Abstract
Biochemical interactions between cis-regulatory DNA sequences and trans-regulatory gene products suggest that cis- and trans-acting polymorphisms may interact genetically. Here we present a strategy to test this hypothesis by comparing the relative cis-regulatory activity of two alleles in different genetic backgrounds. Of the eight genes surveyed in this study, five were affected by trans-acting variation that altered total transcript levels, two of which were also affected by differences in cis-regulation. The presence of trans-acting variation had no effect on relative cis-regulatory activity, showing that cis-regulatory polymorphisms can function independently of trans-regulatory variation. The frequency of such independent interactions on a genomic scale is yet to be determined.
EPISTATIC interactions are a common feature of the genetic architecture underlying quantitative phenotypes (Mackay 2001). Levels of gene expression show patterns of inheritance characteristic of quantitative traits, suggesting that nonadditive interactions may often underlie regulatory variation (Gibson and Weir 2005). Consistent with this hypothesis, over half of the yeast genes for which two quantitative trait loci (QTL) affecting gene expression were identified showed evidence of epistatic interactions (Brem et al. 2005). Epistatic interactions affecting gene expression have also been inferred in Drosophila (Gibson 1996; Gibson et al. 2004; Wayne et al. 2004; Landry et al. 2005; Hughes et al. 2006; Osada et al. 2006), although specific interacting loci have not yet been identified.
The basic molecular mechanisms controlling gene expression provide ample opportunity for epistatic interactions (Alberts et al. 2002; Gibson and Weir 2005; Gjuvsland et al. 2007). Gene expression requires direct binding of trans-acting transcription factors to cis-regulatory sequences as well as protein–protein and protein–RNA interactions among additional, indirect, trans-acting factors. These biochemical interactions suggest that polymorphisms in cis-regulatory sequences and/or in genes encoding trans-acting factors may interact epistatically. Such interactions could occur between two trans-acting loci, between two cis-acting loci, or between cis- and trans-acting loci. The relative importance of each type of interaction remains unknown. (The “cis” or “trans” classification of epistatically interacting regulatory loci identified in yeast was not examined; R. Brem, personal communication). Alternatively, interactions may occur at the molecular level without any sign of statistical (epistatic) interaction among polymorphisms at cis- and trans-acting loci.
Here, we show how the relative activity of two cis-regulatory alleles in different trans-regulatory backgrounds can be compared to test specifically for epistatic interactions between cis- and trans-acting polymorphisms. If cis- and trans-regulatory variants act independently, relative cis-regulatory activity should be the same in the two genetic backgrounds. If, however, cis- and trans-regulatory variants interact epistatically, relative cis-regulatory activity should differ between genetic backgrounds.
trans-regulatory variation affects standing levels of gene expression:
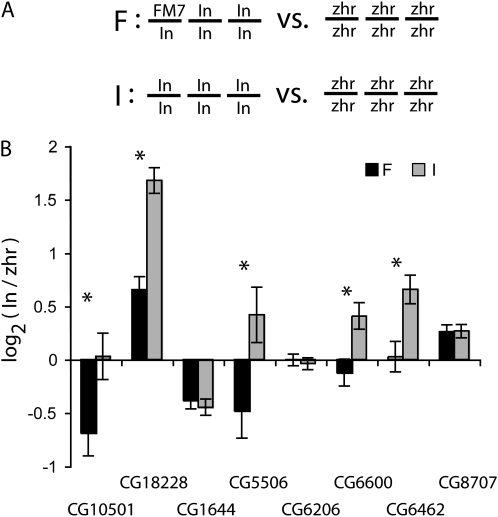
To examine the effects of trans-regulatory variation on transcript abundance, we used pyrosequencing to compare expression of autosomal genes between genotypes that differed by one X chromosome (Figure 1A); X-linked regulatory variants can only have trans-acting effects on autosomal gene expression. An inbred strain of Drosophila melanogaster, In(1)AB, which was segregating the FM7 balancer X chromosome, was used for this work. The FM7 balancer chromosome suppresses recombination, allowing genetic differences to accumulate between the In(1)AB and FM7 X chromosomes. Gene expression was compared between females heterozygous for the FM7 and In(1)AB X chromosomes (“F” in Figure 1A) and females homozygous for the In(1)AB X chromosome (“I” in Figure 1A). All flies were homozygous for the In(1)AB autosomes. A second inbred strain of D. melanogaster, zhr, was included as a common reference point, and four replicate pools of flies were analyzed for each genotypic combination (see Figure 1 legend).
Figure 1.—
trans-regulatory variation affects levels of gene expression. (A) X, second, and third chromosome genotypes present in the F and I pools are shown, with “In” indicating chromosomes from the In(1)AB strain and “zhr” indicating chromosomes from the zhr strain. Note that these genotypic combinations differ only by the presence or absence of the FM7 X chromosome. For both the heterozygous (F) and homozygous (I) X chromosome genotypes, the relative abundance of autosomal transcripts from the zhr and In(1)AB alleles was measured in four replicate pools of adult flies. Each pool contained seven adult females (7–10 days old) from the zhr line and seven females from the In(1)AB line, either with (F) or without (I) the FM7 X chromosome. After sequentially extracting RNA and genomic DNA from each pool, duplicate cDNA pools were synthesized and used to measure expression of eight autosomal genes using pyrosequencing. Allele abundance in the genomic DNA samples was also measured in duplicate and used to normalize measurements as described in Landry et al. (2005). After normalization, the log2 expression ratios of the In(1)AB allele to the zhr allele were fitted to a mixed linear model with a fixed effect of genotype (F and I) and a random effect of replicate fly pools using “proc MIXED” in SAS v.8.2 (Cary, NC). (B) Least-squares means and significance tests from this model are shown. Asterisks indicate cases where P < 0.06 and error bars indicate standard errors. The selection of the nontraditional α = 0.06 is explained in the main text.
Expression of eight genes was examined in this work. These genes are the subset of autosomal genes analyzed in Wittkopp et al. (2004) with nucleotide differences between the In(1)AB and zhr autosomes suitable for pyrosequencing (Ahmadian et al. 2000). Proteins encoded by these genes perform a variety of molecular functions (Table 1). We do not anticipate any bias in this gene set with respect to interactions between cis- and trans-regulatory polymorphisms; however, additional studies of cis- and trans-regulatory interactions are necessary to determine whether these genes are representative of the genome. Furthermore, it remains to be seen whether the types of regulatory polymorphisms that accumulate laboratory lines differ from those segregating in natural populations.
TABLE 1.
Significance tests for differences between parental and hybrid pools
| I vs. Fa
|
B vs. H vs. Rb
|
H vs. Rc
|
H vs. Bd
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Name | Protein function | F-statistice | P-value | F-statistic | P-value | t-value | P-value | t-value | P-value |
| CG10501 | amd | Decarboxylase activity | 5.518 | 0.057 | 1.120 | 0.396 | 1.770 | 0.151 | 0.250 | 0.816 |
| CG1644 | Cyp6t1 | Electron carrier activity | 0.354 | 0.574 | 1.848 | 0.251 | 1.480 | 0.214 | −1.440 | 0.223 |
| CG18228 | — | Unknown | 36.244 | 0.001 | 0.069 | 0.934 | 0.020 | 0.983 | −0.370 | 0.728 |
| CG5506 | — | Unknown | 6.077 | 0.049 | 2.179 | 0.209 | −1.410 | 0.231 | 1.670 | 0.171 |
| CG6206 | — | Mannosidase activity | 10.559 | 0.017 | 1.904 | 0.243 | 1.810 | 0.145 | −0.710 | 0.515 |
| CG6462 | — | Endopeptidase activity | 0.208 | 0.665 | 0.584 | 0.592 | 1.090 | 0.338 | −0.140 | 0.899 |
| CG6600 | — | Cation transporter | 9.043 | 0.024 | 0.206 | 0.821 | −0.610 | 0.576 | −0.070 | 0.950 |
| CG8707 | — | GTPase activity | 0.002 | 0.965 | 1.206 | 0.374 | 0.170 | 0.870 | 1.320 | 0.257 |
I and F are relative expression between parental genotypes, as described in Figure 1; numerator degrees of freedom (d.f.) = 1, denominator d.f. = 6.
B, H, and R are relative allelic expression between hybrid genotypes, as described in Figure 2; numerator d.f. = 2, denominator d.f. = 5.
H and R are genetically identical hybrids that differ by which parent transmitted which allele; d.f. = 4.
H and B hybrid genotypes differ by one X chromosome; d.f. = 4.
F-statistics, t-values, and associated P-values were calculated using the mixed linear model described in Figure legends 1 and 2.
Measurements of relative gene expression (In(1)AB/zhr) were log2 transformed and normalized as described in the legend to Figure 1. A linear model, including replicate pools of flies as a random effect, was used to calculate the relative expression between genotypes for each gene (Table 2) and to test for significant expression differences between the F and I pools (Table 1). Treating replicate pools as a random effect is more conservative than treating them as a fixed effect because it compares the underlying populations from which the replicate pools were drawn rather than comparing only the observed data. With this in mind, we chose a slightly more generous than usual significance threshold of α = 0.06 to infer differences. Using this cutoff, five of the eight genes, including four cases where P < 0.05 and one case where P = 0.057, were deemed to have significant expression differences (Table 1, Figure 1B). The three remaining genes, which were deemed to have no significant expression difference between genotypes, all showed P > 0.5 (Table 1). Gene-specific 95% confidence intervals indicate that this test would reject the null hypothesis at α < 0.05 if substitution of the FM7 chromosome altered expression >3–11% (depending on the gene) in either direction, relative to zhr.
TABLE 2.
Relative expression between strains and between alleles
| Gene | Genotypea | log2 (In/zhr)b | 95% C.I. | t-valuec | d.f. | P-value |
|---|---|---|---|---|---|---|
| CG10501 | F | −0.686 | 0.526 | −3.193 | 6 | 0.019 |
| P | 0.031 | 0.530 | 0.141 | 6 | 0.892 | |
| B | 0.053 | 0.128 | 1.060 | 5 | 0.338 | |
| H | −0.027 | 0.091 | −0.755 | 5 | 0.484 | |
| R | −0.043 | 0.128 | −0.857 | 5 | 0.431 | |
| CG1644 | F | −0.381 | 0.193 | −4.836 | 6 | 0.003 |
| P | −0.447 | 0.189 | −5.797 | 6 | 0.001 | |
| B | −0.443 | 0.281 | −4.053 | 5 | 0.010 | |
| H | −0.661 | 0.199 | −8.556 | 5 | 0.000 | |
| R | −0.460 | 0.281 | −4.208 | 5 | 0.008 | |
| CG18228 | F | 0.656 | 0.295 | 5.440 | 6 | 0.002 |
| P | 1.680 | 0.293 | 14.010 | 6 | 0.000 | |
| B | −0.229 | 0.150 | −3.934 | 5 | 0.011 | |
| H | −0.231 | 0.106 | −5.608 | 5 | 0.002 | |
| R | −0.206 | 0.150 | −3.526 | 5 | 0.017 | |
| CG5506 | F | −0.479 | 0.629 | −1.863 | 6 | 0.112 |
| P | 0.420 | 0.633 | 1.624 | 6 | 0.156 | |
| B | −0.068 | 0.491 | −0.356 | 5 | 0.736 | |
| H | 0.287 | 0.347 | 2.130 | 5 | 0.086 | |
| R | −0.146 | 0.491 | −0.767 | 5 | 0.478 | |
| CG6206 | F | −0.003 | 0.134 | −0.050 | 6 | 0.961 |
| P | −0.038 | 0.137 | −0.686 | 6 | 0.518 | |
| B | −0.048 | 0.119 | −1.031 | 5 | 0.350 | |
| H | −0.107 | 0.084 | −3.287 | 5 | 0.022 | |
| R | −0.100 | 0.119 | −2.156 | 5 | 0.084 | |
| CG6462 | F | 0.028 | 0.346 | 0.198 | 6 | 0.850 |
| P | 0.658 | 0.325 | 4.958 | 6 | 0.003 | |
| B | 0.552 | 0.160 | 8.887 | 5 | 0.000 | |
| H | 0.404 | 0.113 | 9.198 | 5 | 0.000 | |
| R | 0.465 | 0.160 | 7.479 | 5 | 0.001 | |
| CG6600 | F | −0.121 | 0.305 | −0.971 | 6 | 0.369 |
| P | 0.409 | 0.305 | 3.282 | 6 | 0.017 | |
| B | −0.194 | 0.286 | −1.744 | 5 | 0.142 | |
| H | −0.115 | 0.202 | −1.460 | 5 | 0.204 | |
| R | −0.106 | 0.286 | −0.957 | 5 | 0.383 | |
| CG8707 | F | 0.264 | 0.151 | 4.272 | 6 | 0.005 |
| P | 0.268 | 0.151 | 4.336 | 6 | 0.005 | |
| B | 0.201 | 0.235 | 2.197 | 5 | 0.079 | |
| H | 0.179 | 0.166 | 2.772 | 5 | 0.039 | |
| R | 0.024 | 0.235 | 0.260 | 5 | 0.805 |
log2(In/zhr) are the least-squares (LS) means from the mixed linear model described in legends of Figures 1 and 2. The next column shows the 95% confidence intervals for each LS means.
t-values, degrees of freedom (d.f.), and P-values correspond to a t-test (H0:LS means = 0) performed within proc MIXED using SAS v. 8.2.
These data indicate that genetic differences between the In(1)AB and FM7 X chromosomes have trans-acting effects on expression of five of the eight genes examined. The functional polymorphism(s) may lie in the coding or noncoding regions of genes producing direct regulators of the affected gene or within coding or noncoding regions of genes producing indirect regulators that modify the activity of direct regulators. For the three genes that showed no significant expression difference between the F and I pools, the activity of any X-linked trans regulators is functionally equivalent between the FM7 and In(1)AB chromosomes, at least under the conditions assayed.
Relative cis-regulatory activity is independent of trans-regulatory variation:
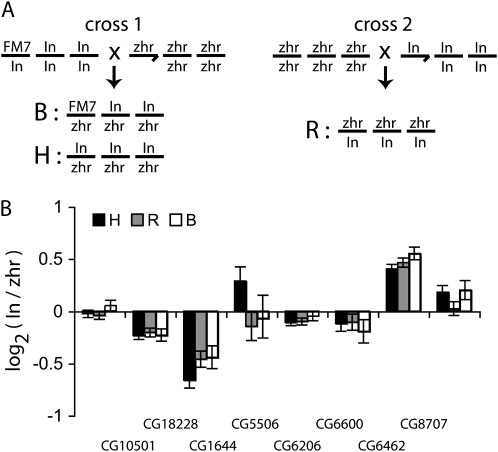
To determine whether trans-acting variation influences the relative activity of cis-regulatory alleles, we used measurements of allele-specific expression to compare cis-regulatory activity in different genetic backgrounds. Relative allelic expression in a heterozygote is a measure of relative cis-regulatory activity (Cowles et al. 2002). In(1)AB females heterozygous for the FM7 chromosome were crossed to zhr males, and allele-specific expression of the eight autosomal genes was measured in F1 hybrids. Reciprocal crosses were also performed to measure parent-of-origin effects on allele-specific gene expression by crossing zhr females to In(1)AB males. These In(1)AB males lacked the FM7 X chromosome, which is lethal in a hemizygous state. Together, these two crosses produced three distinct classes of female offspring (Figure 2A): the “B” class, which had the X chromosome genotype of FM7/zhr and the cytoplasm from the In(1)AB line; the “H” class, which had the X chromosome genotype of In(1)AB/zhr and the cytoplasm from the In(1)AB line; and the “R” class, which had the same genotype as “H” but the cytoplasm derived from the zhr line. These latter two classes also differed by which parent transmitted which allele. All three classes of females were heterozygous for the In(1)AB and zhr autosomes.
Figure 2.—
Relative cis-regulatory activity is independent of trans-regulatory variation. (A) X, second, and third chromosome genotypes of flies crossed to generate different classes of F1 hybrids are shown. “In” indicates chromosomes from the In(1)AB strain and “zhr” indicates chromosomes from the zhr strain. For each of the three hybrid classes, two pools, each containing 14 adult female flies (7–10 days old) were analyzed. RNA and genomic DNA were sequentially extracted from each pool. cDNA was synthesized and used to measure expression of the eight autosomal genes using pyrosequencing. Allele abundance in genomic DNA samples was also measured and used to normalize measurements as described in Landry et al. (2005). After normalization, the log2 ratios of allelic expression were fitted to the mixed linear model with a fixed effect of genotype (B, H, and R) and a random effect of replicate fly pools using proc MIXED in SAS v.8.2. (B) Least-squares means with their standard errors from this model are shown. In all cases, no significant difference was observed among the three hybrid genotypes (P > 0.2).
Relative cis-regulatory activity between the In(1)AB and zhr alleles was measured in all three genotypes using pyrosequencing. A linear mixed model (see Figure 2 legend) was used to calculate least-squares means (Table 2) and to test for significant differences among F1 hybrid genotypes. For all eight genes, no significant difference was observed among the three hybrid classes, between hybrids from reciprocal crosses (H and R) or between hybrids with different X chromosome genotypes (H and B) (Table 1, Figure 2B); relative expression of the zhr and In(1)AB cis-regulatory alleles was the same regardless of the direction of cross or the presence/absence of the FM7 chromosome. Gene-specific 95% confidence intervals indicate that the null hypothesis of equal expression would be rejected at α < 0.05 if expression of the In(1)AB allele differed between hybrids >1–7% (depending on the gene) in either direction, relative to the zhr allele.
The absence of allele-specific, parent-of-origin effects is consistent with prior studies showing no evidence of genomic imprinting in D. melanogaster (Wittkopp et al. 2006). More importantly, the similar allele-specific expression observed between the B and H classes indicates that genetic differences between the FM7 and In(1)AB X chromosomes had no effect on relative cis-regulatory activity. This was true for the three genes unaffected by the substitution of the X chromosome as well as for the five genes affected by trans-acting regulatory differences between the In(1)AB and FM7 X chromosomes, including the two genes with both cis- and trans-regulatory differences.
Discussion:
This study shows how comparisons of allele-specific expression among genetic backgrounds can be used to test for epistasis between cis- and trans-regulatory variation. Although we started with a set of 22 autosomal genes (data not shown), only 8 contained sequence differences suitable for pyrosequencing and, of these, only 2 showed the significant evidence of both cis- and trans-regulatory variation necessary to test for epistasis. Neither of these genes showed evidence of epistatic interactions between cis- and trans-acting regulatory polymorphisms.
Prior studies suggest there may be extensive epistasis among loci underlying differential gene expression (Gibson et al. 2004; Brem et al. 2005), but interactions between cis- and trans-acting factors are only one source of these interactions. Despite our small sample size, we propose that epistatic interactions between cis- and trans-acting factors may be rare in general because they require trans-acting variants to interact differently with alternate cis-regulatory alleles (e.g., a polymorphism in the DNA binding region of a transcription factor may interact epistatically with a polymorphism in the cis-regulatory binding site for this factor). Such combinations of polymorphisms may be rare within species. Interactions among trans-regulatory polymorphisms may be more common: using a crossing design that minimized trans-regulatory variation, Hughes et al. (2006) found less evidence of epistasis among regulatory loci in Drosophila than did studies with other crossing designs (Gibson et al. 2004; Wayne et al. 2004; Osada et al. 2006).
Determining the prevalence of epistatic interactions between cis- and trans-acting loci is important because it affects the way gene expression evolves. In the absence of interactions, cis- and trans-acting regulatory changes can evolve independently. That is, cis-regulatory changes should have similar effects on gene expression, regardless of the genetic background. This may often be true within species, especially within populations harboring little genetic variation. As regulatory divergence increases, however, there may be more opportunities for epistatic interactions and epistatically interacting cis- and trans-regulatory changes may become more common. Consistent with this idea, interactions between cis- and trans-acting changes have been implicated as a source of dysregulation in interspecific hybrids of D. melanogaster and D. simulans (Landry et al. 2005). Determining how cis- and trans-regulatory variants interact with each other over different evolutionary timescales will help us better understand how regulatory polymorphisms segregate within species and become fixed between species.
Acknowledgments
We thank D. Barbash and A. Orr for the In(1)AB and zhr strains of D. melanogaster, respectively, as well as Rachel Brem and Bernardo Lemos for sharing unpublished observations and helpful comments on the manuscript. Funding for this project was provided by National Institutes of Health grants to A.G.C. P.J.W. was a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation, and B.K.H. was funded by a Howard Hughes undergraduate research award.
References
- Ahmadian, A., B. Gharizadeh, A. Gustafsson, F. Sterky, P. Nyrén et al., 2000. Single-nucleotide polymorphism analysis by pyrosequencing. Anal. Biochem. 280 103–110. [DOI] [PubMed] [Google Scholar]
- Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts et al., 2002. Molecular Biology of the Cell. Garland, New York.
- Brem, R., J. Storey, J. Whittle and L. Kruglyak, 2005. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature 436 701–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles, C., J. Hirschhorn, D. Altshuler and E. Lander, 2002. Detection of regulatory variation in mouse genes. Nat. Genet. 32 432–437. [DOI] [PubMed] [Google Scholar]
- Gibson, G., 1996. Epistasis and pleiotropy as natural properties of transcriptional regulation. Theor. Popul. Biol. 49 58–89. [DOI] [PubMed] [Google Scholar]
- Gibson, G., and B. Weir, 2005. The quantitative genetics of transcription. Trends Genet. 21 616–623. [DOI] [PubMed] [Google Scholar]
- Gibson, G., R. Riley-Berger, L. Harshman, A. Kopp, S. Vacha et al., 2004. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics 167 1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjuvsland, A., B. Hayes, S. Omholt and O. Carlborg, 2007. Statistical epistasis is a generic feature of gene regulatory networks. Genetics 175 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, K., J. Ayroles, M. Reedy, J. Drnevich, K. Rowe et al., 2006. Segregating variation in the transcriptome: cis-regulation and additivity of effects. Genetics 173 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, C., P. Wittkopp, C. Taubes, J. Ranz, A. Clark et al., 2005. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171 1813–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, T., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35 303–339. [DOI] [PubMed] [Google Scholar]
- Osada, N., M. Kohn and C. Wu, 2006. Genomic inferences of the cis-regulatory nucleotide polymorphisms underlying gene expression differences between Drosophila melanogaster mating races. Mol. Biol. Evol. 23 1585–1591. [DOI] [PubMed] [Google Scholar]
- Wayne, M., Y. Pan, S. Nuzhdin and L. McIntyre, 2004. Additivity and trans-acting effects on gene expression in male Drosophila simulans. Genetics 168 1413–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp, P., B. Haerum and A. Clark, 2004. Evolutionary changes in cis and trans gene regulation. Nature 430 85–88. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P., B. Haerum and A. Clark, 2006. Parent-of-origin effects on mRNA expression in Drosophila melanogaster not caused by genomic imprinting. Genetics 173 1817–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]