007).
Although the nonsense mutations resulted in a conditionally lethal phenotype, deletion of the entire yejM gene was unconditionally lethal. Together, these results suggest that YejM has two functions. The function carried out by the N-terminal transmembrane domain is essential at all growth temperatures whereas the function of the C-terminal periplasmic domain is dispensable at low-growth temperatures. Both functions may be important for the same cellular process. During our work Baba et al. (2006) reported yejM to be an essential gene in that they were unable to construct a strain carrying a complete deletion of yejM by λ-Red recombinase-mediated gene replacement. In contrast, two groups reported that yejM is not an essential gene on the basis of transposon mutagenesis (Gerdes et al. 2003; Kang et al. 2004). However, in these reports all six of the reported transposon insertions were located downstream of the nonsense mutation of strain LH530 such that the N-terminal transmembrane domain would be translated. These mutant strains would produce YejM proteins at least 60 residues longer than that produced by strain LH530. Hence, the nature of transposon mutagenesis essentially precluded detection of yejM. Growth of the strains carrying the yejM transposon insertion alleles was assayed only at 30° or 37° (Gerdes et al. 2003; Kang et al. 2004), temperatures at which strains LH530 and NRD187 grow reasonably well. It thus remains unclear whether or not the strains carrying yejM transposon insertion alleles are temperature sensitive for growth.
Suppression of the growth defects of strains lacking either the YejM periplasmic domain or the entire protein by overexpression of the cytosolic AcpT phosphopantetheinyl transferase presents a conundrum. How can a mutation resulting in a defective outer membrane be suppressed by overexpression of an enzyme that post-translationally modifies a cytosolic carrier protein? We have shown that the phosphopantetheinyl transferase activity of AcpT is not involved in yejM mutant suppression and thus that AcpT overexpression must act indirectly. One possible explanation based on the properties of AcpT and the nature of the suppression phenomena is that suppression might involve a stress response. In agreement with the results of Lambalot et al. (1996), we found that AcpT accumulates as an insoluble aggregate upon even fairly modest overexpression conditions (data not shown). Aggregation may be due to the fact that AcpT has been evolutionarily isolated from its substrates (De Lay and Cronan 2006) and thus lacks substrates to aid proper protein folding. Moreover, fairly high levels of AcpT overexpression are required for full suppression of the temperature-sensitive phenotype of strain LH530; low levels of overexpression fail to suppress (Hirvas et al. 1997) (data not shown). If misfolding is involved in suppression of YejM mutations, it seems specific to AcpT overexpression since three acpT clones were the only clones isolated from a clone bank of 8000 wild-type E. coli DNA fragments in a high-copy-number vector that allowed growth of strain LH530 at 42° (Hirvas et al. 1997). We also tested the specificity of AcpT overexpression on the temperature-sensitive phenotype of strain LH530 by overexpression of AcpH, another E. coli protein involved in ACP metabolism that aggregates under all of the many overexpression conditions tested (Thomas and Cronan 2005; Thomas et al. 2007). Overexpression of AcpH using the same vector employed in Figure 7 failed to allow growth of strain LH530 at 42° (data not shown). Consistent with the observed and inferred specificity of suppression, AcpT overexpression failed to induce the heat-shock regulon. Induction of the heat-shock regulon by misfolded proteins is frequently observed (Oh and Liao 2000; Lesley et al. 2002; Smith 2007), and thus if induction of heat shock played a major role in the AcpT overproduction suppression phenomenon, a large and diverse set of the Hirvas clones (instead of only acpT clones) would have suppressed the yejM mutation.
Another possible explanation (suggested by a reviewer) is that overexpressed AcpT might specifically bind another normally more abundant protein(s). This protein (or proteins) might then become trapped in AcpT aggregates, thus depleting the cells of the functional AcpT-binding protein(s). The weakness in these and other possible explanations that we have considered is that they fail to provide a plausible mechanism whereby overexpression of a normally cytosolic protein allows growth of E. coli cells that lack an essential membrane protein.
The function of YejM remains obscure. Somehow loss of the periplasmic domain of this inner membrane protein results in an increased permeability of the outer membrane that is exacerbated by increased growth temperature. The reported phenotype of reduced lipid A biosynthesis in strain LH530 (Hirvas et al. 1997; Vaara and Nurminen 1999) is complicated by the possibility of translocation of phospholipids into the outer leaflet of the outer membrane known to occur in strains that carry mutations affecting the organization of lipopolysaccharide on the cell surface (Wu et al. 2006). Hence, since lipid A synthesis was assayed using phospholipid synthesis as a standard, the results previously reported could be at least partially due to increased phospholipid synthesis rather than to decreased lipid A biosynthesis. Indeed, the accumulation of phospholipids in the outer leaflet is thought to render cells sensitive to otherwise impermeable antibiotics (Raetz et al. 2007), a phenotype of strain LH530. Increased sensitivity to such antibiotics is also seen in lipid A mutants. However, all of the known lipid A mutants have marked alterations of the compositions of their lipid A species whereas the yejM mutant strains have essentially normal lipid A compositions (C. Michael Reynolds, personal communication).
Acknowledgments
We thank C. Michael Reynolds and Christian Raetz of the Department of Biochemistry, Duke University, for lipid A analyses and the Coli Genetic Stock Center for strains. This work was supported by National Institutes of Health grant AI15650 from the National Institute of Allergy and Infectious Diseases.
References
- Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura et al., 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell, J. C., K. Tilly, E. Craig, J. King, M. Zylicz et al., 1986. The nucleotide sequence of the Escherichia coli K12 dnaJ+ gene. A gene that encodes a heat shock protein. J. Biol. Chem. 261 1782–1785. [PubMed] [Google Scholar]
- Bjornsson, A., and L. A. Isaksson, 1993. UGA codon context which spans three codons: reversal by ms2i6A37 in tRNA, mutation in rpsD(S4) or streptomycin. J. Mol. Biol. 232 1017–1029. [DOI] [PubMed] [Google Scholar]
- Cowing, D. W., J. C. Bardwell, E. A. Craig, C. Woolford, R. W. Hendrix et al., 1985. Consensus sequence for Escherichia coli heat shock gene promoters. Proc. Natl. Acad. Sci. USA 82 2679–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan, J. E., 2003. Cosmid-based system for transient expression and absolute off-to-on transcriptional control of Escherichia coli genes. J. Bacteriol. 185 6522–6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan, J. E., 2006. A family of arabinose-inducible Escherichia coli expression vectors having pBR322 copy control. Plasmid 55 152–157. [DOI] [PubMed] [Google Scholar]
- Crowell, D. N., M. S. Anderson and C. R. Raetz, 1986. Molecular cloning of the genes for lipid A disaccharide synthase and UDP-N-acetylglucosamine acyltransferase in Escherichia coli. J. Bacteriol. 168 152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley, D. O., M. Rapp, E. Granseth, K. Melen, D. Drew et al., 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308 1321–1323. [DOI] [PubMed] [Google Scholar]
- Datsenko, K. A., and B. L. Wanner, 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay, N. R., and J. E. Cronan, 2006. A genome rearrangement has orphaned the Escherichia coli K-12 AcpT phosphopantetheinyl transferase from its cognate Escherichia coli O157:H7 substrates. Mol. Microbiol. 61 232–242. [DOI] [PubMed] [Google Scholar]
- Ellermeier, C. D., A. Janakiraman and J. M. Slauch, 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290 153–161. [DOI] [PubMed] [Google Scholar]
- Flugel, R. S., Y. Hwangbo, R. H. Lambalot, J. E. Cronan, Jr. and C. T. Walsh, 2000. Holo-(acyl carrier protein) synthase and phosphopantetheinyl transfer in Escherichia coli. J. Biol. Chem. 275 959–968. [DOI] [PubMed] [Google Scholar]
- Galloway, S. M., and C. R. Raetz, 1990. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J. Biol. Chem. 265 6394–6402. [PubMed] [Google Scholar]
- Gerdes, S. Y., M. D. Scholle, J. W. Campbell, G. Balazsi, E. Ravasz et al., 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185 5673–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, C. A., 1996. Function and regulation of heat shock proteins, pp. 1382–1399 in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, edited by F. C. Neidhardt, III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter and H. E. Umbarger. American Society for Microbiology, Washington, DC.
- Gur, E., D. Biran, N. Shechter, P. Genevaux, C. Georgopoulos et al., 2004. The Escherichia coli DjlA and CbpA proteins can substitute for DnaJ in DnaK-mediated protein disaggregation. J. Bacteriol. 186 7236–7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman, L. M., D. Belin, M. J. Carson and J. Beckwith, 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirvas, L., M. Nurminen, I. M. Helander, R. Vuorio and M. Vaara, 1997. The lipid A biosynthesis deficiency of the Escherichia coli antibiotic-supersensitive mutant LH530 is suppressed by a novel locus, ORF195. Microbiology 143 73–81. [DOI] [PubMed] [Google Scholar]
- Kang, Y., T. Durfee, J. D. Glasner, Y. Qiu, D. Frisch et al., 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186 4921–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, T. M., S. A. Stachula, C. R. Raetz and M. S. Anderson, 1993. The firA gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase. The third step of endotoxin biosynthesis. J. Biol. Chem. 268 19866–19874. [PubMed] [Google Scholar]
- Kloser, A. W., M. W. Laird and R. Misra, 1996. asmB, a suppressor locus for assembly-defective OmpF mutants of Escherichia coli, is allelic to envA (lpxC). J. Bacteriol. 178 5138–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle et al., 1996. A new enzyme superfamily: the phosphopantetheinyl transferases. Chem. Biol. 3 923–936. [DOI] [PubMed] [Google Scholar]
- Lesley, S. A., J. Graziano, C. Y. Cho, M. W. Knuth and H. E. Klock, 2002. Gene expression response to misfolded protein as a screen for soluble recombinant protein. Protein Eng. 15 153–160. [DOI] [PubMed] [Google Scholar]
- Miller, J. H., 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Mofid, M. R., M. A. Marahiel, R. Ficner and K. Reuter, 1999. Crystallization and preliminary crystallographic studies of Sfp: a phosphopantetheinyl transferase of modular peptide synthetases. Acta Crystallogr. D Biol. Crystallogr. 55 1098–1100. [DOI] [PubMed] [Google Scholar]
- Mofid, M. R., R. Finking, L. O. Essen and M. A. Marahiel, 2004. Structure-based mutational analysis of the 4′-phosphopantetheinyl transferases Sfp from Bacillus subtilis: carrier protein recognition and reaction mechanism. Biochemistry 43 4128–4136. [DOI] [PubMed] [Google Scholar]
- Mootz, H. D., R. Finking and M. A. Marahiel, 2001. 4′-Phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J. Biol. Chem. 276 37289–37298. [DOI] [PubMed] [Google Scholar]
- Nichols, B. P., O. Shafiq and V. Meiners, 1998. Sequence analysis of Tn10 insertion sites in a collection of Escherichia coli strains used for genetic mapping and strain construction. J. Bacteriol. 180 6408–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark, S., 1970. Genetics of a chain-forming mutant of Escherichia coli: transduction and dominance of the envA gene mediating increased penetration to some antibacterial agents. Genet. Res. 16 63–78. [DOI] [PubMed] [Google Scholar]
- Nurminen, M., L. Hirvas and M. Vaara, 1997. The outer membrane of lipid A-deficient Escherichia coli mutant LH530 has reduced levels of OmpF and leaks periplasmic enzymes. Microbiology 143 1533–1537. [DOI] [PubMed] [Google Scholar]
- Oh, M. K., and J. C. Liao, 2000. DNA microarray detection of metabolic responses to protein overproduction in Escherichia coli. Metab. Eng. 2 201–209. [DOI] [PubMed] [Google Scholar]
- Parris, K. D., L. Lin, A. Tam, R. Mathew, J. Hixon et al., 2000. Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites. Structure 8 883–895. [DOI] [PubMed] [Google Scholar]
- Raetz, C. R., C. M. Reynolds, M. S. Trent and R. E. Bishop, 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76 295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter, K., M. R. Mofid, M. A. Marahiel and R. Ficner, 1999. Crystal structure of the surfactin synthetase-activating enzyme Sfp: a prototype of the 4′-phosphopantetheinyl transferase superfamily. EMBO J. 18 6823–6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, J. R., N. Benson, T. Galitski, K. Haack, J. G. Lawrence et al., 1996. Rearrangements of the bacterial chromosome: formation and applications, pp. 2256–2276 in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, edited by F. C. Neidhardt, III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter and H. E. Umbarger. American Society for Microbiology, Washington, DC.
- Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel et al., 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H. E., 2007. The transcriptional response of Escherichia coli to recombinant protein insolubility. J. Struct. Funct. Genomics 8 27–35. [DOI] [PubMed] [Google Scholar]
- Thomas, J., and J. E. Cronan, 2005. The enigmatic acyl carrier protein phosphodiesterase of Escherichia coli: genetic and enzymological characterization. J. Biol. Chem. 280 34675–34683. [DOI] [PubMed] [Google Scholar]
- Thomas, J., D. J. Rigden and J. E. Cronan, 2007. Acyl carrier protein phosphodiesterase (AcpH) of Escherichia coli is a non-canonical member of the HD phosphatase/phosphodiesterase family. Biochemistry 46 129–136. [DOI] [PubMed] [Google Scholar]
- Tsuruoka, T., M. Ito, S. Tomioka, A. Hirata and M. Matsuhashi, 1988. Thermosensitive omsA mutation of Escherichia coli that causes thermoregulated release of periplasmic proteins. J. Bacteriol. 170 5229–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara, M., and M. Nurminen, 1999. Outer membrane permeability barrier in Escherichia coli mutants that are defective in the late acyltransferases of lipid A biosynthesis. Antimicrob. Agents Chemother. 43 1459–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T., A. C. McCandlish, L. S. Gronenberg, S. S. Chng, T. J. Silhavy et al., 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 103 11754–11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron, C., J. Vieira and J. Messing, 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33 103–119. [DOI] [PubMed] [Google Scholar]
- Young, K., L. L. Silver, D. Bramhill, P. Cameron, S. S. Eveland et al., 1995. The envA permeability/cell division gene of Escherichia coli encodes the second enzyme of lipid A biosynthesis. UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. J. Biol. Chem. 270 30384–30391. [DOI] [PubMed] [Google Scholar]
- Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland et al., 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., S. Lin, R. J. Cotter and C. R. Raetz, 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and palmitate. J. Biol. Chem. 274 18503–18514. [DOI] [PubMed] [Google Scholar]