Abstract
Extensive linkage disequilibrium among classical laboratory strains represents an obstacle in the high-resolution haplotype mapping of mouse quantitative trait loci (QTL). To determine the potential of wild-derived mouse strains for fine QTL mapping, we constructed a haplotype map of a 250-kb region of the t-complex on chromosome 17 containing the Hybrid sterility 1 (Hst1) gene. We resequenced 33 loci from up to 80 chromosomes of five mouse (sub)species. Trans-species single-nucleotide polymorphisms (SNPs) were rare between Mus m. musculus (Mmmu) and Mus m. domesticus (Mmd). The haplotypes in Mmmu and Mmd differed and therefore strains from these subspecies should not be combined for haplotype-associated mapping. The haplotypes of t-chromosomes differed from all non-t Mmmu and Mmd haplotypes. Half of the SNPs and SN indels but only one of seven longer rearrangements found in classical laboratory strains were useful for haplotype mapping in the wild-derived M. m. domesticus. The largest Mmd haplotype block contained three genes of a highly conserved synteny. The lengths of the haplotype blocks deduced from 36 domesticus chromosomes were in tens of kilobases, suggesting that the wild-derived Mmd strains are suitable for fine interval-specific mapping.
THE sequence of the mouse genome (Waterston et al. 2002) is based on the classical laboratory mouse strain C57BL/6J (henceforth B6). A fairly complete sequence based on a few other classical laboratory (“Celera”) strains (129X1/SvJ, 129S1/SvImJ, A/J, and DBA/2J) is also available (Mural et al. 2002). Resequencing of other mouse strains revealed single-nucleotide polymorphisms (SNPs) (Wade et al. 2002; Wiltshire et al. 2003; Frazer et al. 2004, 2007; Zhang et al. 2004). Regions of low (0.5/10 kb) and high (40/10 kb) SNP density were identified, covering roughly two-thirds and one-third of the genome of the laboratory strains, respectively (Wade et al. 2002). The low-density SNP regions were interpreted as coming from the same subspecies, mostly either Mus m. domesticus (Mmd) or Mus m. musculus (Mmmu), and the high-density regions as originating from different subspecies. Detailed analyses of millions of Perlegen SNPs revealed that 65–92% of the genome of classical laboratory strains is of Mmd origin (Frazer et al. 2007; Yang et al. 2007). Mouse haplotypes, the arrangements of alleles along chromosomes, can be described with the help of strain distribution patterns [SDPs; the patterns of allelic differences and similarities among strains at a locus (Grupe et al. 2001; Yalcin et al. 2004)]. The number of SDPs is indirectly proportional to haplotype blocks, regions limited by historical recombination. The length of haplotype blocks was estimated to be a few hundred kilobases to a few megabase pairs in the laboratory mouse (Wade et al. 2002; Yalcin et al. 2004; Liu et al. 2007). A haplotype study that used multiple wild-derived M. musculus strains (Ideraabdullah et al. 2004) has revealed more SDPs than in the laboratory mouse, suggesting shorter haplotype blocks. The length of the haplotype blocks in the wild mouse is currently unknown, although an estimate from a screening of wild Mmd from Arizona places it under 100 kb (Laurie et al. 2007).
Recent studies have shown that the human haplotype blocks usually extend over tens of kilobases (International Hapmap Consortium 2007). The haplotype blocks are defined in human studies as regions of high linkage disequilibrium (LD) and seem to be delineated by recombination hot spots (Jeffreys et al. 2001). LD is the nonrandom association of alleles at tightly linked loci. There are several ways to compute the haplotype blocks (Barrett et al. 2005). Data on human haplotypes and LD across the human genome are of interest in whole-genome association studies.
Working with the mouse, Wade et al. (2002) proposed to use haplotype maps to narrow down the region of interest in positional and quantitative trait locus (QTL) cloning experiments. Positional and QTL cloning use the genome localization information to identify the gene(s) responsible for a particular trait. Haplotype-associated mapping (HAM; also called in silico cloning) can be used to identify QTL loci genomewide by association of haplotypes with phenotypes (Grupe et al. 2001; Pletcher et al. 2004). Interval-specific haplotype analysis has been used to narrow down QTL regions by excluding DNA intervals identical by descent (see Dipetrillo et al. 2005 for a review). However, high-resolution haplotype mapping of mouse traits is precluded by high LD within the classical laboratory strains. Knowledge of the haplotype maps is therefore a useful tool not only to model factors shaping the LD in the human genome, but also to aid in cloning genes of biomedical interest.
Our goal is the positional cloning of the Hybrid sterility 1 (Hst1) gene on mouse chromosome 17 (Forejt and Ivanyi 1975; Trachtulec et al. 1994, 1997a,b, 2005; Gregorova et al. 1996; Trachtulec and Forejt 1999, 2001). The gene participates in a breakdown of spermatogenesis in hybrids between some classical laboratory strains (AKR/J, BALB/c, A/Ph, DBA/1J, and C57BL/10SnPh, henceforth B10) and certain Mmmu mice, e.g., of the PWD/Ph strain. Other classical laboratory strains (CBA/J, P/J, and C3H/DiSnPh, henceforth C3H) produce fertile hybrid males with these Mmmu mice (Forejt and Ivanyi 1975).
The proximal third of chromosome 17, also called the t-complex, was first identified as a tail-length modifier (see Schimenti 2000 for a review). In a wild mouse population, it occurs in two forms, the wild type and the t-haplotype. The t-chromosomes are transmitted from heterozygous males in non-Mendelian ratios (Lyon 2003). The t-haplotypes, which occur in both Mmd and Mmmu, contain at least four large inversions (Herrmann et al. 1986; Artzt et al. 1991). The inversions suppress recombination between the wild type and the t-chromosomes, leading to the accumulation of mutations in the t-haplotypes. Consequently, homozygosity causes embryonic lethality or male sterility (Lyon 2003). However, the t-sterility appears to be distinct from the Hst1-type of hybrid sterility, as the Hst1 allele does not affect the transmission ratio distortion and all t-chromosomes tested produce fertile hybrid males when outcrossed to Mmmu (Forejt and Ivanyi 1975).
The Hst1 gene was mapped by genetic markers to a 360-kb region of the t-complex on our BC1 [(B10-T × C3H)-T × B10] cross (Trachtulec et al. 2005). Large rearrangements in the Hst1 region were excluded by restriction mapping of B6 and C3H yeast artificial chromosomes and genomic DNAs (Trachtulec et al. 1994, 1997b), as well as by mapping and sequencing of 129S1/SvImJ bacterial artificial chromosomes (Trachtulec et al. 2005). Also, no copy number variation of this region was detected by comparative genomic hybridization of 41 mouse strains (including wild derived; Cutler et al. 2007). Sequencing of six genes and tens of conserved stretches from this region has not yet identified the Hst1 candidate mutation, although some differences were found between the B10 and C3H strains. We therefore decided to use these polymorphisms for a haplotype analysis to aid in the cloning of the Hst1 gene and to learn about the properties and history of the surrounding DNA. Our results provide the first detailed haplotype map of wild Mmd. Although the map is limited to the 250-kb region, our conclusions are likely to be useful for other projects carried out in single-copy regions of the mouse genome.
MATERIALS AND METHODS
Mice, tails, and DNAs:
The B10.P, B10.STC77, B10.KPA132, C3H/DiSnPh, C57Bl/10SnPh, FVB/NCrl, 129S2/SvPas, T43H/Ph, and three t-haplotype mice (t0.129, tp4.129, t121tf/t121tf) have been bred in the facility of the Institute of Molecular Genetics. The t121tf is a partial t-haplotype carrying two proximal inversions (wild-type Hst1 region). The PWK/Ph, PWD/Ph, and PWB/Ph strains were established from wild M. m. musculus in our laboratory (Gregorova and Forejt 2000). Principles of laboratory animal care followed the Czech Republic Act on Animal Protection no. 246/92 Sb, fully compatible with the corresponding Directive 806/609/EEC of the Council of Europe Convention ETS123.
The tails of O20/A and STS/A mice were donated by M. Lipoldova from our Institute. The DNAs of DDK/Pas, MAI/Pas, MBT/Pas, STF/Pas, SEG/Pas, WLA/Pas, and WMP/Pas strains were obtained by courtesy of J.-L. Guénet, Institute Pasteur, Paris (Guenet and Bonhomme 2003). The GRS/A tail was kindly provided by E. Lukanidin, Danish Cancer Society (Copenhagen) and I/St DNA by J. Stavnezer, University of Massachusetts (Worcester, MA). The DNAs from the B10.F-H2pb1/(13R), CAST/Ei, CZECHII/Ei, I/LnJ, LEWES/Ei, LPT/Le, MOLF/Ei, MOR/Rk, MSM/Ms, PERA/Ei, PERC/Ei, RBA/Dn, RBB/Dn, SEA/GnJ, SK/CamEi, SM/J, SPRET/Ei, TIRANO/Ei, WSB/Ei, and ZALENDE/Ei were purchased from the Jackson Laboratory, Bar Harbor, Maine. Tails of wild Mmd and wild Mmmu were obtained from J. Pialek, Institute of Vertebrate Biology, Studenec, Czech Republic (Vyskocilova et al. 2005; Pialek et al. 2008). The remaining DNAs (A, AKR, B10.Atf/tf, B10.CAA2, B10.CAS2, B10.STA12, B10.WOA105, B10.WR7, BALB/c, BTBR-Ttf/+tf, CBA/J, DBA/1, DBA/2, DKU 28/97, THF/Tu, and tw12/tw12) were kindly provided by J. Klein and W. Mayer, Max-Planck Institute for Biology, Tuebingen, Germany (Vincek et al. 1990).
Genotyping:
The primers were designed by the program OLIGO, v.6. Primer sequences and annealing temperatures are indicated in the supplementary information. PCR was done in the presence of 100 ng total genomic DNA, 200 nm primers, 50 mm KCl, 10 mm Tris (pH 8.8), 0.08% Nonidet P40, 1.5 mm MgCl2, 0.18 nm dNTPs, and 0.04 units/μl recombinant Taq polymerase (MBI Fermentas) for 37 cycles. Aliquots were checked on agarose gels to ensure the presence of the product of the right size. The PCR reactions were treated by a kit containing exonuclease I and shrimp alkaline phosphatase (ExoSAP-It, USB) to degrade primers and dNTPs, and the enzymes were heat inactivated. Sequencing primer, buffer, polymerase, and a mix of dNTPs and fluorescent di-dNTPs were added, and the sequencing reactions were cycled in a thermocycler. Unincorporated di-dNTPs were removed by ethanol precipitation or by filtration through a column. The reactions were then loaded into a capillary sequencer (ABI or Beckman). The sequences were aligned, the raw data were inspected for differences with the help of the program GeneSkipper followed by manual editing, and the results entered into an MS Excel sheet. Haplotype blocks were computed in the program Haploview (version 3.32; Barrett et al. 2005) with default values. Phylogenetic analysis was performed using Phylo_win (Galtier et al. 1996). Microsatellites were scored on 5% agarose, rearrangements on 1–2% agarose.
RESULTS
Pilot experiment:
We first constructed a longer-range haplotype map of the Hst1 region on mouse chromosome 17. By sequencing genes and other conserved DNA from the region, a total of ∼50 kb, we identified SNP differences between the B10 and C3H strains in seven loci. These loci encompassed 13 SNPs across the 252-kb region, three single-nucleotide insertions/deletions (SN indels), and one deletion of three nucleotides (nt). The two strains thus differed by ∼3 SNPs/10 kb. The average distance between the loci was 40 kb. To obtain a haplotype map, the seven loci polymorphic between B10 and C3H were resequenced from 70 other chromosomes. In addition to the seven SNP loci, two loci polymorphic between B10 and 129S2/SvPas, two polymorphic microsatellites, and five rearrangements from the 252-kb region were typed on our panel, increasing its average resolution to one locus/16 kb. Assays for seven of these loci, including the outermost SNPs, were tested on our [(B10-T × C3H)T × B10] backcross panel (Gregorova et al. 1996). All loci cosegregated with Hst1 in all backcross animals tested.
All mice and strains are listed in materials and methods, and detailed descriptions appear in supplemental Table 1. The DNA samples screened for polymorphisms included 14 classical (Castle's and C57 related) inbreds, 15 nonclassical laboratory strains, 17 wild and recently wild-derived Mmd chromosomes, and controls. The controls included 10 wild-derived Mmmu chromosomes, 2 Mus m. molossinus (Mmmo), 2 Mus m. castaneus (Mmc), 3 Mus spretus wild-derived chromosomes, and 3 previously described t-haplotypes. The results are shown in supplemental Table 1.
There were only three types of haplotypes in the 14 classical laboratory strains used and other strains derived from them. The haplotype of strains FVB, B10.Atf/tf, RBA, and DDK was identical with B6 and B10. The strains A, AKR, DBA/1, DBA/2, THF, SEA, BALB/c, and both 129 substrains used (129S2/SvPas and 129X1/SvJ) were of the same haplotype. Ten tested strains carrying these two haplotypes produced sterile hybrids with Mmmu (A, B10, AKR, BALB/c, and DBA/1: Forejt and Ivanyi 1975; B6, DBA/2, FVB, THF, and 129S2: this article, supplemental Table 1). The strains CBA, B10.P, SM, BTBR-Ttf/+tf, T43H, and t121tf had the same sequences as C3H. Five tested strains of this third haplotype produced fertile hybrids with Mmmu (CBA and C3H: Forejt and Ivanyi 1975; B10.P, BTBR-Ttf/+tf, and t121tf: this article, supplemental Table 1). Seven nonclassical laboratory strains displayed four haplotypes distinct from the classical strains: I/St (the same haplotype as I/LnJ), GRS/A (the same as STS and SJL), O20, and B10.F(13R). In contrast to classical laboratory strains, most wild-derived Mmd strains, lines, and mice included in our set displayed haplotype breakpoints.
Resequencing results:
To obtain a haplotype map, polymorphisms with a sufficient minor allele frequency (MAF) in the wild mouse are necessary. Due to the low number of these differences in our pilot set, we performed additional sequencing of the region from the C3H strain, this time from randomly selected subclones. Loci polymorphic between C3H and B6 or Celera strains were then resequenced in other selected strains and wild-derived mice. In total, we resequenced 13 kb of sequence in 33 loci from a mean of 42 chromosomes. Twenty-four new SNPs were found in nonclassical laboratory and wild(-derived) Mmd mice (∼2/kilobase of sequence) and they had a low MAF. Altogether, half of the SNPs polymorphic among laboratory strains had a MAF >15% in wild Mmd mice.
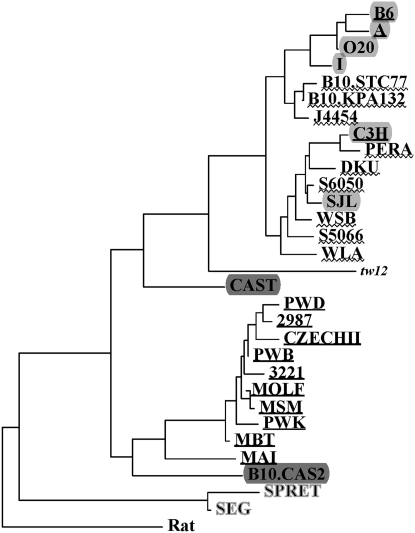
In total, we obtained >500 kb of sequence in almost 1400 reads. We discovered a total of 346 SNPs, five microsatellite variants, 14 SN indels, and 14 rearrangements of >2 nt. There were 2.8 times more transitions (Ti) than transversions (Tv), a ratio similar to previously published results (Ti/Tv = 2.5; Ideraabdullah et al. 2004). We reconstructed the history of the Hst1 region by phylogenetic analysis (Figure 1). Laboratory strains resided in the same part of our tree as wild Mmd. Eighty SNPs were identified in M. spretus only (8.3/kilobase, Ti/Tv = 3). Sixteen of 43 (37%) M. spretus-specific SNPs resequenced from two or more M. spretus strains were polymorphic in these strains (Ti/Tv = 1.4). Our data assigned strains derived from M. spretus to a branch different from all M. musculus in our tree (Figure 1). Cases of a likely introgression of Mmd DNA into the MAI/Pas, B10.CAS2, and CAST/Ei strains were found in some loci (see Figure 1 and supplemental Tables 1 and 2).
Figure 1.—
Phylogenetic history of the Hst1 region. A neighbor-joining tree of selected mice and strains was constructed from 339 SNPs (pairwise gap removal, observed divergence). The tree was rooted by genotypes derived from the rat assembly. All laboratory strains are shown on a lightly shaded background; classical laboratory strains are underlined. Wild-derived mice and strains are indicated: M. m. domesticus, wavy underlining; t-haplotype tw12, italics; M. m. musculus and M. m. molossinus, underlining; M. m. castaneus, darkly shaded background; M. spretus, shaded letters.
Distinct haplotypes for some mouse subspecies and t-haplotypes:
To construct a haplotype map, we needed to know whether we could pool data from different M. m. subspecies. We found only two SNPs that distinguish Mmmu from Mmmo in our region, which comprise only a few percent of SNPs segregating in Mmmu (henceforth Mmm designates both M. m. musculus and M. m. molossinus). When comparing Mmm to Mmd, we found about eight fixed and two segregating SNPs per kilobase of sequence. Trans-species SNPs (the same differences polymorphic in two or more subspecies) were rare (6%) between wild-derived Mmd (average of 25 sequenced chromosomes) and Mmm (8 chromosomes). Moreover, the structure of even very tightly linked SNPs was mosaic (as there would be no LD) in these inter-subspecific comparisons (supplemental Tables 1 and 2). These results indicate that the haplotypes in Mmmu and Mmd are different from each other in the Hst1 region.
The investigated region of chromosome 17 is located in the third inversion of the t-haplotypes (Artzt et al. 1991; Trachtulec et al. 1994). We found 28 t-specific SNPs (2.2/kilobase, Ti/Tv = 1.8). Most SNPs segregating in either Mmm or Mmd were fixed in the three t-chromosomes tested and the structure of these polymorphisms was mostly mosaic compared to both Mmd and Mmm. The sequence of the t-haplotypes had significantly (P < 0.0001, t-test) less identity to Mmm (98.98% ± 0.07) than to Mmd (99.32% ± 0.05). These features also apply to the tp4 chromosome that was isolated from a Mmmu population (Forejt et al. 1988). Thus, the t-chromosomes have a structure distinct from both Mmm and Mmd wild-type (non-t) haplotypes.
Haplotypes of wild(-derived) M. m. domesticus:
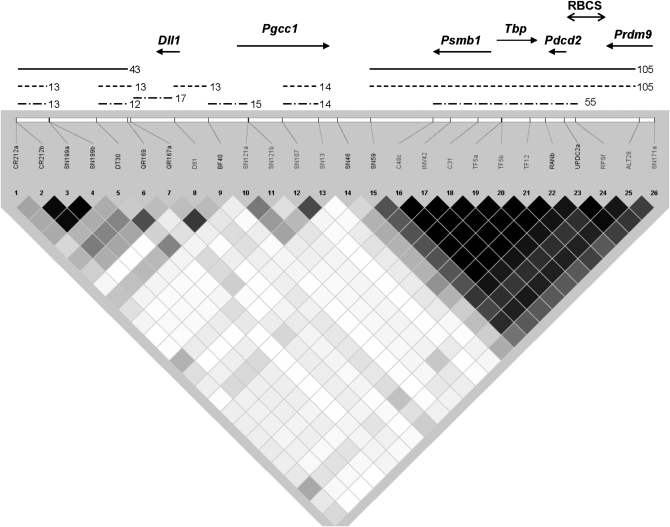
To obtain a fine haplotype map that would also include wild(-derived) Mmd mice, we used 43 SNPs with MAF >15%. The SNPs were contained in 26 loci of the 250-kb region. We utilized information from 22 loci on six wild Mmd mice, 18 wild Mmd-derived strains, and one Mmd line and we included seven laboratory strains showing distinct haplotypes (Figure 2 and supplemental Table 2). Because the remaining 4 loci mapped close to some of the 22 loci, they were resequenced from only a limited number of chromosomes to confirm or refine the haplotype blocks (supplemental Table 2). The number of new haplotypes in wild-derived Mmd increased, while the number of haplotypes in laboratory strains remained the same.
Figure 2.—
Haplotype map of M. m. domesticus in the 250-kb Hst1 region of chromosome 17. The map was constructed using 22 loci encompassing differences with a MAF >15% from 36 Mmd chromosomes. Genes are shown as arrows pointing in the direction of transcription. RBCS, reused breakpoint of conserved synteny. The squared correlation coefficient between two loci r2 = 1 (perfect LD) is indicated by solid diamonds; r2 = 0, open diamonds; 0 < r2 < 1, various shading. Haplotype blocks are indicated as lines with their length indicated to the right (in kilobases); they were computed using three methods: confidence intervals (solid line), solid spine of LD (dashed), and four-gamete rule (dot-and-dash).
To compute haplotype blocks, we employed three methods currently used in human genetics and included in the program Haploview (Barrett et al. 2005). The first method, confidence intervals (Gabriel et al. 2002), counts 95% confidence bounds on LD and each comparison is called “strong LD,” “inconclusive,” or “strong recombination.” A block is created if 95% of informative (i.e., noninconclusive) comparisons are “strong LD.” In the “four-gamete rule” method (Wang et al. 2002), the population frequencies of the four possible two-marker haplotypes are computed for each marker pair. If all four are observed with a frequency ≥0.01, a recombination event is assumed. Blocks are formed by consecutive markers where only three gametes are observed. The third method, solid spine of LD (Barrett et al. 2005), searches for a “spine” of strong LD running from one marker to another along the legs of the triangle in the LD chart.
The computed lengths of the haplotype blocks ranged from 12 to 105 kb (Figure 2). The region of the Pgcc1 gene (for PPARgamma constitutive coactivator 1, also called D17Ph4e or 4932442K08Rik), 37 kb in length, contained many haplotype breakpoints. The largest nonrecombining block, 45–76 kb in length, involved three genes of a highly conserved synteny (Trachtulec and Forejt 2001). The genes are proteasome subunit β1 (Psmb1), TATA-binding protein (Tbp), and programmed cell death 2 (Pdcd2). The region distal to Pdcd2 containing a multiple times utilized break of conserved synteny (Trachtulec et al. 2004) was depleted of haplotype breakpoints.
Rearrangements and M. m. domesticus haplotypes:
To determine the usefulness of rearrangements for haplotype mapping, we inspected our data for insertions, deletions, and inversions. We excluded microsatellite variation, defining microsatellites as described previously (Ideraabdullah et al. 2004). Of nine SN indels found in the laboratory strains, four had a MAF >10% in the wild-derived Mmd, with a rate 44% similar to the rate for SNPs, and corresponded well with the SNP-based haplotype map. Our sequences also encompassed two Mmd rearrangements of three or more nt. We also found five rearrangements of >3 nt by representational difference analysis (Lisitsyn and Wigler 1993) using B6 and C3H clones covering the Hst1 region. We then screened the rearrangements by PCR in other strains and mice (supplemental Table 1). Four of seven (57%) rearrangements had a MAF <15% in the wild Mmd and six of seven (86%) rearrangements did not correspond to haplotypes determined by SNP variation. The only rearrangement of >3 nt corresponding to Mmd haplotypes was an inversion of ∼1 kb in an intron of the Psmb1 proteasome subunit gene. The remaining six rearrangements were indels; three of them were >100 nt and included repetitive elements (L1, B2, and IAP).
Common features of human and M. m. domesticus haplotypes:
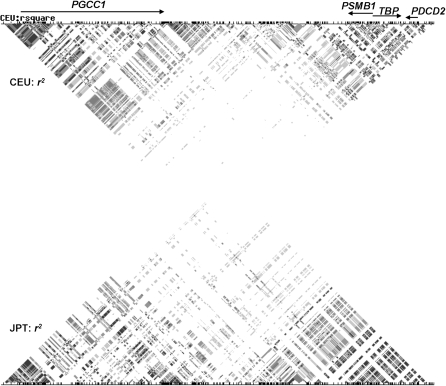
To compare the Mmd haplotypes with the human region of conserved synteny, we used HapMap data (International HapMap Consortium 2007) from the q-terminal end of human chromosome 6. The PGCC1 (FAM120B, KIAA1838)-PSMB1 intergenic region contains recombination hot spots. The haplotype block encompassing the genes of the highly conserved synteny PSMB1, TBP, and PDCD2 was depleted of breakpoints in two populations (Figure 3) as in the mouse region (Figure 2).
Figure 3.—
Haplotype map of human Chr6qter. Genes are shown as arrows pointing in the direction of transcription. CEU, Caucasian population; JPT, Japanese population; 0 < r2 < 1, various shading; r2 = 1, solid.
DISCUSSION
Our Mmd mouse haplotype map indicates that the long haplotype blocks in the classical laboratory strains are due to a limited number of chromosomes that had entered the inbreeding process of these strains, as has been suggested (Wade et al. 2002; Wiltshire et al. 2003; Ideraabdullah et al. 2004). However, the number of chromosomes in the initial stock was probably higher than previously indicated (Wade et al. 2002), as in the 14 classical strains that we analyzed there are three haplotypes in the Hst1 region and seven H2 haplotypes unlikely to have arisen by recombination. Complex haplotypes also help explain the distribution of SNPs along chromosome 1 (Yalcin et al. 2004) and other regions (Frazer et al. 2004). The length of haplotype blocks is much shorter in most of the wild(-derived) mice and some nonclassical laboratory strains, resembling the size of haplotype blocks found in humans. The wild mice and wild-derived strains can therefore serve as a model for investigating the properties of human haplotypes.
Ten mouse classical laboratory strains that produce sterile hybrid males with Mmmu share two haplotypes in the cosegregating region, while five strains with the third haplotype produce fertile hybrids. The haplotype analysis of laboratory strains thus confirms our backcross data. The Hst1 region could not be reduced by typing other classical strains, as there is a lack of haplotype breakpoints in the 252-kb region. We are therefore testing the nonclassical and wild-derived Mmd strains for Hst1 in an attempt to narrow the Hst1 candidate region by interval-specific haplotype analysis beyond the resolution achieved with 1500 backcross mice.
The resequencing of multiple mice of different (sub)species allowed us to determine the origin of the alleles in all mouse strains. All haplotypes of the classical strains, as well as of most nonclassical inbreds, were of Mmd origin. Half of the SNPs polymorphic among laboratory strains had a MAF >15% in wild Mmd mice. In wild Mmd mice from Arizona, one-half to two-thirds of classical strain SNPs had a MAF >5% (Laurie et al. 2007).
The comparison of Mmd and Mmm revealed a density of ∼10 SNPs/kilobase. This number agrees with previous studies that compared the B6 sequence pairwise with random sequences of one of two wild-derived Mmm strains, MSM/Ms (Abe et al. 2004) or PWD/Ph (Jansa et al. 2005). We also show that up to 80% of the differences are fixed. This result suggests that genotyping of mouse strains without sufficiently deep resequencing of wild mice of multiple subspecies can still be useful in discerning the Mmmu and Mmd origin, but not the particular subspecific haplotype. Thus, many SNPs discovered in laboratory strains will be useless for typing other mouse (sub)species and vice versa (Yang et al. 2007). Excluding Mmmu-derived strains from haplotype and in silico/HAM analysis results in improved signal (Liu et al. 2007; Payseur and Place 2007), suggesting that not just our region carries haplotypes distinct for Mmm and Mmd. In natural conditions, Mmmu and Mmd have occupied distinct geographical areas for a long time, and therefore unique haplotypes for both subspecies are expected.
Our data carry other important implications for projects of positional and QTL cloning in the mouse. Only some of the classical laboratory strains have megabase-sized haplotype blocks, and thus more detailed SNP mapping will be required for other strains, especially for the recently derived wild strains, to reach a resolution useful for QTL cloning studies. Typing of the strains with long haplotypes can be used to exclude large regions of a chromosome, while typing of wild-derived strains may help in narrowing down the defined candidate region beyond thousands of recombinants. Although whole-genome haplotype maps useful for wild-Mmd-derived mouse strains would require many more SNPs than are now affordable, these analyses could be made feasible by resequencing of only the particular region from the relevant (phenotyped) strains. These approaches apply only if the same single gene in the given chromosomal region is responsible for the phenotype of interest. Thus, the use of mouse crosses cannot be omitted.
The suitability of a wild Mmd mouse population for whole-genome HAM has been suggested recently (Laurie et al. 2007). While this approach could improve the mapping resolution reached with laboratory strains, one of its disadvantages is that only a limited number of phenotypes can be obtained per single genotyped animal. Laurie et al. (2007) estimated that about half of the traits found in the classical laboratory strains are also present in their population of Arizona mice and a similar number is expected for wild-Mmd-derived strains. While the number may seem low, for many QTL it is desirable to determine whether they also occur in the wild (e.g., traits affecting fitness).
Trans-species polymorphisms were rare in our region (∼6% between Mmm and Mmd). In a recent study (Ideraabdullah et al. 2004), 12–22% of variant alleles were estimated to segregate simultaneously in two different M. musculus subspecies by resequencing 14 wild-derived strains of M. m. subspecies. Some of the ancestral differences reported by Ideraabdullah et al. (2004) can be subspecific introgressions of larger regions, but our results can also be attributed to the uniqueness of the Hst1 region, which may be less efficiently transmitted to other subspecies.
Knowledge of the properties of nonmicrosatellite rearrangements is also important for QTL cloning. While SN indels had similar properties as SNPs in our region, only one of seven rearrangements of >3 nt corresponded to SNP-based haplotypes in the wild Mmd. Our data suggest that longer rearrangements could be useful as markers for haplotype analysis only in classical laboratory strains, a conclusion previously reached for microsatellites (Pletcher et al. 2004). However, many copy number variations in the genomes of the laboratory strains are the results of recurrent mutations (Egan et al. 2007).
Although there is a little overlap between the location of human and chimpanzee recombination hot spots, there are homologous regions of strong LD in both humans and chimps (Ptak et al. 2005; Winckler et al. 2005). A comparison of haplotype blocks of a megabase region from humans, rats, and laboratory mice revealed a tendency to encompass entire genes (Guryev et al. 2006). This feature appears to be general in the human genome (Eberle et al. 2006). In the Hst1 region, the lengths of the haplotype blocks deduced from 36 domesticus chromosomes of independent origin are on the order of tens of kilobases. The largest Mmd haplotype block encompasses three genes of the highly conserved synteny Psmb1, Tbp, and Pdcd2 (Trachtulec et al. 1997a, 2004; Trachtulec and Forejt 2001; Mihola et al. 2007). The human region on 6qter carrying the orthologs of these three genes in the same order and orientation also has a high LD (Chistiakov et al. 2005; Payne et al. 2005; International HapMap Consortium 2007). These three genes map to the same domain carrying a unique pattern of histone modifications in both humans (Barski et al. 2007) and mice (Mikkelsen et al. 2007). An antisense overlap of alternative mRNAs of Tbp and Pdcd2 is conserved in chicken and may regulate the transcription of the Pdcd2 gene, suggesting a function for the conserved linkage (Mihola et al. 2007). It remains to be investigated whether a strong LD in regions carrying multiple genes of conserved synteny applies to mammalian genomes in general.
Acknowledgments
We thank all contributors of mouse DNAs and tails listed in materials and methods, P. Jansa and J. Felsberg for running sequences, P. Divina for help with bioinformatics, and M. Landikova for technical assistance. We are grateful to F. P.-M. de Villena and J. C. Schimenti for helpful comments and S. Takacova for improving the manuscript. This work was supported by grant no. A5052406 from the Grant Agency of the Academy of Sciences of the Czech Republic, no. 301/05/0738 from the Czech Science Foundation, and nos. AV0Z50520514 and 1M6837805002 from the Ministry of Education, Youth and Sports of the Czech Republic.
References
- Abe, K., H. Noguchi, K. Tagawa, M. Yuzuriha, A. Toyoda et al., 2004. Contribution of Asian mouse subspecies Mus musculus molossinus to genomic constitution of strain C57BL/6J, as defined by BAC-end sequence-SNP analysis. Genome Res. 14 2439–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artzt, K., D. Barlow, W. F. Dove, K. Fischer-Lindahl, J. Klein et al., 1991. Mouse chromosome 17. Mamm. Genome 1 (Spec No): S280–S300. [DOI] [PubMed] [Google Scholar]
- Barrett, J. C., B. Fry, J. Maller and M. J. Daly, 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21 263–265. [DOI] [PubMed] [Google Scholar]
- Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones et al., 2007. High-resolution profiling of histone methylations in the human genome. Cell 129 823–837. [DOI] [PubMed] [Google Scholar]
- Chistiakov, D. A., Y. A. Seryogin, R. I. Turakulov, K. V. Savost'anov, E. V. Titovich et al., 2005. Evaluation of IDDM8 susceptibility locus in a Russian simplex family data set. J. Autoimmun. 24 243–250. [DOI] [PubMed] [Google Scholar]
- Cutler, G., L. A. Marshall, N. Chin, H. Baribault and P. D. Kassner, 2007. Significant gene content variation characterizes the genomes of inbred mouse strains. Genome Res. 17 1743–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipetrillo, K., X. Wang, I. M. Stylianou and B. Paigen, 2005. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 21 683–692. [DOI] [PubMed] [Google Scholar]
- Eberle, M. A., M. J. Rieder, L. Kruglyak and D. A. Nickerson, 2006. Allele frequency matching between SNPs reveals an excess of linkage disequilibrium in genic regions of the human genome. PLoS Genet. 2 e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan, C. M., S. Sridhar, M. Wigler and I. M. Hall, 2007. Recurrent DNA copy number variation in the laboratory mouse. Nat. Genet. 39 1384–1389. [DOI] [PubMed] [Google Scholar]
- Forejt, J., and P. Ivanyi, 1975. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.). Genet. Res. 24 189–206. [DOI] [PubMed] [Google Scholar]
- Forejt, J., S. Gregorova and P. Jansa, 1988. Three new t-haplotypes of Mus musculus reveal structural similarities to t-haplotypes of Mus domesticus. Genet. Res. 51 111–119. [DOI] [PubMed] [Google Scholar]
- Frazer, K. A., C. M. Wade, D. A. Hinds, N. Patil, D. R. Cox et al., 2004. Segmental phylogenetic relationships of inbred mouse strains revealed by fine-scale analysis of sequence variation across 4.6 mb of mouse genome. Genome Res. 14 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer, K. A., E. Eskin, H. M. Kang, M. A. Bogue, D. A. Hinds et al., 2007. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature 448 1050–1053. [DOI] [PubMed] [Google Scholar]
- Gabriel, S. B., S. F. Schaffner, H. Nguyen, J. M. Moore, J. Roy et al., 2002. The structure of haplotype blocks in the human genome. Science 296 2225–2229. [DOI] [PubMed] [Google Scholar]
- Galtier, N., M. Gouy and C. Gautier, 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12 543–548. [DOI] [PubMed] [Google Scholar]
- Gregorova, S., and J. Forejt, 2000. PWD/Ph and PWK/Ph inbred mouse strains of Mus m. musculus subspecies: a valuable resource of phenotypic variations and genomic polymorphisms. Folia Biol. (Praha) 46 31–41. [PubMed] [Google Scholar]
- Gregorova, S., M. Mnukova-Fajdelova, Z. Trachtulec, J. Capkova, M. Loudova et al., 1996. Sub-milliMorgan map of the proximal part of mouse chromosome 17 including the hybrid sterility 1 gene. Mamm. Genome 7 107–113. [DOI] [PubMed] [Google Scholar]
- Grupe, A., S. Germer, J. Usuka, D. Aud, J. K. Belknap et al., 2001. In silico mapping of complex disease-related traits in mice. Science 292 1915–1918. [DOI] [PubMed] [Google Scholar]
- Guenet, J. L., and F. Bonhomme, 2003. Wild mice: an ever-increasing contribution to a popular mammalian model. Trends Genet. 19 24–31. [DOI] [PubMed] [Google Scholar]
- Guryev, V., B. M. Smits, J. Van De Belt, M. Verheul, N. Hubner et al., 2006. Haplotype block structure is conserved across mammals. PLoS Genet. 2 e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, B., M. Bucan, P. E. Mains, A. M. Frischauf, L. M. Silver et al., 1986. Genetic analysis of the proximal portion of the mouse t complex: evidence for a second inversion within t haplotypes. Cell 44 469–476. [DOI] [PubMed] [Google Scholar]
- Ideraabdullah, F. Y., E. De La Casa-Esperon, T. A. Bell, D. A. Detwiler, T. Magnuson et al., 2004. Genetic and haplotype diversity among wild-derived mouse inbred strains. Genome Res. 14 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium, 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansa, P., P. Divina and J. Forejt, 2005. Construction and characterization of a genomic BAC library for the Mus m. musculus mouse subspecies (PWD/Ph inbred strain). BMC Genomics 6: 161. [DOI] [PMC free article] [PubMed]
- Jeffreys, A. J., L. Kauppi and R. Neumann, 2001. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29 217–222. [DOI] [PubMed] [Google Scholar]
- Laurie, C. C., D. A. Nickerson, A. D. Anderson, B. S. Weir, R. J. Livingston et al., 2007. Linkage disequilibrium in wild mice. PLoS Genet. 3 e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsyn, N., and M. Wigler, 1993. Cloning the differences between two complex genomes. Science 259 946–951. [DOI] [PubMed] [Google Scholar]
- Liu, P., H. Vikis, Y. Lu, D. Wang and M. You, 2007. Large-scale in silico mapping of complex quantitative traits in inbred mice. PLoS ONE 2 e651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon, M. F., 2003. Transmission ratio distortion in mice. Annu. Rev. Genet. 37 393–408. [DOI] [PubMed] [Google Scholar]
- Mihola, O., J. Forejt and Z. Trachtulec, 2007. Conserved alternative and antisense transcripts at the programmed cell death 2 locus. BMC Genomics 8 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, T. S., M. Ku, D. B. Jaffe, B. Issac, E. Lieberman et al., 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mural, R. J., M. D. Adams, E. W. Myers, H. O. Smith, G. L. Miklos et al., 2002. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296 1661–1671. [DOI] [PubMed] [Google Scholar]
- Payne, F., D. J. Smyth, R. Pask, J. D. Cooper, J. Masters et al., 2005. No evidence for association of the TATA-box binding protein glutamine repeat sequence or the flanking chromosome 6q27 region with type 1 diabetes. Biochem. Biophys. Res. Commun. 331 435–441. [DOI] [PubMed] [Google Scholar]
- Payseur, B. A., and M. Place, 2007. Prospects for association mapping in classical inbred mouse strains. Genetics 175 1999–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pialek, J., M. Vyskocilova, B. Bimova, D. Havelkova, J. Pialkova et al., 2008. Development of unique house mouse resources suitable for evolutionary studies of speciation. J. Hered. 99 34–44. [DOI] [PubMed] [Google Scholar]
- Pletcher, M. T., P. McClurg, S. Batalov, A. I. Su, S. W. Barnes et al., 2004. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2 e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptak, S. E., D. A. Hinds, K. Koehler, B. Nickel, N. Patil et al., 2005. Fine-scale recombination patterns differ between chimpanzees and humans. Nat. Genet. 37 429–434. [DOI] [PubMed] [Google Scholar]
- Schimenti, J., 2000. Segregation distortion of mouse t haplotypes: the molecular basis emerges. Trends Genet. 16 240–243. [DOI] [PubMed] [Google Scholar]
- Trachtulec, Z., and J. Forejt, 1999. Transcription and RNA processing of mammalian genes in Saccharomyces cerevisiae. Nucleic Acids Res. 27 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtulec, Z., and J. Forejt, 2001. Synteny of orthologous genes conserved in mammals, snake, fly, nematode, and fission yeast. Mamm. Genome 12 227–231. [DOI] [PubMed] [Google Scholar]
- Trachtulec, Z., V. Vincek, R. M. Hamvas, J. Forejt, H. Lehrach et al., 1994. Physical map of mouse chromosome 17 in the region relevant for positional cloning of the hybrid sterility 1 gene. Genomics 23 132–137. [DOI] [PubMed] [Google Scholar]
- Trachtulec, Z., R. M. Hamvas, J. Forejt, H. R. Lehrach, V. Vincek et al., 1997. a Linkage of TATA-binding protein and proteasome subunit C5 genes in mice and humans reveals synteny conserved between mammals and invertebrates. Genomics 44 1–7. [DOI] [PubMed] [Google Scholar]
- Trachtulec, Z., M. Mnukova-Fajdelova, R. M. Hamvas, S. Gregorova, W. E. Mayer et al., 1997. b Isolation of candidate hybrid sterility 1 genes by cDNA selection in a 1.1 megabase pair region on mouse chromosome 17. Mamm. Genome 8 312–316. [DOI] [PubMed] [Google Scholar]
- Trachtulec, Z., C. Vlcek, O. Mihola and J. Forejt, 2004. Comparative analysis of the PDCD2-TBP-PSMB1 region in vertebrates. Gene 335 151–157. [DOI] [PubMed] [Google Scholar]
- Trachtulec, Z., O. Mihola, C. Vlcek, H. Himmelbauer, V. Paces et al., 2005. Positional cloning of the hybrid sterility 1 gene: fine genetic mapping and evaluation of two candidate genes. Biol. J. Linn. Soc. Lond. 84 637–641. [Google Scholar]
- Vincek, V., J. Sertic, Z. Zaleska-Rutzynska, F. Figueroa and J. Klein, 1990. Characterization of H-2 congenic strains using DNA markers. Immunogenetics 31 45–51. [DOI] [PubMed] [Google Scholar]
- Vyskocilova, J., Z. Trachtulec, J. Forejt and J. Pialek, 2005. Does geography matter in hybrid sterility in house mice? Biol. J. Linn. Soc. Lond. 84 663–674. [Google Scholar]
- Wade, C. M., E. J. Kulbokas, III, A. W. Kirby, M. C. Zody, J. C. Mullikin et al., 2002. The mosaic structure of variation in the laboratory mouse genome. Nature 420 574–578. [DOI] [PubMed] [Google Scholar]
- Wang, N., J. M. Akey, K. Zhang, R. Chakraborty and L. Jin, 2002. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am. J. Hum. Genet. 71 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril et al., 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420 520–562. [DOI] [PubMed] [Google Scholar]
- Wiltshire, T., M. T. Pletcher, S. Batalov, S. W. Barnes, L. M. Tarantino et al., 2003. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc. Natl. Acad. Sci. USA 100 3380–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler, W., S. R. Myers, D. J. Richter, R. C. Onofrio, G. J. McDonald et al., 2005. Comparison of fine-scale recombination rates in humans and chimpanzees. Science 308 107–111. [DOI] [PubMed] [Google Scholar]
- Yalcin, B., J. Fullerton, S. Miller, D. A. Keays, S. Brady et al., 2004. Unexpected complexity in the haplotypes of commonly used inbred strains of laboratory mice. Proc. Natl. Acad. Sci. USA 101 9734–9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., T. A. Bell, G. A. Churchill and F. Pardo-Manuel De Villena, 2007. On the subspecific origin of the laboratory mouse. Nat. Genet. 39 1100–1107. [DOI] [PubMed] [Google Scholar]
- Zhang, K., Z. S. Qin, J. S. Liu, T. Chen, M. S. Waterman et al., 2004. Haplotype block partitioning and tag SNP selection using genotype data and their applications to association studies. Genome Res. 14 908–916. [DOI] [PMC free article] [PubMed] [Google Scholar]