Abstract
The larval body wall muscles of Drosophila melanogaster arise by fusion of founder myoblasts (FMs) and fusion-competent myoblasts (FCMs). Sticks-and-Stones (SNS) is expressed on the surface of all FCMs and mediates adhesion with FMs and developing syncytia. Intracellular components essential for myoblast fusion are then recruited to these adhesive contacts. In the studies herein, a functional analysis of the SNS cytodomain using the GAL4/UAS system identified sequences that direct myoblast fusion, presumably through recruitment of these intracellular components. An extensive series of deletion and site-directed mutations were evaluated for their ability to rescue the myoblast fusion defects of sns mutant embryos. Deletion studies revealed redundant functional domains within SNS. Surprisingly, highly conserved consensus sites for binding post-synaptic density-95/discs large/zonula occludens-1-domain-containing (PDZ) proteins and serines with a high probability of phosphorylation play no significant role in myoblast fusion. Biochemical studies establish that the SNS cytodomain is phosphorylated at multiple tyrosines and their site-directed mutagenesis compromises the ability of the corresponding transgenes to rescue myoblast fusion. Similar mutagenesis revealed a requirement for conserved proline-rich regions. This complexity and redundancy of multiple critical sequences within the SNS cytodomain suggest that it functions through a complex array of interactions that likely includes both phosphotyrosine-binding and SH3-domain-containing proteins.
IN a Drosophila embryo, founder myoblasts (FMs) specify the ultimate pattern of multinucleate syncytia and seed the fusion process, while fusion-competent myoblasts (FCMs) recognize, adhere to, and fuse with these FMs. The initial fusion event between the FM and the FCM is followed by multiple rounds of fusion between FCMs and the developing syncytia until the final muscle size is achieved (reviewed in Abmayr and Kocherlakota 2005). The FMs must express one of two functionally redundant cell adhesion molecules, either Dumbfounded/Kin-of-IrreC (Duf/Kirre) or Irregular Chiasm-C/Roughest (IrreC/Rst) (Ruiz-Gomez et al. 2000; Strunkelnberg et al. 2001). These proteins have been shown to act as attractants for the FCMs (Ruiz-Gomez et al. 2000; Strunkelnberg et al. 2001). The FCMs must, in turn, express the Sticks-and-Stones (SNS) cell adhesion molecule (Bour et al. 2000). Mutants in sns phenocopy the myoblast fusion defect seen in Df(1)w67k30 embryos, which lack both Duf/Kirre and IrreC/Rst, and are characterized by the complete absence of differentiated muscle fibers (Bour et al. 2000; Ruiz-Gomez et al. 2000).
SNS, Duf/Kirre, and IrreC/Rst are all members of the immunoglobulin superfamily (IgSF), encoding single-pass transmembrane proteins with cytoplasmic tails of 370, 358, and 210 amino acids, respectively. They function as ligand-receptor pairs that become localized to points of cell-cell contact (Galletta et al. 2004), whereupon they recruit and/or activate downstream components that lead to myoblast fusion. Duf/Kirre is thought to act upstream of the monomeric guanosine triphosphatase (GTPase) Rac1 in the FM. In this pathway, the adaptor molecule Antisocial/Rolling pebbles (Ants/Rols) (Chen and Olson 2001) is recruited to Duf/Kirre via interaction of its TPR repeats with the intracellular domain of Duf/Kirre (Kreiskother et al. 2006). Ants/Rols then appears to recruit Myoblast city (MBC) (Erickson et al. 1997; Chen and Olson 2001), an unconventional guanine nucleotide exchange factor (GEF) for Rac1 (Nolan et al. 1998; Geisbrecht et al. 2008). In a pathway that appears to be parallel, Duf/Kirre recruits Loner, a GEF for the small GTPase Arf6 (Chen et al. 2003). While the molecular targets of each pathway have not been defined, they are both thought to converge on the actin cytoskeleton (Chen and Olson 2004).
Since Ants/Rols is unique to the FMs, alternative mechanisms must direct changes in the actin cytoskeleton at points of cell contact in the FCMs. However, much less is known about these interactions. MBC, which is also required in these cells (Balagopalan et al. 2006), must be recruited to SNS and/or points of cell contact by an adaptor that is distinct from Ants/Rols. The adaptor protein Crk is one candidate for this purpose, since it has been shown to interact both biochemically and genetically with MBC in other systems. However, the observation that the Crk binding sites in MBC are expendable for myoblast fusion argues against this simple model (Ishimaru et al. 2004; Balagopalan et al. 2006). Intriguingly, recent studies implicate Crk in direct interactions with SNS and with the FCM-specific protein, Solitary (Sltr) (Kim et al. 2007), also reported as the Drosophila ortholog of Wasp Interacting protein (D-WIP) (Massarwa et al. 2007). In the FCMs, the formation of F-actin foci appears to be dependent on the ability of SNS to recruit Sltr/D-WIP to sites of cell contact, likely via Crk (Kim et al. 2007). Sltr/D-WIP, in turn, directs changes in the actin cytoskeleton via Wasp and the Arp 2/3 pathway (Kim et al. 2007; Massarwa et al. 2007; Schafer et al. 2007). F-actin foci have been implicated in cytoskeletal reorganization (Richardson et al. 2007) and may be associated with the electron-dense fusion vesicles (Doberstein et al. 1997; Kim et al. 2007). These vesicles do not appear to accumulate at sites of adhesion in either sns or sltr/D-wip mutant embryos (Doberstein et al. 1997; Kim et al. 2007). Consistent with these observations, SNS is present at sites of cell contact in a ring-shaped structure termed a fusion-restricted myogenic-adhesive structure (FuRMAS) throughout myoblast fusion (Kesper et al. 2007). This structure is thought to link cell adhesion and local F-actin assembly and dynamics to downstream events that ultimately lead to plasma membrane fusion (Kesper et al. 2007).
Nephrin and Syg-2, structural orthologs of SNS, have been identified in mammals and Caenorhabditis elegans, respectively (Lenkkeri et al. 1999; Shen et al. 2004). Nephrin serves as a key component of the slit diaphragm in the kidney glomerulus. The cytodomain of nephrin shares 25% identity with that of SNS and is essential for its function. Individuals with a truncated form of nephrin that lacks the cytodomain suffer from severe proteinuria, as a consequence of defects in filtration through the slit diaphragm (Lenkkeri et al. 1999). Analysis of the nephrin cytoplasmic region has placed it upstream of multiple intracellular signaling pathways that lead to actin polymerization. Nephrin interacts directly with two cytoplasmic proteins, the Src homology 3 (SH3) domain containing CD2-associated protein (CD2AP) and the Src homology 2 (SH2)–SH3 adaptor protein, Nck (Huber et al. 2003a; Jones et al. 2006). The latter interaction is enhanced by nephrin phosphorylation on tyrosine residues (Jones et al. 2006). Interaction with Nck in turn facilitates localized actin reorganization in cell culture (Jones et al. 2006). Nephrin also colocalizes with the PDZ-domain-containing protein zona occludens 1 (ZO-1) in kidney cells, a protein that interacts directly with the Duf/Kirre ortholog Neph1 (Ruotsalainen et al. 2000; Huber et al. 2003b).
With the expectation that SNS may, like nephrin, direct signaling events that modify cell structure and behavior, we have carried out a detailed functional dissection of the SNS cytodomain. Mutagenized forms of SNS have been expressed in the mesoderm of sns mutant embryos using the GAL4/UAS system (Brand and Perrimon 1993) and assayed for their ability to rescue the myoblast fusion phenotype. Deletion studies have revealed a region of 166 amino acids that is essential for SNS function during myoblast fusion, and smaller domains within this region function redundantly to fulfill this requirement. In an effort to better define these sequences, we have carried out extensive site-directed mutagenesis. We have also established that SNS is phosphorylated specifically on tyrosine residues and that these residues play an important role in the ability of SNS to restore the wild-type muscle pattern in sns mutant embryos. In addition, proline-rich regions that form consensus binding sites for SH3-domain-containing proteins also play a role in the ability of SNS to direct myoblast fusion. These data show that specific sequences of the SNS cytodomain are required for its function during myoblast fusion and support the notion that, like Duf/Kirre, multiple proteins may interact with SNS via different sequences.
MATERIALS AND METHODS
Fly stocks:
All stocks used in this study were maintained on standard cornmeal media at 18° or 25° as necessary. Oregon R was used as a wild-type strain. Stocks P{(w[+mC]=mef2-GAL4)}, snsZf1.4, and snsXB3 have been described (Ranganayakulu et al. 1998; Bour et al. 2000). sns20-5 was obtained from R. Renkawitz-Pohl (Paululat et al. 1995). To determine the molecular lesion, sns20-5 was balanced with CyO, P{(w[+mC]=GAL4-twi.G)}2.2, P(UAS-2xEGFP)AH2.2 (Bloomington Stock Center), and homozygous mutant embryos were identified by the absence of green fluorescence protein (GFP). The sns sequence was amplified from total RNA by RT–polymerase chain reaction (PCR) using Superscript III (Invitrogen, San Diego) and high-fidelity polymerase (Roche, Indianapolis). The products were sequenced at the Stowers Institute Molecular Biology Facility. Transgenic stocks were generated by Genetic Services. The transgenes were recombined into snsZf1.4 or snsXB3 genetic background, as indicated, and balanced with CyO P[w+wgen11en-LacZ]. In all cases, at least two independent transgenic stocks were analyzed.
Cloning and constructs:
The full-length sns cDNA isolated by Bour et al. (2000) was subcloned into pRmHa3 (Bunch et al. 1988), using the EcoRI linker from the bacteriophage library at the 5′ end and an NheI site at position 5612 of the cDNA sequence (Galletta et al. 2004). A single hemagglutinin (HA) tag was added immediately following the last amino acid of the SNS coding sequence and cloned into pUAST to generate UAS-sns-HA. Deletions were generated using PCR with mismatch oligonucleotides. For the HA-tagged membrane proximal deletions, the corresponding region in pUAST-sns-HA was replaced using XhoI and XbaI restriction sites. The untagged membrane distal deletion (UAS-snsΔ1279-1482) was similarly generated by PCR and replaced into pUAST-sns (Galletta et al. 2004), using AflII and KpnI. To make UAS-sns5xPDZ-HA, UAS-sns2xPXXP, UAS-snsA17-HA, and UAS-snsF14-HA, site-directed mutagenesis was employed and the region amplified by PCR using mismatch oligonucleotides, in several rounds until all the desired sites were mutagenized. The region was then replaced into pUAST-sns for UAS-sns2xPXXP using restriction sites AflII and RsrII and into pUAST-sns-HA for the HA-tagged constructs using restriction sites XhoI and XbaI. The final sequence of the entire cDNA in all constructs was confirmed prior to injection to ensure lack of second-site unwanted errors. The snsGal4 construct was made by cloning a 5-kb fragment including 2 kb upstream of the sns genomic region into pPTGAL vector (Sharma et al. 2002). sns-LacZ was generated by introducing the same region into pH-pelican (Barolo et al. 2000).
Immunohistochemistry:
Embryos were collected on agar–apple juice plates at 25°, aged at 25° or 18° as needed, and fixed as described (Erickson et al. 1997). Homozygous mutant embryos were identified by the absence of β-galactosidase (β-gal) activity and verified to be 25% of the population. Primary antibodies included a monoclonal antibody to myosin heavy chain (MHC) (1:1000, D. Kiehart), rabbit polyclonal anti-β-gal (1:1000, MP Biomedicals), and affinity-purified rabbit anti-SNS antisera at 1:100 (Galletta et al. 2004). Colorimetric detection was performed using the Vectastain ABC elite kit (Vector Laboratories, Burlingame, CA) according to manufacturer's instructions with biotinyl-tyramide enhancement for anti-SNS. Fluorescent detection was performed using Alexa-labeled secondary antibodies (1:200; Molecular Probes, Eugene, OR). Colorimetrically stained embryos were visualized and imaged using a Zeiss Axioplan2; fluorescently stained embryos were imaged using a Zeiss LSM confocal microscope and analyzed using the corresponding software.
Quantitation and statistical comparison:
The presence of muscles LT1–3 (Bate and Rushton 1993) was quantitated for abdominal segments A2–A5 in myosin-stained stage 16 embryos in which snsGal4 directed expression of the transgene. The hemisegment was considered defective if one or more of LT1–3 was missing. The defective hemisegments were calculated as a percentage of total hemisegments analyzed and compared across genotypes. The n value indicates the total number of hemisegments analyzed for each genotype. Pairwise comparisons were performed using a one-way analysis of variance (ANOVA) model followed by a Tukey's honestly significant differences (HSD) test and the resulting P-values used to ascertain statistical significance.
The number of unfused myoblasts was quantitated in rescued embryos fluorescently labeled for β-gal. Embryos were first visualized by differential interference contrast (DIC) and staged according to Campos-Ortega to ensure comparison of matched early stage-16 embryos (Campos-Ortega and Hartenstein 1997). The ventral musculature of abdominal hemisegments A2–A4 was imaged in serial 1-μm sections, and the images were imported into Imaris (Bitplane). The cell diameter was set to 4 μm to distinguish unfused myoblasts and eliminate faintly stained myotubes. Computationally counted cells were verified visually. The number of cells across hemisegments A2–A4 was determined for each genotype and the average compared with that obtained with the full-length cDNA (n indicates the number of embryos analyzed). The comparison was done in pairwise fashion and P-values were calculated using a one-way ANOVA model followed by a Tukey's HSD test.
Analysis of phosphorylation by immunoprecipitation and Western blotting:
Schneider line 2 (S2) cells were grown as described (Cherbas and Cherbas 1998) and transiently transfected using calcium phosphate (Ashburner 1989). SNS expression was induced from the metallothionein promoter in pRmHa3 (Bunch et al. 1998), using 700 μm copper sulfate. Cells were resuspended in lysis buffer (2% Triton X-100, 60 mm Tris-HCl pH 7.4, 6 mm EDTA, and 300 mm NaCl) in the presence of protease inhibitors (2 mm leupeptin and pepstatin, 1 mm PMSF) and sodium orthovanadate (Na3VO4, 1 mm) as a phosphatase inhibitor. Lysates were passaged through a 25-gauge needle for 10 strokes. For embryo lysates, UAS-snsHA/mef2Gal4 embryos were collected at 25° for 6 hr and aged at 18° for 16 hr. The embryos were dechorionated in 50% bleach and homogenized in the above lysis buffer at a concentration of 0.1 mg wet weight/ml. The lysates were centrifuged at 20,000 × g to remove debris.
Immunoprecipitations utilized 1 mg total protein from S2 cell lysates or 2–5 mg total protein from embryo lysates. Lysates were incubated with 20 μl anti-HA affinity matrix (Roche) overnight at 4°. The beads were washed in lysis buffer three times and bound material was eluted by boiling in 40 μl Laemmli buffer unless treated with phosphatases (see below). The eluted proteins were analyzed by SDS–PAGE on a 5% acrylamide gel and transferred to PVDF membranes. Membranes were probed with anti-phosphotyrosine (1:1000; Upstate Biotechnology) and anti-SNS at 1:500 (Galletta et al. 2004) or anti-HA antibodies (1:1000, Roche). The immunoblots were developed using chemiluminescence by ECL reagents (Amersham GE) and detected by scanning on a Typhoon 9400 (Amersham GE).
Phosphatase treatment:
Following immunoprecipitation, the resin was washed in phosphatase buffer (50 mm Tris-HCl pH 8.0, 0.1 mm EDTA pH 8.0, 1 mm EDTA, 0.01% NP-40, and 2 mm MnCl2) three times and incubated with 1 μl protein tyrosine phosphatase 1b (3 units/μl, Upstate Biotechnology) or λ-phosphatase (400 units/μl; New England Biolabs, Beverly, MA) at 30° for 30 min in 50 μl of phosphatase buffer in the absence of Na3VO4. The resin was washed three times with lysis buffer to remove phosphatase, and the bound protein was eluted by boiling in Laemmli buffer. Samples were analyzed on Western blots as described above.
Isolation of membrane-bound proteins:
Membrane fractions were isolated as previously described (Hortsch 1994). Briefly, embryos expressing the transgene of interest were aged as above, dechorionated, and homogenized in hypotonic buffer (10 mm Tris-HCl pH 8.0, 1 mm EDTA pH 8.0) with protease inhibitor cocktail (Roche) at 0.1 mg wet weight/ml. Lysates were centrifuged at 20,000 × g to remove debris and the supernatant was spun through a 50-mm sucrose cushion in a 70.1Ti Beckman (Fullerton, CA) rotor at 100,000 × g for 1 hr at 4°. The supernatant (cytosolic fraction) was collected. The pellet (membrane fraction) was resuspended in an equal volume of hypotonic buffer. Embryo equivalents of membrane and cytosolic fractions were resolved by 4–12% SDS–PAGE and compared to evaluate membrane localization of the protein of interest. Mouse anti-tubulin (1:25,000; Sigma, St. Louis) antibody ensured that cytosolic proteins are not in the membrane fractions.
RESULTS
The cytodomain is essential for SNS function in embryonic myoblasts:
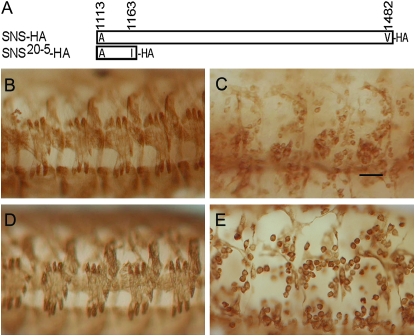
We have previously shown that the IgSF proteins SNS and either Duf/Kirre or IrreC/Rst direct heterotypic cell interactions when transfected into S2 cells. We have also shown that neither the SNS nor the Duf/Kirre transmembrane and cytoplasmic regions are necessary in this assay, since proteins anchored to the membrane via a GPI linkage can still direct cell interactions. However, the GPI-anchored form of SNS is unable to rescue myoblast fusion in the embryo, implicating the transmembrane and/or cytoplasmic domains in SNS function (Galletta et al. 2004). We have now investigated whether the cytodomain of SNS is specifically required in myoblasts. The first evidence that the SNS cytodomain is essential was provided by the EMS-induced allele sns20-5, in which the splice donor site in intron 18 is mutated (supplemental Figure 1A at http://www.genetics.org/supplemental/). Two transcripts are present in these mutants. Intron 18 is not removed in the first of these transcripts, such that it encodes a protein in which the normal SNS sequence ends at amino acid I1163 and is followed by 46 additional amino acids that result from translation of intronic sequence. A second transcript results from the use of a cryptic splice donor site that is three nucleotides after the correct splice site and encodes a protein in which the SNS sequence also diverges at I1163. Protein is clearly detected upon immunostaining of sns20-5 embryos using an antibody to the extracellular region of SNS (supplemental Figure 1C). Despite the presence of this protein, the myoblast fusion defects in these embryos are similar to those observed in mutant alleles that are completely lacking SNS (Figure 1C) (Bour et al. 2000), indicating that the cytodomain beyond I1163 is essential for SNS function. To ensure that the loss-of-function phenotype of sns20-5 is not due to interference from the extra amino acids added as a consequence of the splice site lesion, we engineered a transgene with a stop codon after amino acid I1163 of the SNS coding sequence (Figure 1A). We confirmed expression of the resulting epitope-tagged transgene in a Western blot using HA antibody (data not shown) and then examined its ability to rescue the sns mutant phenotype. As shown in Figure 1E, mesodermal expression of UAS-sns20-5-HA in sns mutant embryos using mef2Gal4 failed to rescue the myoblast fusion defect. On the basis of these results, we conclude that the SNS cytodomain C-terminal to I1163 is essential.
Figure 1.—
The region following I1163 is essential for SNS function. (A) Schematic representation of the SNS cytodomain regions present in UAS-sns-HA and UAS-sns20-5HA. (B–E) Stage-16 embryos in which the musculature has been visualized by immunostaining with an antibody against muscle myosin. All are lateral views in which anterior is to the left and dorsal at the top. (B) Wild-type, (C) sns20-5/sns20-5, and (D) sns mutant embryos in which myoblast fusion has been rescued by expression of the wild-type cDNA under control of mef2Gal4, with the genotype snsZf1.4,UAS-sns-HA/snsXB3; mef2Gal4/+. (E) sns mutant embryo in which myoblast fusion has been rescued by expression of the sns20-5 cDNA under control of mef2Gal4, with the genotype UAS-sns20-5HA, snsZf1.4/snsXB3; mef2Gal4/+. In these embryos, the sns transgene produces the truncated protein shown schematically in A. The lack of fusion is apparent in C and E, indicating a requirement for the region of the SNS cytodomain after I1163. Bar, 20 μm.
Highly conserved motifs for PDZ-domain binding and serine phosphorylation are not necessary for SNS-directed myoblast fusion:
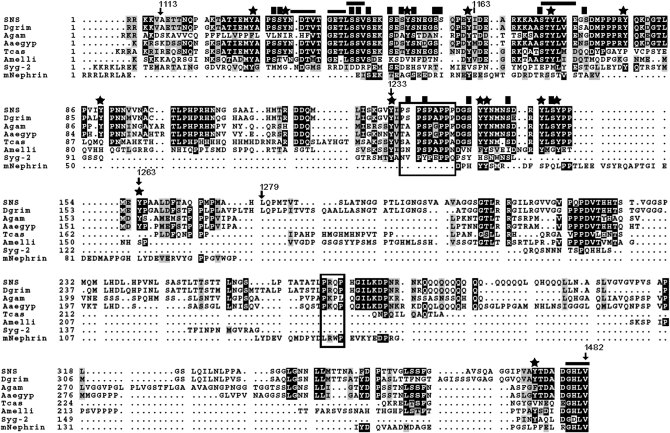
To identify specific residues within the cytodomain that direct myoblast fusion in the embryo, we first examined sequences that are highly conserved in SNS orthologs. A higher level of sequence conservation is apparent between insect orthologs, while the C. elegans and vertebrate orthologs are more divergent (Figure 2). We noted the four C-terminal amino acids, which form a strikingly conserved motif for binding to PDZ-domain-containing proteins. Aside from this C-terminal motif, the homology between SNS and its orthologs is much greater in the membrane proximal half of the cytodomain. Included in this region are potential sites for serine phosphorylation (Figure 2) that are also conserved in the SNS paralog Hibris (Hbs) (Artero et al. 2001).
Figure 2.—
Sequence conservation of the SNS cytodomain among various orthologs. Orthologs are shown from the following organisms: Drosophila grimshawi (Dgrim), Anopheles gambiae (Agam), Aedes aegypti (Aaeg), Tribolium castaneum (Tcas), Apis mellifera (Amelli), Caenorhabditis elegans (Syg-2), and mus musculus (mNephrin). Numbered amino acid positions mark the regions of deletion constructs discussed in the text. Stars mark the positions of tyrosine residues and solid squares highlight the serine residues analyzed in this study. Horizontal lines demarcate the putative PDZ-domain-binding sites and boxes surround the putative SH3-domain-binding sites.
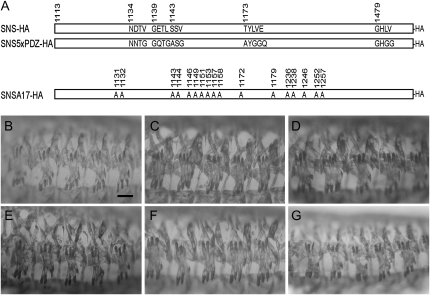
We first examined the importance of the conserved PDZ-domain-binding motif in SNS-mediated myoblast fusion using the GAL4/UAS system. Surprisingly, mutation of these four C-terminal amino acids to alanines had no apparent impact on the ability of the resulting SNS transgene to rescue myoblast fusion (data not shown). Although PDZ-domain-binding sites are most commonly found at the C-terminal ends of proteins, they have also been identified in more internal positions (Songyang et al. 1997). We therefore identified and mutated four additional sites with the potential to bind PDZ-domain-containing proteins. The positions and conservation of each of these sites are highlighted in the alignment in Figure 2, with the mutated residues shown in Figure 3A. A single transgene with conservative amino acid substitutions in all five sites (UAS-5xPDZHA) is capable of rescuing the sns mutant phenotype when expressed under the control of mef2Gal4 at 25° (Figure 3C). To temper the level of SNS, we have utilized snsGal4, an FCM-specific sns-promoter Gal4 transgene (data not shown) that drives expression at a lower level than mef2Gal4 (supplemental Figure 2A at http://www.genetics.org/supplemental/). We have also taken advantage of the observation that Gal4 activity is reduced at lower temperatures. As shown in Figure 3E, snsGal4-directed expression of UAS-sns-HA rescues myoblast fusion in sns mutants at 18° with few defects. UAS-5xPDZ-HA rescues myoblast fusion under the same conditions (Figure 3F), indicating that these sequences are not essential for SNS function.
Figure 3.—
Functional analysis of PDZ-domain binding motifs and conserved serines that are candidates for phosphorylation. (A) Schematic of the UAS-sns5xPDZ-HA and UAS-snsA17-HA transgenes. The mutated sites and their amino acid substitutions are indicated. (B–G) Lateral views of representative embryos in which the musculature has been visualized by immunostaining with an antibody to muscle myosin. (B–D) Expression of the transgene was directed by mef2Gal4 in a snsZf1.4/snsXB3 mutant embryo, with individual transgenes: (B) UAS-sns-HA, (C) UAS-sns5xPDZ-HA, and (D) UAS-snsA17-HA. (E–G) Expression of the transgene was directed by snsGal4 in a snsZf1.4/snsZf1.4 mutant embryo, with individual transgenes: (E) UAS-sns-HA, (F) UAS-sns5xPDZ-HA, and (G) UAS-snsA17-HA. There is no apparent difference in the ability of full-length and mutagenized transgenes to rescue the myoblast fusion defect of sns mutants when driven by mef2Gal4 and snsGal4. Bar, 20 μm.
Like amino acids that bind PDZ-domain-containing proteins, phosphorylated serines are known for their role as docking sites for proteins that mediate downstream signal transduction events. With this in mind, candidate sites for phosphorylation by CKII and PKC were previously noted in the cytodomains of SNS and its paralog, Hbs (Artero et al. 2001). Consensus sites for phosphorylation by GSK-3, CKI, and PKA are also present. In total, the SNS cytodomain contains 35 serine residues. The online prediction tools Netphos2.0 (Blom et al. 1999), Disphos 1.3 (Iakoucheva et al. 2004), PONDR DEPP (Romero et al. 2004), PredPhospho (Kim et al. 2004), and NetphosK (Blom et al. 1999) were used to evaluate the potential of each serine to be phosphorylated. A total of 17 serines were selected for site-directed mutagenesis on the basis of these predictions (supplemental Table 1 at http://www.genetics.org/supplemental/) and their evolutionary conservation (Figure 2). These 17 serine residues were mutated to alanines in UAS-snsA17-HA (Figure 3A) and examined for their ability to rescue the myoblast fusion defects of sns mutant embryos using mef2Gal4 at 25° and snsGal4 at 18° as above. Surprisingly, all muscle fibers were formed in each hemisegment and the number of unfused myoblasts was not significantly different from that observed with the unmutated construct (Figure 3, D and G). Thus, it does not appear that the most highly conserved serines, and best candidates for phosphorylation, play a critical role in SNS function.
Specific regions within the SNS cytodomain play redundant, but essential, roles in SNS-directed myoblast fusion:
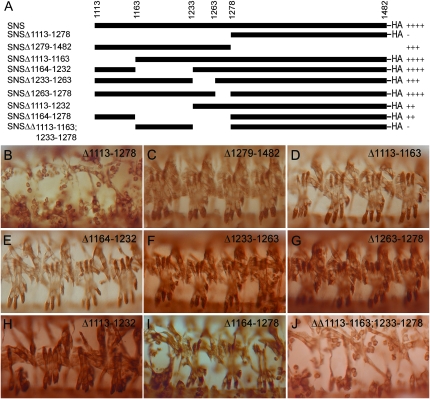
With the finding that highly conserved PDZ-binding motifs and candidate sites for serine phosphorylation are nonessential in SNS, we generated a series of transgenes in which various regions of the SNS cytodomain were deleted without bias toward conserved regions (Figure 4A). Two independent transgenic lines of each construct were examined for their ability to rescue the sns mutant phenotype when expressed under control of mef2Gal4. As shown in Figure 4, B and C, the membrane proximal half of the cytodomain is required to rescue myoblast fusion, while the carboxy-terminal half of the cytodomain is largely dispensable. We confirmed that two transgenes expressing the A1113-H1278 deletion are expressed and enriched at the plasma membrane, one of which is shown in supplemental Figure 2B. To identify the minimal critical region of SNS, transgenes were generated that removed four distinct nonoverlapping regions of the membrane proximal half of SNS. While the ability to rescue sns mutant embryos was slightly compromised in the absence of Y1233–Y1263, transgenic flies bearing any of these constructs were able to rescue a substantial amount of myoblast fusion and give rise to embryos with a fairly normal pattern of muscle fibers (Figure 4, D–G). Thus, the inability of the A1113–H1278 deletion to rescue SNS-directed myoblast fusion does not result from the absence of a single motif or site but, rather, seems to reflect either additive effects or functionally redundant regions. This hypothesis is supported by results obtained with three additional transgenes. The first of these removes A1113–V1232 (Figure 4H), a span of 120 amino acids that includes the two regions deleted individually in Figure 4, D and E. A second transgene removes D1164–H1278 (Figure 4I), the entire 115-amino-acid region covered in three smaller deletions in Figure 4, E–G. Although muscles are occasionally missing in embryos rescued with these transgenes, the number and organization of multinucleate syncytia is near wild type. By contrast, a third transgene in which A1113–I1163 and Y1233–H1278 have been removed drastically reduces the ability of the transgene to rescue myoblast fusion (Figure 4J). Again, we confirmed that two transgenes expressing this deletion are expressed and enriched at the plasma membrane, one of which is shown in supplemental Figure 2B. From these data, we conclude that multiple regions of the SNS cytodomain function redundantly to direct myoblast fusion.
Figure 4.—
The region A1113–H1278 is critical for SNS function. (A) Schematic of deletion constructs in which the deleted amino acids are indicated as open spaces. The presence of C-terminal HA tags is as indicated. (B–J) Lateral views in which the musculature has been visualized by immunostaining with an antibody to muscle myosin. The embryo is representative of results obtained using two independent transgenic lines. In all analyses, expression of the transgene was directed by mef2Gal4 in a snsZf1.4/snsXB3 mutant embryo, with individual transgenes: (B) UAS-Δ1113–1278HA, (C) UAS-Δ1279–1482, (D) UAS-Δ1113–1163HA, (E) UAS-Δ1164–1232HA, (F) UAS-Δ1233–1263HA, (G) UAS-Δ1263–1278HA, (H) UAS-Δ1113–1232HA, (I) UAS-Δ1164–1278HA, and (J) UAS-ΔΔ1113–1163 + 1233–1278HA. The absence of muscle fibers in J is comparable to that seen in B. With all other transgenes, the pattern of differentiated muscle fibers resembles that seen with the full-length sns cDNA in Figures 1 and 2.
The SNS cytodomain is phosphorylated on tyrosines:
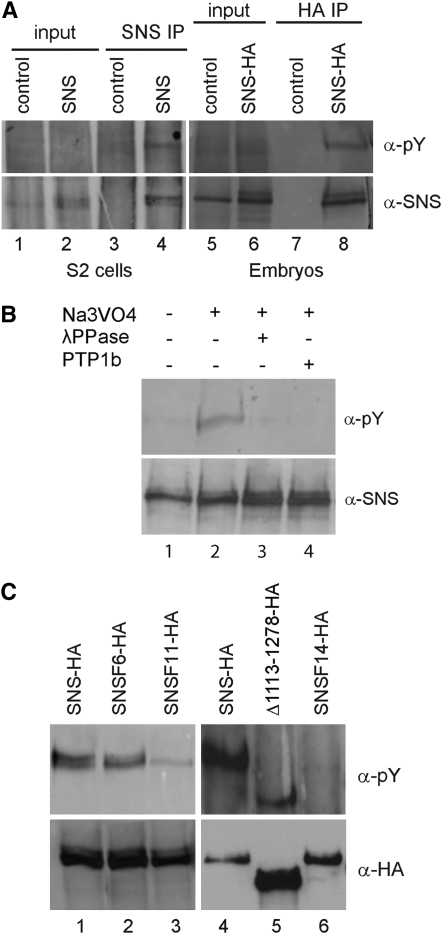
The above results are consistent with the hypothesis that multiple independent motifs contribute to the full functionality of the SNS cytodomain. Noting the presence of conserved tyrosine residues sprinkled throughout the cytodomain (Figure 2), and that phosphorylated tyrosines function in many signal transduction cascades, it was of interest to determine whether SNS is phosphorylated on tyrosine. Initially, an HA-tagged SNS construct was transiently transfected into Drosophila S2 cells and lysates were prepared in the presence of the phosphatase inhibitor sodium orthovanadate. SNS was immunoprecipitated as described in materials and methods, and the immunoprecipitates were analyzed for the presence of phosphorylated SNS. A single band of 165 kDa, the approximate predicted molecular weight of SNS, was detected with antisera directed against SNS and against phosphorylated tyrosine (Figure 5A, lane 4). To confirm tyrosine phosphorylation of SNS in the embryonic musculature, we expressed the functional UAS-sns-HA transgene in the embryonic musculature of wild-type embryos using mef2Gal4. Lysates were prepared from appropriately staged embryos and analyzed as above. As for SNS-transfected S2 cells, a single band of ∼165 kDa was detected with anti-SNS and anti-phosphotyrosine antisera (Figure 5A, lane 8). No phosphotyrosine immunoreactivity was detected in the absence of orthovanadate or upon treatment of the lysate with λ-phosphatase or protein tyrosine phosphatase 1b, confirming that the signal specifically reflected phosphotyrosine (Figure 5B).
Figure 5.—
The SNS cytodomain is phosphorylated on tyrosines. (A) Immunoblots of crude lysates and immunoprecipitates from untransfected control S2 cells (lanes 1 and 3), RmHa3-sns transfected S2 cells (lanes 2 and 4), control embryos (lanes 5 and 7), and embryos expressing UAS-sns-HA under control of mef2Gal4 (lanes 6 and 8). A tyrosine phosphorylated protein of the size of SNS is visible in the immunoprecipitate from transfected but not untransfected S2 cells (compare lanes 3 and 4). Similarly, a band detected by the phosphotyrosine-specific antisera is enriched upon immunoprecipitation of the HA-tagged SNS from embryos (compare lanes 7 and 8). (B) The phosphotyrosine-specific band is not detected in the absence of Na3VO4 (lane 1) or when the immunoprecipitates are treated with λ-phosphatase (lane 3) or protein tyrosine phosphatase 1b (lane 4). The total amount of SNS does not change appreciably, as shown by SNS immunoblot. (C) Changes in the level of phosphorylation are apparent upon substitution of cytodomain tyrosines in sns transgene constructs. Lysates were prepared from embryos expressing various mutant forms of SNS under the control of mef2Gal4 and immunoprecipitated with anti-HA antisera. Lanes 1 and 4, control immunoprecipitates from embryos expressing full-length SNS; lane 2, UAS-snsF6-HA in which six candidate sites for phosphorylation on tyrosine have been mutated; lane 3, UAS-snsF11-HA, in which five additional tyrosines have been mutated to phenylalanine, shows a substantially reduced level of phosphorylation; lane 5, UAS-Δ1113–1278HA, which deletes all but one of the tyrosines in the cytodomain and has a very reduced level of phosphorylation when normalized to the level of SNS protein detected by the HA tag; and lane 6, UAS-snsF14-HA in which all cytodomain tyrosines have been mutated and phosphorylation is no longer detected.
Prior to site-directed mutagenesis, we sought to determine which tyrosines were the most likely candidates for phosphorylation and analyzed all 14 tyrosines in the SNS cytodomain using the phosphorylation prediction programs Netphos 2.0, Disphos 1.3, and PONDR (supplemental Table 2 at http://www.genetics.org/supplemental/). These predictions were then used as the basis for site-directed mutagenesis in which tyrosines were systematically replaced with phenylalanine in the UAS-sns-HA construct. Transgenes from the resulting constructs were expressed in the mesoderm of wild-type embryos and analyzed for the presence of phosphorylated SNS as above. Surprisingly, tyrosine phosphorylation was not eliminated until all 14 sites were mutated (Figure 5C, lane 6), despite the prediction that several of these tyrosines were not candidates for such modification. Nevertheless, these results clearly establish that SNS is phosphorylated on tyrosine.
SNS function is compromised in the absence of cytodomain phosphotyrosines:
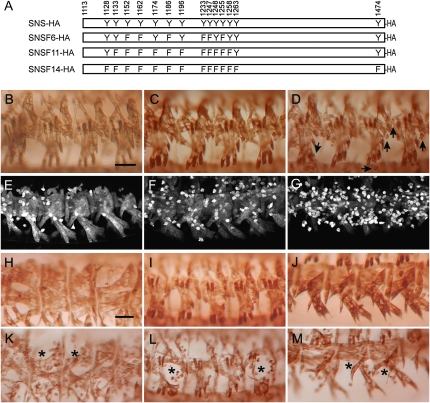
To test whether phosphotyrosines in the SNS cytodomain play a critical role in its function, we examined the ability of three transgenes in which select tyrosines were mutated to phenylalanines to rescue the myoblast fusion defects in sns mutant embryos when expressed under control of the mef2Gal4 promoter. Multinucleate muscle fibers are clearly visible in embryos rescued by expression of transgenes in which the strongest predicted candidates for phosphorylation were altered (Figure 6A, supplemental Table 2), including UAS-snsF6-HA and UAS-snsF11-HA (Figure 6, B and C). Muscle fibers are also apparent in embryos rescued by mef2Gal4-directed expression of UAS-snsF14-HA, in which all tyrosines have been mutated in the same molecule (Figure 6D). In this latter case, the number of unfused myoblasts is significantly higher than that observed with the UAS-sns-HA control and has been quantified using sns-LacZ, an FCM-specific reporter that is expressed at high levels in unfused myoblasts (Figure 6, E–G; see below for quantitation). To address the possibility that higher levels of SNS may, in some way, drive interactions and compensate for mutant forms of the protein, we also used snsGal4 at 18°, to direct a lower level of expression of the transgene. We then more closely examined the dorsal, lateral, and ventral muscle groups. As shown in Figure 6, K–M, a substantial number of unfused myoblasts are apparent compared to rescue with the wild-type construct (Figure 6, H–J; see below for quantitation). Thus, while some myoblast fusion occurs in the absence of tyrosine residues in the SNS cytodomain, and higher levels of mef2Gal4-driven expression provide some compensation, these data establish the importance of the tyrosines in myoblast fusion. We therefore conclude that phosphorylated tyrosines in the SNS cytodomain play a key role in downstream events that direct myoblast fusion.
Figure 6.—
Tyrosine residues in the SNS cytodomain are required for efficient myoblast fusion. (A) Schematic of the transgenes, with the positions of the substituted tyrosine residues indicated. In all cases, tyrosines were mutated to phenylalanine. (B–M) Stage-16 embryos in which the musculature has been visualized by immunostaining with an antibody to muscle myosin (B–D and H–M) and β-galactosidase (E–G). All rescued embryos are representative of results obtained using two independent transgenic lines. (B–D, F, and G) Expression of the transgene was directed to snsZf1.4/snsXB3 mutant embryos using mef2Gal4 at 25° using the transgenic lines (B) UAS-snsF6-HA, (C) UAS-snsF11-HA, (D and G) UAS-snsF14-HA, and (F) UAS-sns-HA. (E) Wild type. (E–G) Embryos also contained an sns-LacZ transgene that is expressed at a high level in unfused myoblasts and facilitates their quantitation. While these rescued embryos exhibit more unfused myoblasts than a wild-type embryo (compare F and E), significantly more unfused myoblasts are seen in embryos “rescued” with a transgene in which all cytodomain tyrosines have been mutated (G). (H–M) Embryos in which expression of the transgene was directed to snsZf1.4/snsZf1.4 mutant embryos using snsGal4 at 18°, using the transgenes (H–J) UAS-sns-HA and (K–M) UAS-snsF14-HA. The inefficiency of rescue is apparent at lower levels of expression, when large numbers of unfused myoblasts are seen in dorsolateral, lateral, and ventrolateral views of mutant embryos expressing the mutant transgene but not its wild-type counterpart. Arrows point to unfused myoblasts in D and asterisks in K–M mark regions of missing muscles. Bar, 20 μm.
Proline-rich regions play an essential role in SNS-directed myoblast fusion:
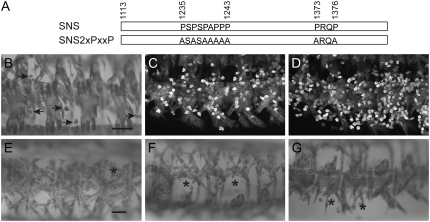
The SNS cytodomain contains two consensus PXXP SH3 binding motifs that are conserved among the examined Drosophila orthologs (Figure 2). The first of these sites lies between P1235 and P1243 while the other is located between P1373 and P1376 (Figure 7A). Given these locations, the proline-rich regions are unlikely to account for the functional redundancy within A1113–H1278. However, we reasoned that the apparent expendability of the carboxy-terminal half of the cytodomain (L1279–V1482) might reflect further functional redundancy with sites in the membrane proximal half. To test the requirement for these SH3 binding motifs during myoblast fusion, we mutated the sites as indicated in Figure 7A. Flies transgenic for this construct were then examined for their ability to rescue sns mutants when driven with mef2Gal4 at 25° or snsGal4 at 18°. Transgenes lacking the consensus PXXP sites are capable of rescuing some myoblast fusion, particularly when expression is driven with mef2Gal4 (Figure 7B). However, the number of unfused myoblasts that remain is significantly higher than that in the control rescued embryos (arrows in Figure 7B). This number has been quantitated using sns-LacZ to mark the unfused myoblasts, as in Figure 6. Representative embryos for rescue with control UAS-sns-HA and mutant UAS-sns2xPXXP transgenes are shown in Figure 7, C and D, respectively. Perturbations in the dorsal, lateral, and ventral muscle groups are also apparent when lower levels of expression are directed by snsGal4 (Figure 7, E–G). Thus, as with the tyrosine residues, although their requirement can be partially compensated by increased levels of protein, the consensus PXXP motifs in the SNS cytodomain play an important role during myoblast fusion.
Figure 7.—
SH3-domain binding motifs in the SNS cytodomain are required for proper myoblast fusion. (A) Schematic of the transgenes, with the positions of the SH3 binding motifs and their amino acid substitutions indicated. (B and E–G) Stage 16 embryos in which the musculature has been visualized by immunostaining with an antibody to muscle myosin. Rescued embryos are representative of results obtained using two independent transgenic lines. (B–D) Expression of the transgene was directed to snsZf1.4/snsXB3 mutant embryos using mef2Gal4 at 25°, using the transgenic lines (B and D) UAS-sns2xPXXP and (C) UAS-snsHA. (C and D) Embryos express sns-LacZ and are stained fluorescently for β-galactosidase to highlight the unfused myoblasts. (E–G) Dorsal, lateral, and ventral views, in which the UAS-sns2xPXXP transgene was directed to snsZf1.4/snszf1.4 mutant embryos using snsGal4 at 18°. Arrows mark unfused myoblasts in D and asterisks mark regions of missing muscles in E–G. Bar, 20 μm.
Quantitative comparisons of the extent of myoblast fusion:
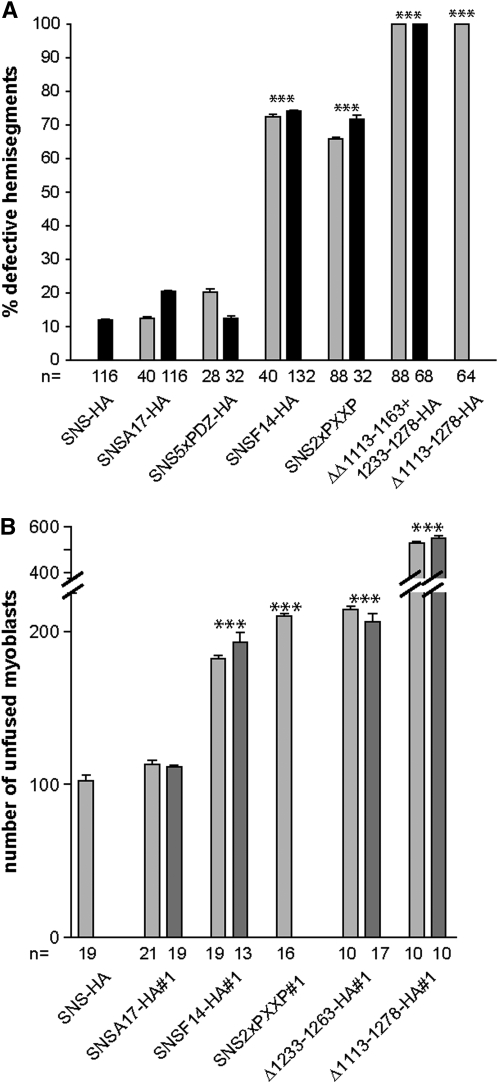
In an effort to quantitate the extent of rescue using each of the transgenic lines, we first counted the number of hemisegments that were defective in embryos stained with muscle myosin, using two independent lines for mutagenized transgenes. The results shown in Figure 8A corroborate our empirical conclusions from examination of individual embryos. Specifically, neither the five PDZ-domain-binding sites nor the 17 consensus serines for phosphorylation appear to be required for SNS function. By contrast, removal of either all 14 tyrosines in the cytodomain or the two proline-rich PXXP sites negatively affected SNS function. Neither of the latter had as severe an impact, however, as removal of the membrane proximal half of the cytodomain. Of note, removal of two regions of ∼50 amino acids within this domain, neither of which results in defects on its own, has a similar impact on muscle development.
Figure 8.—
Quantitation of efficiency of transgenes to rescue the sns mutant phenotypes. (A) Quantitation of percentage of hemisegments with defects in muscle pattern of the lateral transverse muscles in rescued embryos analyzed by MHC staining where expression of indicated transgenes is driven using snsGal4. (B) Quantitation of average number of unfused myoblasts by staining for LacZ in rescued embryos where expression of indicated transgenes is driven using mef2Gal4. Increased numbers of unfused myoblasts are observed in rescue using UAS-snsF14-HA, UAS-sns2xPXXP, UAS-Δ1233–1263HA, and UAS-Δ1113–1278HA in comparison to UAS-sns-HA and UAS-snsA17-HA. The n value represents the number of embryos analyzed in A and the number of hemisegments analyzed in B. Pairwise P-values were calculated by a one-way ANOVA model combined with Tukey's HSD tests. ***P-value < 0.001. Error bars correspond to the standard errors of means for each group.
We also sought to compare the actual degree of myoblast fusion achieved with each of the rescue constructs described above quantitatively. While visualization of the muscle pattern using antisera directed against muscle myosin has revealed clear and reproducible defects in the ability of various SNS deletions to rescue myoblast fusion, we have noted that unfused myoblasts express a relatively lower level of myosin than the resulting syncytia. This feature makes quantitation of unfused myoblasts problematic and more prone to error. We therefore developed a promoter-reporter construct using the sns promoter that is expressed strongly in the unfused FCMs. Independent analyses have confirmed the specific expression of this transgene (sns-LacZ) in FCMs and its absence on FMs (data not shown). This transgene was therefore incorporated into the snsZf1.4, sns-LacZ; mef2Gal4 stock used for rescue, as described above, and the musculature of the resulting embryos visualized with antisera directed against β-galactosidase. Discrete unfused myoblasts are readily apparent (Figure 6, E–G; Figure 7, C and D) and can be quantitated in a confocal Z-series using appropriate software (materials and methods). The results of this analysis, shown in Figure 8B, corroborate impressions obtained upon visual inspection of the rescued embryos. The tyrosine residues and proline-rich regions play a significant role in the ability of SNS to direct myoblast fusion but neither the loss of the PDZ sites nor the loss of the most conserved serines negatively influences SNS function, consistent with the results from Figure 8A. Moreover, the regions within SNS that contain the bulk of the tyrosines and one PXXP site play redundant roles that are independently expendable.
DISCUSSION
The process of myoblast fusion in Drosophila embryos requires intracellular events that include the formation of F-actin-rich foci and recruitment of electron-dense vesicles to sites of cell–cell contact (Doberstein et al. 1997; Kim et al. 2007). In the fusion-competent myoblast, actin polymerization occurs coincident with transport of fusion-related electron-dense vesicles and is suggested to aid in the targeting of the latter (Kim et al. 2007; Massarwa et al. 2007). The vesicles appear to become actin coated as they move through the actin-rich foci close to the cell surface, where they undergo exocytosis (Kim et al. 2007). Separate studies have identified discrete actin-rich complexes at the cell surface that are composed of proteins essential for myoblast fusion, including SNS (Kesper et al. 2007). These structures, termed FuRMAS, have been implicated in determining the exact site of fusion and the position and size of the fusion pore (Kesper et al. 2007). Thus, signal transduction pathways that direct actin polymerization and mediate cytoskeletal reorganization appear to be guided by cell-adhesion molecules to points of cell contact prior to membrane breakdown at these sites (Chen and Olson 2001; Chen et al. 2003; Kesper et al. 2007; Kim et al. 2007). The cytodomains of these cell-adhesion molecules therefore likely serve critical roles in signaling pathways that lead to myoblast fusion.
Our studies establish the importance of a highly conserved membrane proximal region of 166 amino acids in the SNS cytodomain. Surprisingly, smaller deletions within this region do not have a profound impact on SNS function, with the deletions A1113–I1163, Y1233–Y1263, and Y1263–H1278 each restoring the wild-type muscle pattern in sns mutants. However, a combination of these deletions represented by UAS-ΔA1113–I1163 + Y1233–H1278 is unable to rescue the fusion defects in sns mutant embryos. We conclude from these results that the SNS cytodomain houses significant functional redundancy and is rendered nonfunctional only when multiple sites that serve the same purpose are removed. Examination of the sequences within each of these regions reveals a high number of predicted and conserved sites for phosphorylation on both serine and tyrosine. We cannot conclude that serines are unimportant in SNS-mediated signal transduction since all sites have not been mutated. Nevertheless we can conclude that loss of the 17 most conserved, best candidate sites for phosphorylation on serine does not affect SNS function. Moreover, the remaining serines must be capable of fulfilling any role that these serines play in SNS-mediated signal transduction. Phosphotyrosines can also be critical intermediates in signal transduction pathways and mediate interactions with proteins containing SH2, WW, or phosphotyrosine-binding (PTB) domains. Tyrosine phosphorylation is a key modification in the SNS ortholog, nephrin (Li et al. 2004; Jones et al. 2006), and regulates signaling to two downstream pathways: one that activates Akt by phosphoinositide 3-kinase (PI3K) and another that directs actin rearrangements via Nck binding. Both the Akt signaling and Nck pathways are critical for nephrin function (Huber et al. 2003a; Jones et al. 2006).
We have demonstrated herein that SNS is phosphorylated on tyrosine and that mutagenesis of these sites impairs SNS function during myoblast fusion. Surprisingly, even residues with a low probability of modification in SNS are nonetheless phosphorylated in the SNS cytodomain. It is not possible with the methods available to distinguish between residues that are phosphorylated in the wild-type protein and residues that are phosphorylated in the mutant proteins in the absence of preferred targets. Nevertheless, all sites must be removed to affect SNS function. Proteins that may interact with these phosphotyrosines include SH2–SH3 domain-containing adaptor proteins that have similar roles downstream of other membrane receptors (Birge et al. 1996; Mccarty 1998; Sarmay et al. 2006). Candidates include Crk, Dreadlocks (Dock)/Nck, and Downstream of receptor kinase (Drk), the Drosophila ortholog of Grb2. SNS has been shown to recruit Sltr/D-Wip to the membrane of cotransfected S2 cells via Crk, supporting it as a pathway intermediate in the FCMs (Kim et al. 2007), and Nck plays a critical role downstream of nephrin as cited above.
While the importance of phosphorylated tyrosines is clear, removal of all tyrosines does not render SNS completely nonfunctional. Our data also demonstrate a requirement for two proline-rich regions with core PXXP motifs. These sites are common to transmembrane receptors and, for example, the IgSF proteins Robo and DSCAM utilize such sites for interactions that lead to directional migration in the Drosophila nervous system (Bashaw et al. 2000; Fan et al. 2003). PXXP sites frequently interact with SH3 domains, which are found in signaling proteins that include adaptors, kinases, and GTPase activator proteins (Sudol 1998). In SNS, the simultaneous mutation of both PXXP sites (as in UAS-sns2xPXXP) impairs the ability of SNS to function during myoblast fusion. Although a direct requirement for a conserved motif in nephrin has not been described, the nephrin cytodomain interacts with the third SH3 domain of CD2AP to activate Akt signaling and for association with the actin cytoskeleton in mouse kidney cells (Yuan et al. 2002; Huber et al. 2003a). Moreover, the loss of CD2AP in the mouse is associated with kidney defects reminiscent of those observed in the absence of nephrin (Shih et al. 1999), suggesting that CD2AP–nephrin interactions are critical to maintaining slit diaphragm function.
One muscle-specific candidate for interaction with the SNS PXXP site is MBC, which is essential in both FMs and FCMs and requires its SH3 domain for function during myoblast fusion (Balagopalan et al. 2006). While this SH3 domain seems more likely to mediate interaction of MBC with ELMO to form a bipartite guanine nucleotide exchange factor, it is a formal possibility that it interacts directly with SNS. Alternatively, if the interaction is indirect, one might anticipate the presence of factors in the FCMs that recruit MBC to SNS in a manner reminiscent of the relationship between MBC and Ants/Rols downstream of Duf/Kirre in the FMs. Other candidates for interaction with the SNS PXXP sites that are ubiquitous during embryogenesis or expressed specifically during myogenesis are D-Crk and CG31012, the apparent ortholog of CD2AP (Galletta et al. 1999; Balagopalan et al. 2006), as well as the SH2–SH3-domain-containing adaptor proteins previously mentioned.
This study has revealed two critical sequences in the SNS cytodomain. However, neither phosphotyrosines nor consensus PXXP motifs alone can account for the full repertoire of interactions necessary for SNS function. It is unclear if removal of both signaling motifs might account for the more drastic effect of the double deletion on the ability of SNS to direct myoblast fusion. It also remains to be determined whether these two motifs function in related interactions that converge on a single pathway or reflect independent interactions that direct distinct downstream events. Each motif seems most likely to bind a different target protein, as observed for domains within the neuronal cell adhesion molecules NCAM, L1CAM, Robo, and DSCAM (Skaper et al. 2001). Multiple protein interaction domains are also found in the more closely related nephrin and Duf/Kirre proteins (Bashaw et al. 2000; Crossin and Krushel 2000; Patrakka and Tryggvason 2007). Another formal possibility is that both the phosphotyrosines and the PXXP motifs serve the same function by actually binding to the same protein. This type of mechanism, albeit unlikely and unprecedented to our knowledge, seems feasible given the large number of adaptor proteins with multiple SH2 and/or SH3 domains. In this case, interaction of SNS with its target adaptor might be compromised but not completely lost by mutagenesis of either phosphotyrosine- or proline-rich regions, as observed in the results herein. In summary, current models support the presence of at least two signaling pathways downstream of SNS, one leading from SNS via Crk and Sltr/D-Wip to Wasp and another involving MBC-mediated activation of Rac and its subsequent activation of Arp2/3 via Kette/WAVE (Abmayr and Kocherlakota 2005; Kim et al. 2007). Other SNS-associated pathways may yet be identified that play distinct roles in directional migration of FCMs to FMs, accurate alignment prior to fusion, and cytoskeletal or membrane turnover. The SNS cytodomain seems a likely site for interactions with proteins that mediate many of these events.
Acknowledgments
We thank D. Kiehart for the generous gift of the anti-myosin heavy chain antibody and R. Renkawitz-Pohl for the sns20-5 allele. We thank the Stowers Institute Molecular Biology and Imaging core facilities for their assistance with mutagenesis and confocal analysis, respectively. We are grateful to Dongxiao Zhu for help with the statistical analysis and Sue-Jean Hong, Rakhee Banerji, and Mei-Hui Chen for technical assistance. This work was supported by the Stowers Institute for Medical Research and by National Institutes of Health award RO1 AR44274 to S.M.A.
References
- Abmayr, S. M., and K. S. Kocherlakota, 2005. Muscle morphogenesis: the process of embryonic myoblast fusion, pp. 92–103 in Muscle Development in Drosophila, edited by H. Sink. Springer Science and Business Media, New York.
- Artero, R. D., I. Castanon and M. K. Baylies, 2001. The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. Development 128 4251–4264. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., 1989. Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Balagopalan, L., M. H. Chen, E. R. Geisbrecht and S. M. Abmayr, 2006. The CDM superfamily protein MBC directs myoblast fusion through a mechanism that requires phosphatidylinositol 3,4,5-triphosphate binding but is independent of direct interaction with DCrk. Mol. Cell. Biol. 26 9442–9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo, S., L. A. Carver and J. W. Posakony, 2000. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29 726, 728, 730, 732. [DOI] [PubMed] [Google Scholar]
- Bashaw, G. J., T. Kidd, D. Murray, T. Pawson and C. S. Goodman, 2000. Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the roundabout receptor. Cell 101 703–715. [DOI] [PubMed] [Google Scholar]
- Bate, M., and E. Rushton, 1993. Myogenesis and muscle patterning in Drosophila. C. R. Acad. Sci. III 316 1047–1061. [PubMed] [Google Scholar]
- Birge, R. B., B. S. Knudsen, D. Besser and H. Hanafusa, 1996. SH2 and SH3-containing adaptor proteins: redundant or independent mediators of intracellular signal transduction. Genes Cells 1 595–613. [DOI] [PubMed] [Google Scholar]
- Blom, N., S. Gammeltoft and S. Brunak, 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294 1351–1362. [DOI] [PubMed] [Google Scholar]
- Bour, B. A., M. Chakravarti, J. M. West and S. M. Abmayr, 2000. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 14 1498–1511. [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Bunch, T. A., Y. Grinblat and L. S. Goldstein, 1988. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 16 1043–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch, T. A., M. W. Graner, L. I. Fessler, J. H. Fessler, K. D. Schneider et al., 1998. The PS2 integrin ligand tiggrin is required for proper muscle function in Drosophila. Development 125 1679–1689. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega, J. A., and V. Hartenstein, 1997. The Embryonic Development of Drosophila melanogaster. Springer-Verlag, Berlin.
- Chen, E. H., and E. N. Olson, 2001. Antisocial, an intracellular adaptor protein, is required for myoblast fusion in Drosophila. Dev. Cell 1 705–715. [DOI] [PubMed] [Google Scholar]
- Chen, E. H., and E. N. Olson, 2004. Towards a molecular pathway for myoblast fusion in Drosophila. Trends Cell Biol. 14 452–460. [DOI] [PubMed] [Google Scholar]
- Chen, E. H., B. A. Pryce, J. A. Tzeng, G. A. Gonzalez and E. N. Olson, 2003. Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell 114 751–762. [DOI] [PubMed] [Google Scholar]
- Cherbas, L., and P. Cherbas, 1998. Cell culture, pp. 319–346 in Drosophila: A Practical Approach, Ed. 2, edited by D. B. Roberts. IRL Press, New York.
- Crossin, K. L., and L. A. Krushel, 2000. Cellular signaling by neural cell adhesion molecules of the immunoglobulin superfamily. Dev. Dyn. 218 260–279. [DOI] [PubMed] [Google Scholar]
- Doberstein, S. K., R. D. Fetter, A. Y. Mehta and C. S. Goodman, 1997. Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J. Cell Biol. 136 1249–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson, M. R. S., B. J. Galletta and S. M. Abmayr, 1997. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure and cytoskeletal organization. J. Cell Biol. 138 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, X., J. P. Labrador, H. Hing and G. J. Bashaw, 2003. Slit stimulation recruits Dock and Pak to the roundabout receptor and increases Rac activity to regulate axon repulsion at the CNS midline. Neuron 40 113–127. [DOI] [PubMed] [Google Scholar]
- Galletta, B. J., X. P. Niu, M. R. Erickson and S. M. Abmayr, 1999. Identification of a Drosophila homologue to vertebrate Crk by interaction with MBC. Gene 228 243–252. [DOI] [PubMed] [Google Scholar]
- Galletta, B. J., M. Chakravarti, R. Banerjee and S. M. Abmayr, 2004. SNS: adhesive properties, localization requirements and ectodomain dependence in S2 cells and embryonic myoblasts. Mech. Dev. 121 1455–1468. [DOI] [PubMed] [Google Scholar]
- Geisbrecht, E. R., S. Haralalka, S. K. Swandson, L. Florens, M. P. Washburn et al., 2008. Drosophila ELMO/CED-12 interacts with Myoblast city to direct myoblast fusion and ommatidial organization. Dev. Biol. 314(1): 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortsch, M., 1994. Preparation and analysis of membranes and membrane proteins from Drosophila, pp. 289–301 in Drosophila melanogaster: Practical Uses in Cell and Molecular Biology, edited by L. S. B. Goldstein and E. A. Fyrberg. Academic Press, San Diego. [DOI] [PubMed]
- Huber, T. B., B. Hartleben, J. Kim, M. Schmidts, B. Schermer et al., 2003. a Nephrin and CD2AP associate with phosphoinositide 3-OH kinase and stimulate AKT-dependent signaling. Mol. Cell. Biol. 23 4917–4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, T. B., M. Schmidts, P. Gerke, B. Schermer, A. Zahn et al., 2003. b The carboxyl terminus of Neph family members binds to the PDZ domain protein zonula occludens-1. J. Biol. Chem. 278 13417–13421. [DOI] [PubMed] [Google Scholar]
- Iakoucheva, L. M., P. Radivojac, C. J. Brown, T. R. O'Connor, J. G. Sikes et al., 2004. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 32 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru, S., R. Ueda, Y. Hinohara, M. Ohtani and H. Hanafusa, 2004. PVR plays a critical role via JNK activation in thorax closure during Drosophila metamorphosis. EMBO J. 23 3984–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, N., I. M. Blasutig, V. Eremina, J. M. Ruston, F. Bladt et al., 2006. Nck adaptor proteins link nephrin to the actin cytoskeleton of kidney podocytes. Nature 440 818–823. [DOI] [PubMed] [Google Scholar]
- Kesper, D. A., C. Stute, D. Buttgereit, N. Kreiskother, S. Vishnu et al., 2007. Myoblast fusion in Drosophila melanogaster is mediated through a fusion-restricted myogenic-adhesive structure (FuRMAS). Dev. Dyn. 236 404–415. [DOI] [PubMed] [Google Scholar]
- Kim, J. H., J. Lee, B. Oh, K. Kimm and I. Koh, 2004. Prediction of phosphorylation sites using SVMs. Bioinformatics 20 3179–3184. [DOI] [PubMed] [Google Scholar]
- Kim, S., K. Shilagardi, S. Zhang, S. N. Hong, K. L. Sens et al., 2007. A critical function for the actin cytoskeleton in targeted exocytosis of prefusion vesicles during myoblast fusion. Dev. Cell 12 571–586. [DOI] [PubMed] [Google Scholar]
- Kreiskother, N., N. Reichert, D. Buttgereit, A. Hertenstein, K. F. Fischbach et al., 2006. Drosophila rolling pebbles colocalises and putatively interacts with alpha-Actinin and the Sls isoform Zormin in the Z-discs of the sarcomere and with Dumbfounded/Kirre, alpha-Actinin and Zormin in the terminal Z-discs. J. Muscle Res. Cell Motil. 27 93–106. [DOI] [PubMed] [Google Scholar]
- Lenkkeri, U., M. Mannikko, P. McCready, J. Lamerdin, O. Gribouval et al., 1999. Structure of the gene for congenital nephrotic syndrome of the Finnish type (NPHS1) and characterization of mutations. Am. J. Hum. Genet. 64 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., S. Lemay, L. Aoudjit, H. Kawachi and T. Takano, 2004. SRC-family kinase Fyn phosphorylates the cytoplasmic domain of nephrin and modulates its interaction with podocin. J. Am. Soc. Nephrol. 15 3006–3015. [DOI] [PubMed] [Google Scholar]
- Massarwa, R., S. Carmon, B. Z. Shilo and E. D. Schejter, 2007. WIP/WASp-based actin-polymerization machinery is essential for myoblast fusion in Drosophila. Dev. Cell 12 557–569. [DOI] [PubMed] [Google Scholar]
- McCarty, J. H., 1998. The Nck SH2/SH3 adaptor protein: a regulator of multiple intracellular signal transduction events. BioEssays 20 913–921. [DOI] [PubMed] [Google Scholar]
- Nolan, K. M., K. Barrett, Y. Lu, K. Q. Hu, S. Vincent et al., 1998. Myoblast city, the Drosophila homolog of DOCK180/CED-5, is required in a Rac signaling pathway utilized for multiple developmental processes. Genes Dev. 12 3337–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrakka, J., and K. Tryggvason, 2007. Nephrin—a unique structural and signaling protein of the kidney filter. Trends Mol. Med. 13 396–403. [DOI] [PubMed] [Google Scholar]
- Paululat, A., S. Burchard and R. Renkawitz-Pohl, 1995. Fusion from myoblasts to myotubes is dependent on the rolling stone gene (rost) of Drosophila. Development 121 2611–2620. [DOI] [PubMed] [Google Scholar]
- Ranganayakulu, G., D. A. Elliott, R. P. Harvey and E. N. Olson, 1998. Divergent roles for NK-2 class homeobox genes in cardiogenesis in flies and mice. Development 125 3037–3048. [DOI] [PubMed] [Google Scholar]
- Richardson, B. E., K. Beckett, S. J. Nowak and M. K. Baylies, 2007. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development 134 4357–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, P., Z. Obradovic and A. K. Dunker, 2004. Natively disordered proteins: functions and predictions. Appl. Bioinformatics 3 105–113. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez, M., N. Coutts, A. Price, M. V. Taylor and M. Bate, 2000. Drosophila Dumbfounded: a myoblast attractant essential for fusion. Cell 102 189–198. [DOI] [PubMed] [Google Scholar]
- Ruotsalainen, V., J. Patrakka, P. Tissari, P. Reponen, M. Hess et al., 2000. Role of Nephrin in cell junction formation in human nephrogenesis. Am. J. Pathol. 157 1905–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmay, G., A. Angyal, A. Kertesz, M. Maus and D. Medgyesi, 2006. The multiple function of Grb2 associated binder (Gab) adaptor/scaffolding protein in immune cell signaling. Immunol. Lett. 104 76–82. [DOI] [PubMed] [Google Scholar]
- Schafer, G., S. Weber, A. Holz, S. Bogdan, S. Schumacher et al., 2007. The Wiskott-Aldrich syndrome protein (WASP) is essential for myoblast fusion in Drosophila. Dev. Biol. 304 664–674. [DOI] [PubMed] [Google Scholar]
- Sharma, Y., U. Cheung, E. W. Larsen and D. F. Eberl, 2002. PPTGAL, a convenient Gal4 P-element vector for testing expression of enhancer fragments in drosophila. Genesis 34 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, K., R. D. Fetter and C. I. Bargmann, 2004. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell 116 869–881. [DOI] [PubMed] [Google Scholar]
- Shih, N. Y., J. Li, V. Karpitskii, A. Nguyen, M. L. Dustin et al., 1999. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science 286 312–315. [DOI] [PubMed] [Google Scholar]
- Skaper, S. D., S. E. Moore and F. S. Walsh, 2001. Cell signalling cascades regulating neuronal growth-promoting and inhibitory cues. Prog. Neurobiol. 65 593–608. [DOI] [PubMed] [Google Scholar]
- Songyang, Z., A. S. Fanning, C. Fu, J. Xu, S. M. Marfatia et al., 1997. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275 73–77. [DOI] [PubMed] [Google Scholar]
- Strunkelnberg, M., B. Bonengel, L. M. Moda, A. Hertenstein, H. G. de Couet et al., 2001. rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development 128 4229–4239. [DOI] [PubMed] [Google Scholar]
- Sudol, M., 1998. From Src homology domains to other signaling modules: proposal of the ‘protein recognition code’. Oncogene 17 1469–1474. [DOI] [PubMed] [Google Scholar]
- Yuan, H., E. Takeuchi, G. A. Taylor, M. McLaughlin, D. Brown et al., 2002. Nephrin dissociates from actin, and its expression is reduced in early experimental membranous nephropathy. J. Am. Soc. Nephrol. 13 946–956. [DOI] [PubMed] [Google Scholar]