Abstract
Esa1 is the only essential histone acetyltransferase (HAT) in budding yeast. It is the catalytic subunit of at least two multiprotein complexes, NuA4 and Piccolo NuA4 (picNuA4), and its essential function is believed to be its catalytic HAT activity. To examine the role of Esa1 in DNA damage repair, we isolated viable esa1 mutants with a range of hypersensitivities to the toposide camptothecin. Here we show that the sensitivity of these mutants to a variety of stresses is inversely proportional to their level of histone H4 acetylation, demonstrating the importance of Esa1 catalytic activity for resistance to genotoxic stress. Surprisingly, individual mutations in two residues directly involved in catalysis were not lethal even though the mutant enzymes appear catalytically inactive both in vivo and in vitro. However, the double-point mutant is lethal, demonstrating that the essential function of Esa1 relies on residues within the catalytic pocket but not catalysis. We propose that the essential function of Esa1 may be to bind acetyl-CoA or lysine substrates and positively regulate the activities of NuA4 and Piccolo NuA4.
MYST histone acetyltransferases (HAT) comprise an evolutionarily conserved family of eukaryotic enzymes with important roles in many aspects of cell and developmental biology and with significant implications for our understanding of oncogenesis (Lafon et al. 2007). ESA1 encodes the only essential HAT in Saccharomyces cerevisiae, a 445-amino-acid enzyme featuring an N-terminal chromodomain and a C-terminal MYST domain (Smith et al. 1998). Esa1 is the primary HAT for histone H4 acetylation in vivo and can also acetylate histones H2A and H2AZ (Smith et al. 1998; Allard et al. 1999; Clarke et al. 1999; Babiarz et al. 2006; Keogh et al. 2006; Millar et al. 2006). It is the catalytic subunit of at least two complexes, NuA4 and Piccolo NuA4 (picNuA4) (Allard et al. 1999; Boudreault et al. 2003; Doyon and Côté 2004). NuA4 is a 1.3-MDa multisubunit HAT complex made up of at least 13 polypeptides including the essential proteins Act1, Arp4, Epl1, Esa1, Swc4, and Tra1 (Allard et al. 1999). A subset of the larger NuA4 complex, picNuA4 is composed of only three polypeptides: Esa1, Yng2, and Epl1 (Boudreault et al. 2003). NuA4 is thought to provide the locus-specific targeted histone acetylation associated with transcriptional activation and DNA repair, while picNuA4 controls the global levels of histone H4 acetylation in the cell (Boudreault et al. 2003). The yeast NuA4 HAT complex has been highly conserved during evolution and shares significant structural and functional similarities with its human and Drosophila counterparts hNuA4/Tip60 and DmTip60, respectively; these metazoan complexes also contain related MYST family HAT enzymes as their catalytic subunits (Doyon et al. 2004; Doyon and Côté 2004; Kusch et al. 2004).
Esa1 and the NuA4 complex have been implicated in multiple nuclear processes. Like its GNAT family (Gcn5-related N-acetyltransferase) paralog Gcn5, Esa1 is responsible for histone acetylation at the promoters of a subset of yeast genes, facilitating recruitment of the transcription machinery and subsequent gene activation (Galarneau et al. 2000; Reid et al. 2000; Nourani et al. 2004; Durant and Pugh 2006, 2007). Esa1-catalyzed acetylation of histone H4 near DNA double-strand-break (DSB) sites is a critical step in the repair process through its role in recruiting the INO80 and SWR complexes to modify chromatin structure at the break (Bird et al. 2002; Downs et al. 2004). In addition, NuA4 may play a role in kinetochore assembly and chromosome segregation, as NuA4 has been found to be localized to the centromere (Ogiwara et al. 2007), and multiple mutants of NuA4 subunits are sensitive to the microtubule depolymerizing agent benomyl (Le Masson et al. 2003; Kobor et al. 2004; Krogan et al. 2004; Ogiwara et al. 2007). There is also evidence that Esa1 participates in DNA replication by regulating Start (Early et al. 2004), although this role has not been well characterized. Finally, NuA4 is involved in at least two aspects of ribosome biogenesis through its roles in regulating transcription of ribosomal protein genes (Reid et al. 2000; Rohde and Cardenas 2003) and rDNA transcription/silencing (Clarke et al. 2006).
At present, Esa1 is the only subunit of the NuA4 and picNuA4 complexes known to have enzymatic activity. The catalytic mechanism of Esa1 is controversial (Yan et al. 2000, 2002; Berndsen et al. 2007a). Initial structural and biochemical studies of an active fragment of Esa1, favorable for crystallography, suggested a ping-pong mechanism involving residues Cys304 and Glu338 in catalysis (Yan et al. 2000, 2002). According to this model the first step in the HAT reaction is deprotonation of Cys304 by Glu338 and nucleophilic attack transferring the acetyl moiety from acetyl-coenzyme A (Ac-CoA) to Cys304, creating an acetyl-Cys304 enzyme intermediate. Subsequently, Glu338 deprotonates the histone lysine residue, mediating the transfer of the acetyl group from acetyl-Cys304 to the ɛ-amino group of the lysine substrate. In support of this model, individual mutations of both Cys304 and Glu338 were found to eliminate the HAT activity of the recombinant Esa1 fragment in vitro (Yan et al. 2000, 2002). Furthermore, cocrystals of Esa1 and Ac-CoA revealed the transfer of the acetyl group from Ac-CoA to Cys304, and this transfer failed to occur in cocrystals of Esa1-E338Q and Ac-CoA. When purified, this acetyl-C304 enzyme intermediate was then able to acetylate lysines on H3 and H4 peptides in vitro in the absence of cofactor. Finally, bisubstrate kinetic analysis also supported the ping-pong model. The invariant conservation of the Glu and Cys residues in the sequences of MYST enzymes suggests that this ping-pong mechanism is characteristic of the family (Yan et al. 2002) and is distinct from the direct transfer mechanism utilized by members of the GNAT family (Tanner et al. 1999, 2000).
Recently, this ping-pong model has been called into question. Berndsen et al. (2007a) have examined the biochemistry of full-length Esa1, alone and in the context of reconstituted picNuA4 complex. In agreement with previous results, the Esa1-E338Q mutant was catalytically inactive. Furthermore, the requirement for E338 in catalysis could be bypassed by carrying out the reaction at elevated pH where the substrate lysine residue is already deprotonated. However, in contrast to the results of Yan et al. (2002), Esa1-C304S was fully functional both as full-length enzyme and as picNuA4 complex, arguing against a ping-pong catalytic mechanism. Autoacetylation rates of picNuA4 were found to be low and the stability of the autoacetylated products was inconsistent with an acetyl-cysteine intermediate. Finally the bisubstrate kinetics of the picNuA4 reaction supported a direct Glu338-mediated transfer of the acetyl moiety to the histone lysine substrate. Regardless of the precise catalytic mechanism, however, there is universal agreement that E338 is essential for catalysis and that its replacement with glutamine destroys the ability to deprotonate the ɛ-amino group of lysine substrates (Yan et al. 2000, 2002; Berndsen et al. 2007a).
It is currently thought that the essential function of Esa1 resides in its protein acetyltransferase activity. In plasmid shuffle experiments, the esa1-E338Q mutant allele was unable to support viability and actually conferred a dominant growth inhibition (Yan et al. 2000). Overexpression of this allele also causes defects in silencing at the rDNA and telomeres (Clarke et al. 2006). Thus, current experimental results are consistent with an essential role for the catalytic activity of Esa1, NuA4, and picNuA4.
We have previously characterized esa1-1851, a mutant allele that specifically phenocopies the DNA damage sensitivity of an H4 mutant lacking all four N-terminal tail lysines subject to reversible acetylation (Megee et al. 1995; Bird et al. 2002). Surprisingly, this allele resulted in a dramatic decrease in histone H4 lysine acetylation, suggesting a defect in catalytic activity of Esa1-1851. To better understand the role of Esa1 in DNA damage repair, we isolated and characterized additional esa1 mutants with a range of sensitivity to DNA damage. Here we report that the sensitivity to genotoxic damage in these mutants is inversely proportional to their HAT activity, providing strong support for the causative role of H4 acetylation by Esa1 in DNA DSB repair. Surprisingly, we find that residues directly involved in the mechanism of catalysis are not required for cell viability and therefore the essential function of Esa1 does not involve protein acetylation.
MATERIALS AND METHODS
Yeast manipulations:
The strains used in these experiments are listed in Table 1. Standard media and yeast manipulations were used throughout (Amberg et al. 2005).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303-1B | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Wallis et al. (1989) |
| MSY2594 | W303-1B esa1-D343V∷K.l.URA3 | This study |
| MSY2666 | W303-1B esa1-W66R∷K.l.URA3 | This study |
| MSY2711 | W303-1B esa1-L357H∷K.l.URA3 | This study |
| MSY2890 | W303-1B esa1-C304S∷K.l.URA3 | This study |
| MSY3141 | W303-1B esa1-E338Q∷K.l.URA3 | This study |
| MSY3855 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 ESA1-6HA∷S.p.his5/ESA1 | This study |
| MSY4040 | MATa/MATα ADE2/ade2-1 his3/his3 leu2/leu2 TRP1/trp1-1 ESA1-6HA∷S.p.his5/ESA1 eaf3Δ∷KanMX/eaf3Δ∷KanMX | This study |
| MSY3853 | MSY3855 esa1-C304S-6HA∷S.p.his5/ESA1 | This study |
| MSY3852 | MSY3855 esa1-E338Q-6HA∷S.p.his5/ESA1 | This study |
| MSY3851 | MSY3855 esa1-C304S,E338Q-6HA∷S.p.his5/ESA1 | This study |
| MSY3792 | MSY3855 esa1-C304S,E338Q∷K.l.URA3/ESA1 | This study |
| MSY2563 | MATα his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 esa1∷KanMX pQQ48[ESA1 URA3 CEN ARS] | This study |
Mutagenesis:
All gene knockouts and epitope tagging were done using methods and constructs described previously (Longtine et al. 1998). ESA1 was mutagenized by PCR misincorporation and targeted to its chromosomal location by one-step cotransformation linked with the Kluyveromyces lactis PYRF_KLULA (K.l.URA3) gene as a selectable marker. Mutagenic amplification of ESA1 was carried out in reactions with 7 mm MgCl2, 0.2 mm dATP and dGTP, and 1 mm of dTTP and dCTP using primer 1 (TCCCATGACGGAAAAGAAGAACCTGG) and primer 2 (CATGGCAAGTCCCGTGGATCCTGTATATCTTAAGTAAGAGTATTAACTTACAGGAATACTG). The K.l.URA3 marker was amplified in standard fidelity reactions using primer 3 (GATCCACGGGACTTGCCATGTGTGATTCTGGGTAGAAGATC) and primer 4 (GCTTTTACATTAGAAGTTGTTTGAATGTAAGTTTAGGAAAGCACTACCGATGATGTAGTTTCTGGTT). Primers 1 and 4 have homology with the ESA1 locus, targeting the two fragments to the chromosome, while primers 2 and 3 have complementary homology with each other, linking the esa1 and K.l.URA3 fragments by recombination. A total of 299 Ura+ colonies were screened for growth at 28° on YPD medium, with and without 30 μg/ml CPT, and for growth on YPD at 37°. Primary screening identified 42 candidate mutants, which were then tested by serial spot dilution plating. Subsequently, the three alleles with the most severe drug sensitivity were selected for further study. Limited characterizations of esa1-1851 and esa1-L357H were reported previously (Bird et al. 2002). Molecular modeling was carried out using PyMol (Delano Scientific, Palo Alto, CA; http://www.pymol.org).
Cell extract preparation:
Logarithmically growing cultures were diluted in YPD to a final volume of 20 ml. After 1 hr at 28°, 10 ml of each culture were shifted to 37° for 4 hr. Cells were centrifuged, washed with dH2O, and resuspended in 250 μl HSB buffer (45 mm HEPES-KOH, pH 7.4, 150 mm NaCl, 10% glycerol, 1 mm EDTA, 20 mm sodium butyrate, 2 μm DTT, 0.5% NP-40) with protease inhibitors (100 μm PMSF, 1 μg/ml each of aprotinin, leupeptin, chymostatin, and pepstatin A). Cells were lysed with 425–600 μm acid-washed glass beads in a Mini-BeadBeater (Biospec, Bartlesville, OK). Recovered lysate was centrifuged to clear cell debris.
Antibodies:
A goat polyclonal anti-Esa1 peptide antibody was purchased from Santa Cruz Biotechnology [yL-20; catalog (cat.) no. SC-12155] and was used for Western blot analysis at a 1:250 dilution overnight at 4°. Rabbit polyclonal antibodies against full-length Esa1 and Eaf3 were purchased from Abcam (cat. nos. ab4466 and ab4467, respectively) and used for immunoprecipitation. Rabbit polyclonal anti-histone H4 antibody was purchased from Cell Signaling (cat. no. 2592) and used at a 1:1000 dilution overnight at 4° in the presence of 3% chicken egg albumin as a blocking agent. Rabbit polyclonal antibodies directed against acetylated histones H4 and H3 were purchased from Upstate (cat. nos. 06-946 and 06-599, respectively) and used at a 1:1000 dilution for 1 hr at room temperature. An anti-glucose-6-phosphate dehydrogenase (GPDH) rabbit polyclonal antibody was purchased from Sigma (St. Louis; cat. no. A9521) and used at a 1:20,000 dilution for 1 hr at room temperature. An anti-HA epitope tag mouse monoclonal antibody (12CA5) was used at a 1:2000 dilution for 1 hr at room temperature. All Western blot secondary incubations were carried out for 1 hr at room temperature with a 1:3000 or 1:5000 dilution of goat anti-rabbit (Bio-Rad, Hercules, CA; cat. no. 170-6515), bovine anti-goat (Santa Cruz Biotechnology; cat. no. SC-2378), or sheep anti-mouse (Amersham, Piscataway, NJ; cat. no. NA9310V) antibody. To reprobe, the blots were stripped using the Restore Western blot stripping buffer (Pierce Chemical, Rockford, IL; cat. no. 21059).
Immunoprecipitations and HAT assays:
Log-phase yeast cultures were prepared, pelleted, and lysed by bead-beating in HSB buffer as above. Cleared extracts (4000 μg) were incubated with rabbit polyclonal anti-Esa1 antibody (Abcam) (6 μl) and Protein A sepharose beads (Amersham) at 4°, followed by washing four times in HSB buffer. The immunoprecipitates were assayed for HAT activity as described previously (Iizuka and Stillman 1999), using purified chicken core histones. Immunoblotting against Esa1 was with yL-20 anti-Esa1 antibody (1:250). Immunoprecipitations of NuA4 in the HA-tagged heterozygous diploid yeast strains were carried out as above except that an anti-Eaf3 antibody (Abcam) was used and the immunoprecipitates were resolved by SDS–PAGE.
Preparation of recombinant Esa1 and HAT assays:
HIS6-ESA1, HIS6-esa1-E338Q, HIS6-esa1-C304S, and HIS6-esa1-C304S,E338Q were cloned into the pET-15b vector (Novagen, San Diego) and expressed in Escherichia coli strain BL21-CodonPlus(DE3)-RIL cells (Stratagene, La Jolla, CA) by inducing expression with 1 mm IPTG at 28° for 2 hr. After centrifugation and flash freezing of cell pellets in a dry ice/ethanol bath, cell lysates were prepared under native conditions and incubated with Ni-NTA beads (QIAGEN, Valencia, CA; cat. no. 1018244) essentially according to the manufacturer's recommended protocol with the following modifications: lysis buffer was 50 mm NaH2PO4, 500 mm NaCl, 10 mm imidazole, 10% glycerol, 1% Triton X-100, 10 mm β-mercaptoethanol, pH 8.0; His6-Esa1 was eluted in 100 μl fractions, and the peak His6-Esa1 fraction (determined by SDS–PAGE and Coomassie staining) was used as described below; and all steps were performed at room temperature due to aggregation of Esa1 at 4°. His6-Esa1 (2 μg) was then incubated with chicken core histones and [3H]-acetyl CoA for 30 min at room temperature followed by separation by SDS–PAGE and fluorography to determine incorporation of [3H]-acetyl groups into histones.
RESULTS
The mutation of histone H4 N-terminal domain lysines causes increased constitutive DNA damage, activation of the DNA damage checkpoint pathway, and hypersensitivity to DNA damaging agents such as CPT and MMS (Megee et al. 1995; Bird et al. 2002). Consistent with its role as the major H4 HAT in the cell, we have previously reported that Esa1 function is essential for protection against replication-coupled DNA damage (Bird et al. 2002). In that study we isolated an allele of ESA1, esa1-1851, that specifically phenocopies the DNA damage sensitivity of the H4 mutant. This allele resulted in a decrease in total histone H4 lysine acetylation to near background levels, providing a mechanistic explanation for the DNA damage sensitivity of the mutant. However, this result also suggested a severe defect in the catalytic activity of Esa1-1851. Since Esa1's catalytic HAT activity was thought to be essential, this apparent loss of activity posed an intriguing paradox.
New esa1 alleles:
To explore the catalytic activity of Esa1 in more detail, we isolated, characterized, and sequenced additional CPT-hypersensitive esa1 alleles. ESA1 was mutagenized in vitro by low-fidelity PCR and targeted to the chromosome by homologous recombination using the K. lactis (PYRF_KLULA) gene (K.l.URA3) as a linked selectable marker (see materials and methods). We focused on three isolates with the strongest phenotypes, esa1-1851, esa1-1852, and esa1-1853, which cause a graded range of hypersensitivity to CPT at 28°. The CPT phenotype was recessive in all three mutants, segregated 2:2 in backcrosses, and was linked to the K.l.URA3 marker. Furthermore, each was complemented by the wild-type ESA1 gene on a single-copy plasmid.
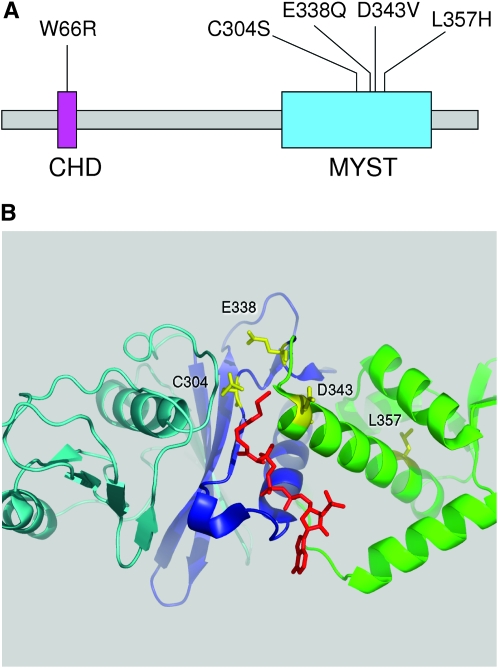
The ESA1 gene was sequenced from each mutant to identify the amino acid residues responsible for the defect. The results are summarized in Figure 1. The esa1-1852 allele contains a single-base change resulting in the substitution of Asp343 with valine (D343V). The sequences of esa1-1853 and esa1-1851 revealed mutations resulting in two- and three-amino-acid substitutions, respectively. By selective transformation of these mutations individually, we were able to map the responsible mutation in esa1-1853 to a single-amino-acid replacement of Trp66 with arginine (W66R), and in esa1-1851 to a single replacement of Cys304 with serine (C304S).
Figure 1.—
Panel of esa1 mutants. (A) Mutants shown on the primary structure of Esa1. W66R lies in the chromodomain (CHD) while the others fall within the MYST family HAT domain. (B) Relevant residues (yellow) on the crystal structure of Esa1(160–435) in complex with coenzyme A (red).
Mutations in the catalytic residues of Esa1 are viable:
The recovery of esa1-C304S was unexpected since, at the time, Cys304 was implicated directly in the transfer of the acetyl group to histone H4 and recombinant Esa1-C304S was catalytically dead in vitro (Yan et al. 2002). By both one-step transformation and genetic crosses, we were able to replace ESA1 with esa1-C304S in both W303 and S288C background strains without lethality. We considered two interpretations of these results: either Esa1-C304S retains catalytic acetyltransferase activity using the Glu338 residue or the HAT activity of Esa1 is not essential for growth. To distinguish between these possibilities, we constructed the esa1-E338Q allele by site-directed mutagenesis and characterized it in vivo.
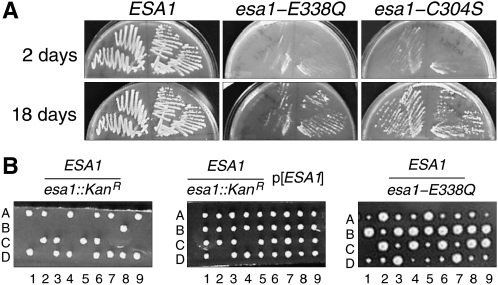
First we tested esa1-E338Q by plasmid shuffle. The BY4743-derived hemizygous esa1∷Kan/ESA1 diploid strain, from the systematic gene-deletion collection (Winzeler et al. 1999), was sporulated and tetrads were dissected. As expected, the tetrads gave uniformly two live and two dead spores in which all the viable segregants were ESA1. The inviability of the esa1∷Kan haploids was rescued by a CEN ARS URA3 plasmid containing the wild-type ESA1 gene. The rescued spore colonies were G418 resistant, because of the esa1∷Kan allele, but were unable to grow on media containing 5-FOA, because loss of the URA3 ESA1 plasmid was lethal. We then transformed one of the plasmid-rescued haploid strains with CEN ARS LEU2 plasmids containing ESA1, esa1-C304S, or esa1-E338Q. The primary transformants grew well without noticeable dominant growth phenotypes. When plated on media containing 5-FOA to select against the URA3 ESA1 plasmid, both esa1-C304S and esa1-E338Q plasmids supported growth after 14–18 days (Figure 2A). The growth of the mutant shuffle strains was slower than those with the wild-type ESA1 gene. The plasmids from both mutant shuffle strains were recovered and the presence of the mutant alleles was confirmed by DNA sequence analysis.
Figure 2.—
Viability of esa1 catalytic-site mutants. (A) LEU2-marked plasmids bearing ESA1, esa1-E338Q, or esa1-C304S were transformed into a BY4743-derived esa1∷kan strain carrying an ESA1 URA3 plasmid. Resulting strains were then streaked onto 5-FOA to counterselect the ESA1 URA3 plasmid. (B) An esa1-E338Q allele was knocked into the genomic ESA1 locus in a W303 diploid strain, which was then sporulated and dissected. The esa1-E338Q allele supports viability (right, small colonies), while an esa1∷kan allele does not (left) unless covered with an ESA1 plasmid (middle).
It was formally possible that the viability of the plasmid shuffle strains was due to suppressor mutations in the background. Therefore, we integrated the esa1-C304S and esa1-E338Q alleles into the chromosome of a W303 diploid by one-step gene replacement linked with the K.l.URA3 selectable marker. In both cases, dissections of the sporulated diploids produced four viable spore colonies with two large colonies and two small colonies (Figure 2B). All of the small colonies were Ura+ indicating the presence of the mutant allele. We confirmed the presence of the esa1-C304S and esa1-E338Q alleles in the Ura+ haploid segregants by DNA sequencing. To ensure that the esa1-E338Q mutant did not carry unknown suppressors, it was backcrossed to wild-type W303-1A twice. Tetrads continued to produce two large and two small spore colonies with esa1-E338Q linked to the small colony size. We observed 92% spore viability in dissections of 45 tetrads from the second backcross and the low level of inviability was unlinked to esa1-E338Q. We confirmed the presence of the E338Q mutation in the final mutant segregants by DNA sequence analysis. These results show that the catalytic residues specifically required for the acetyltransferase activity of Esa1 are not essential for cell viability.
Phenotypes of esa1 mutants:
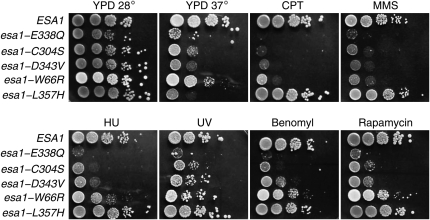
Phenotypic characterization of the esa1 mutant panel is shown in Figure 3. We also included esa1-L357H in these studies, a previously described temperature-sensitive lethal allele that is otherwise wild type at permissive temperatures (Bird et al. 2002) The mutants show a graded response to DNA damage-inducing agents CPT and MMS, with the catalytically dead mutants esa1-E338Q and esa1-C304S being the most sensitive and esa1-L357H showing wild-type resistance. Interestingly, the esa1 mutants were also sensitive to UV irradiation, suggesting a previously unidentified role for NuA4 in DNA mismatch repair. The same pattern of sensitivity is observed on plates containing benomyl, hydroxyurea, and rapamycin, indicative of defects in chromosome segregation, replication, and TOR signaling, respectively. The exception to the pattern of esa1 mutant sensitivity is seen on YPD plates incubated at 37°: esa1-L357H is markedly sensitive while the other mutants show various intermediate degrees of temperature sensitivity. All the phenotypes shown are recessive and complemented by a plasmid-containing wild-type ESA1 (data not shown).
Figure 3.—
Phenotypes of the esa1 panel. Serial dilutions of the esa1 panel were spotted on YPD-based plates containing no drug or 30 μg/ml camptothecin (CPT); 0.025% methylmethane sulfonate (MMS); 12.5 μg/ml benomyl; 0.1 m hydroxyurea; or 25 nm rapamycin. For the UV treatment, the YPD plate was irradiated with 0.015 J/cm2 of 254 nm UV light in a Stratalinker.
We next examined the levels of histone H4 acetylation in the esa1 mutant strains. Whole-cell extracts were separated by SDS–PAGE and analyzed by Western blot using antibodies against acetylated H4 to measure HAT activity and against bulk unmodified H4 as control. As shown in Figure 4A, the mutants exhibit a range of H4 acetylation, with the two catalytic-site mutants esa1-E338Q and esa1-C304S showing the least H4 acetylation. Normalized for the bulk unmodified histone H4 in the blots, acetylated histone H4 is reduced to <0.5% and 2.6% of wild-type levels in esa1-E338Q and esa1-C304S, respectively. This increases to ∼59% for esa1-L357H. Across the panel, the level of histone H4 acetylation is inversely proportional to the sensitivity of the mutants to each of the drugs tested above (compare Figures 3 and 4).
Figure 4.—
Characterization of esa1 mutants by Western blot. (A) The indicated strains were grown to mid-log phase and whole-cell extracts were prepared, separated by SDS–PAGE, and probed by Western blot for acetylated H4 (H4ac) and bulk histone H4 (H4). (B) The indicated strains were grown to mid-log phase, diluted, and then grown for 4 hr at either 28° or 37°. Whole-cell extracts were then prepared, separated by SDS–PAGE, and then probed by Western blot for Esa1 protein, histone H4 and H3 acetylation, and glucose-6-phosphate dehydrogenase (GPDH) as a loading control. The hhf1-ΔN allele encodes histone H4 missing N-terminal residues 1–27.
Next we characterized the levels of Esa1 protein present in the mutants and the state of histone H3 and H4 acetylation at both 28° and 37° (Figure 4B). Whole-cell extracts were again separated by SDS–PAGE and analyzed by Western blot using antibodies against Esa1, acetylated H3, acetylated H4, and glucose-6-phosphate dehydrogenase (GPDH) as a loading control. At both 28° and 37°, the levels of Esa1 protein present in esa1-E338Q, esa1-C304S, and esa1-D343V cells are similar to that of wild type. The level of Esa1 in the Ts− mutant esa1-L357H is reduced at 28° relative to wild type and is further reduced after incubation for 4 hr at 37°. In addition, the Esa1 Western blot signal from the esa1-W66R mutant is relatively weak at both temperatures. At 28°, the gradient of acetylated H4 is similar to that observed previously while at 37° H4 acetylation is lost in the temperature-sensitive esa1-L357H as expected. Acetylation of histone H3 was prominent in all of the esa1 mutants, demonstrating that they did not markedly affect other histone acetylation activities. The amount of acetylated H4 in esa1-L357H at 37°, where cells are inviable, is similar to that in esa1-E338Q and esa1-C304S at 28°, where cells are viable, suggesting that this residual acetylation cannot account for the essential function of Esa1 and is likely catalyzed by another H4 HAT enzyme.
HAT activity of Esa1 immunocomplexes:
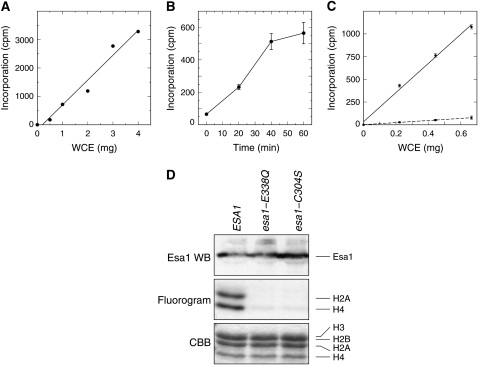
To help place an upper bound on the activity of mutant Esa1 enzymes, we immunoprecipitated wild-type or catalytic-site mutant Esa1-containing complexes from whole-cell extracts and measured their HAT activity using purified histones as substrate. The HAT activity of the Esa1 complexes increased linearly with the amount of whole-cell extract immunoprecipitated, up to 4 mg of protein (Figure 5A), and the incorporation of 3H-acetate was linear for ∼40 min when using 0.67 mg of protein (Figure 5B). However, as shown in Figure 5C, the activity of anti-Esa1 immunoprecipitates from esa1-E338Q is <8% of that from wild-type ESA1 cells.
Figure 5.—
In vitro HAT activity of esa1 mutants in the context of their native complexes. (A) Increasing amounts of whole-cell extracts from ESA1 cells were immunoprecipitated with a constant 5 μl of anti-Esa1 antibody (Abcam). The immunoprecipitates were subjected to HAT assays by incubating them with [3H]-acetyl CoA and purified chicken core histone substrate for 60 min. The reactions were collected on filters, extensively washed, and measured by scintillation counting. Subsequent experiments used 5 μl of anti-Esa1 antibody per 2.0 mg of whole-cell extract protein. (B) Whole-cell extracts were prepared from ESA1 cells and assayed for HAT activity for increasing lengths of time. Each point represents the average and standard deviation of triplicate assays of 0.67 mg of starting whole-cell extract protein. Subsequent experiments were carried out for 40 min. (C) Whole-cell extracts were prepared from ESA1 (circles, solid line) and esa1-E338Q cells (squares, dashed line) and increasing amounts of protein were immunoprecipitated with anti-Esa1 antibody. Each point represents the average and standard deviation of triplicate assays carried out for 40 min. (D) Esa1-containing complexes were immunoprecipitated using an Esa1 antibody, and immune complexes were incubated with [3H]-acetyl CoA and chicken core histones. After reaction, SDS–PAGE loading buffer was added to 1× concentration and samples were boiled prior to SDS–PAGE. The bottom portion of the gel was then stained with Coomassie brilliant blue (CBB; bottom) and subjected to fluorography (middle) to determine incorporation of [3H]-acetyl groups into histones, while the top portion of the gel was transferred to nitrocellulose and probed for Esa1 protein with an Esa1 antibody (top).
We also assayed the IP HAT assays by fluorography and Western blot analysis. As seen in Figure 5D, we recovered similar levels of Esa1 protein in the immunocomplexes from the wild-type and mutant strains, with slightly increased amounts for esa1-C304S. The immunoprecipitated protein complexes from wild-type ESA1 were able to efficiently acetylate histones H2A and H4 as expected. However, acetylation of H2A was below the limits of detection for complexes from both the esa1-E338Q and esa1-C304S mutants, while acetylation of H4 was <6% of wild-type levels. Together, the results of these HAT assays show that the catalytic activity of Esa1 complexes immunoprecipitated from mutant cells is severely impaired by the E338Q and C304S mutations. At present, it is difficult to determine whether the low residual activity seen in the esa1-E338Q and esa1-C304S complexes is due to activity of the mutant Esa1 enzyme itself or to other weakly crossreacting activities brought down by the anti-Esa1 antibody.
A catalytic-site double-point mutant is inviable:
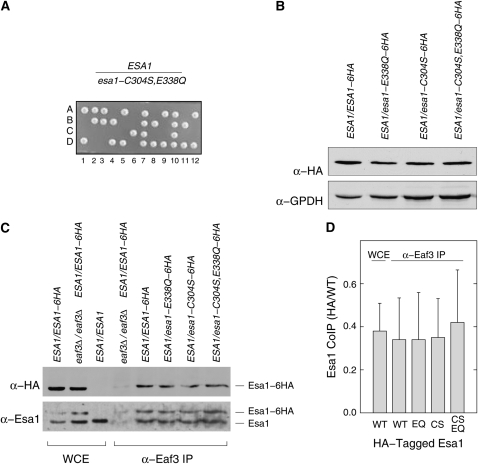
These results were consistent with a requirement for both C304 and E338 in histone acetylation in cells. Since single mutations in these residues each block histone acetylation, it would be expected that they would be functionally equivalent. However, this was not the case and the phenotype of E338Q was consistently more severe than that of C304S (see Figure 3). We reasoned that understanding the epistasis of these two mutations might shed light on the catalytic pathway. Therefore, we constructed the esa1-C304S,E338Q allele mutated at both residues, expecting the phenotype to match that of either C304S or E338Q. We integrated esa1-C304S,E338Q∷K.l.URA3 at the chromosomal locus in a W303 diploid as before and confirmed proper allele construction by DNA sequencing. Surprisingly, the phenotype of the double-point mutant failed to match that of either single mutant. When we sporulated this strain and dissected tetrads, we observed a 2:2 pattern of viable Ura− to inviable (inferred to be Ura+) spores (Figure 6A). To confirm that the mutant protein was expressed, we created diploid strains in which one chromosome carried the wild-type ESA1 gene and the other carried wild-type ESA1, esa1-E338Q, esa1-C304S, or esa1-C304S,E338Q C-terminally tagged with six copies of the HA epitope tag. Whole-cell extracts from these strains were separated by SDS–PAGE and probed with anti-HA antibody to measure the expression of the tagged protein and anti-GPDH antibody to serve as a loading control. We found that Esa1-HA is expressed at roughly equal levels in all four strains, precluding the possibility that the mutant Esa1-C304S,E338Q protein was intrinsically unstable (Figure 6B).
Figure 6.—
Characterization of the esa1-C304S,E338Q allele. (A) The esa1-C304S,E338Q double-point mutant is inviable. A heterozygous ESA1/esa1-C304S,E338Q diploid strain was sporulated and the resulting tetrads were dissected. The growth of the four spore colonies (A–D) from 12 tetrads are shown. Tetrads gave predominantly a 2:2 pattern of viable:inviable spores, with all viable spores being Ura−. In this example, two tetrads (7 and 10) produced four viable spore colonies, but in both cases all four spores were Ura− and homozygous for the wild-type ESA1 allele presumably as a result of recombination. (B) Esa1-C304S,E338Q protein is expressed normally. Whole-cell extracts were prepared from four heterozygous diploid strains in which one ESA1 allele was wild type and encoded untagged protein, while the other allele was either wild type or mutant and expressed protein tagged with a C-terminal 6HA epitope. The extracts were then probed by Western blot with an anti-HA antibody to determine the level of expression of the different proteins. The anti-GPDH probing served to control for protein loading. (C) Esa1 proteins carrying catalytic-site mutations co-immunoprecipitate with the Eaf3 subunit of NuA4. Extracts were prepared from the heterozygous diploid strains indicated and immunoprecipitated with an anti-Eaf3 antibody. Aliquots of the whole-cell extracts (WCE) or immunoprecipitates (α-Eaf3 IP) were separated by PAGE. The gel was blotted and probed with anti-HA antibody to score the tagged Esa1 protein (α-HA) and with anti-Esa1 antibody (α-Esa1) to score both the tagged and the untagged proteins. (D) Relative recovery of tagged and untagged Esa1 proteins in anti-Eaf3 immunoprecipitates. The relative co-IP recoveries were determined for the data in C and two other independent experiments. Western blots probed with anti-Esa1 antibody were digitized and the intensities of the Esa1-6HA and Esa1 proteins present in the whole-cell extracts and recovered in the Eaf3 IPs were quantified for each mutant. The average ratio and standard deviation of HA-tagged to untagged protein is shown.
Catalytic-site mutants associate with NuA4:
Since Esa1 functions in the context of NuA4 and picNuA4 complexes, we reasoned that the phenotypes of the catalytic-site mutants, particularly esa1-C304S,E338Q, might be due to a failure of the mutant enzymes to associate with NuA4. To test this possibility, we prepared whole-cell extracts from heterozygous diploid strains expressing untagged wild-type Esa1 together with each of the different HA-tagged Esa1 wild-type and catalytic-site mutant proteins. These extracts were then immunoprecipitated with an anti-Eaf3 antibody to pull down the endogenous NuA4 complexes. Although Eaf3 is specific for the NuA4 complex, Esa1 is recruited to NuA4 through its association with the picNuA4 subcomplex (Boudreault et al. 2003), and thus this assay monitors association of Esa1 with both complexes. The immunoprecipitates were fractionated by SDS–PAGE and assayed by Western blot for the co-immunoprecipitation of Esa1 proteins. We found that wild-type Esa1 and all three catalytic-site mutant enzymes associated with Eaf3 immunoprecipitates (Figure 6C). Quantification of the relative amounts of tagged and untagged Esa1 shows that all four tagged proteins associate with NuA4 equivalently and with the same ratio as is present in the whole-cell extracts (Figure 6D). These results are consistent with previous biochemical experiments in which Esa1-C304S and Esa1-E338Q proteins were successfully reconstituted into stable picNuA4 complexes in vitro (Berndsen et al. 2007a). Thus, mutations in the catalytic residues of Esa1 do not block its association with NuA4. Nevertheless, Esa1-C304S and Esa1-E338Q can carry out functions essential for cell viability, whereas the double-point mutant cannot. These results show that the essential function of Esa1 relies on residues within the MYST domain catalytic pocket but not on catalysis itself.
HAT activity of recombinant Esa1:
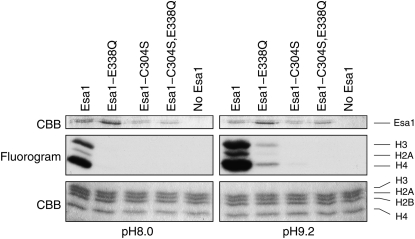
Given the uncertainty regarding the role of C304S in catalysis, we revisited the in vitro HAT activity of wild-type and mutant Esa1 proteins. We purified full-length hexahistidine-tagged Esa1, Esa1-E338Q, Esa1-C304S, or Esa1-C304S,E338Q and carried out in vitro HAT assays (see materials and methods). As expected, wild-type Esa1 had robust activity toward histones H4, H2A, and H3 (Figure 7, left), consistent with the reported in vitro preference of recombinant Esa1 toward core histones (Clarke et al. 1999). However, the Esa1-E338Q, Esa1-C304S, and Esa1-C304S,E338Q proteins lacked any detectable HAT activity (Figure 7, left) consistent with the results of Yan et al. (2002) with a truncated recombinant Esa1 fragment. Berndsen et al. (2007a) were able to bypass the requirement for E338 in catalysis by carrying out reactions at pH 9.2 where substrate lysines are already deprotonated. Therefore, we repeated our HAT assays at pH 9.2 (Figure 7, right). Although much weaker than the wild-type enzyme, recombinant Esa1-E338Q displayed detectable HAT activity at elevated pH consistent with Berndsen et al. (2007a). Esa1-C304S gave barely detectable activity at pH 9.2 (<2% of wild-type Esa1) and we were unable to detect any activity at all in the double-mutant protein Esa1-C304S,E338Q.
Figure 7.—
In vitro HAT activity of recombinant Esa1. The wild-type and catalytic-site mutants were N-terminally tagged with a His6 epitope tag and expressed in E. coli. After purification on Ni-NTA resin, relatively equal amounts (top, Esa1 CBB; note that the Esa1-E338Q samples have slightly more protein present) of Esa1 (or no Esa1 as a control) were combined with chicken core histones and [3H]-acetyl CoA at either pH 8.0 or pH 9.2 and incubated at room temperature for 30 min. To assay incorporation of [3H]-acetyl groups into the histones, we ran the reaction on SDS–PAGE, dried the gel, and subjected it to fluorography (middle). A Coomassie stain of the same region of the gel is shown to confirm equal loading of histones (bottom).
DISCUSSION
The amino acid substitutions recovered in our mutant collection provide insights into Esa1 function. Cells expressing esa1-L357H mutant are temperature-sensitive lethal. They retain H4 acetylation activity and drug resistance at permissive temperature, but fail to grow at restrictive temperature. Consistent with this, the amount of Esa1-L357H protein present in cells is severely reduced at restrictive temperature. These phenotypic properties fit with the structure of the MYST domain. In the Esa1 crystal structure Leu 357 is buried in the center of a set of stacked α-helices (Figure 1). The replacement of this residue with His 357 is likely to destabilize this structure at restrictive temperature leading to its degradation. The mutations in the damage-sensitive alleles all occur in one of two protein domains: the chromodomain or the MYST domain near the catalytic site (Figure 1). The chromodomain mutant, esa1-W66R, has impaired levels of H4 acetylation in vivo, supporting the site-directed mutational results of Selleck et al. (2005), which showed that chromodomain mutations Y56A, E65R, and E65L inhibit the ability of recombinant Piccolo complex to acetylate nucleosomal substrates in vitro. The esa1-D343V mutant is particularly interesting. The Esa1-D343V protein is stable but cells expressing this allele have low levels of H4 acetylation and are hypersensitive to CPT, HU, and benomyl. In the Esa1 crystal structure Asp 343 is surface exposed and directed away from the catalytic pocket. Thus, it is likely that this residue is important for binding nucleosomal substrates and/or other NuA4 components.
The most surprising result from our studies is the finding that the essential function of Esa1 may not be its histone acetyltransferase activity. Several lines of evidence support this interpretation: (1) cells expressing esa1-E338Q, an allele mutated for an essential catalytic residue, are viable in both S288C and W303 strain backgrounds; (2) full-length recombinant Esa1-E338Q is catalytically inactive in vitro (Figure 7; Berndsen et al. 2007a); (3) Esa1-E338Q protein complexes immunoprecipitated from whole-cell extracts are severely impaired in HAT activity; and (4) cells expressing esa1-E338Q have very low levels of global histone H4 acetylation in vivo. Our in vivo results for esa1-E338Q appear to differ from those of Yan et al. (2000). In that work a plasmid shuffle assay was used to evaluate the phenotype of esa1-E338Q. However, we found that colonies of esa1-E338Q and esa1-C304S appear only after prolonged incubation in plasmid shuffle assays and, thus, it is possible that these were missed in the previous experiments. Alternatively, the background genotype of the strains used by Yan et al. (2000) may have carried alleles of one or more unknown genes that confer synthetic lethality in combination with catalytically dead esa1.
Our results also suggest that Cys304 is important for histone acetyltransferase activity, an interpretation more consistent with a ping-pong model of catalysis than direct transfer (Yan et al. 2002; Berndsen et al. 2007a). We find that full-length recombinant Esa1-C304S enzyme is inactive in vitro, that immunoprecipitates of Esa1-C304S complexes from cells are severely impaired in HAT assays, and that cells expressing esa1-C304S as their sole source of Esa1 protein have very low levels of histone H4 acetylation in vivo. These results differ from those recently reported by Berndsen et al. (2007a). It is possible that Esa1-C304S is particularly labile and, in our hands, may be specifically inactivated during the purification of the recombinant protein. If so, this instability might also contribute to a loss of activity during immunoprecipitation of Esa1-C304S complexes from cells. However, it is difficult to reconcile wild-type HAT activity of Esa1-C304S in vivo with the very low level of global histone H4 acetylation observed in esa1-C304S mutants. Further experimentation is required to understand and resolve these apparent discrepancies.
Important clues to the essential function of Esa1 follow from our finding that the esa1-C304S,E338Q allele is lethal. This result demonstrates that residues C304 and E338 in the catalytic pocket provide partially overlapping contributions to the essential function of Esa1. There are at least three models consistent with these observations. First, it is formally possible that the essential function of Esa1 is actually protein acetylation, provided that both Esa1-C304S and Esa1-E338Q retained residual HAT activity above an essential threshold, which is then lost in the Esa1-C304S,E338Q double mutant. However, it is unlikely that Esa1-E338Q retains HAT activity since the Gln 338 residue is chemically incapable of deprotonating the ɛ-amino group of substrate lysines; this attack would need to be provided by an unknown extrinsic acidic moiety operating within the relatively tight confines of the catalytic pocket (Yan et al. 2000, 2002).
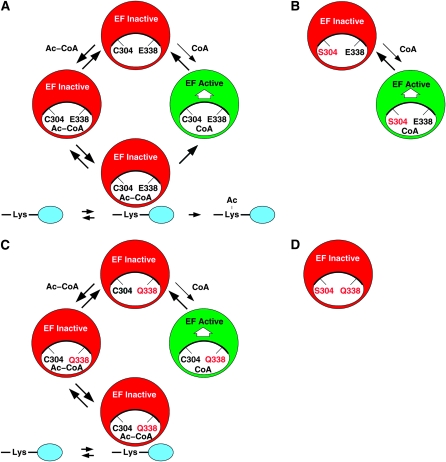
Alternatively, the essential function of Esa1 may not be protein acetylation, per se, but rather Ac-CoA cofactor or lysine substrate binding for the purpose of positively regulating essential chromatin functions of NuA4 and picNuA4. We speculate that Esa1-C304S and Esa1-E338Q may each retain partial cofactor binding and NuA4 regulatory activity, Esa1-C304S more so than Esa1-E338Q, but that the Esa1-C304S,E338Q double mutant is synthetically defective for binding and regulation, resulting in a severe loss of NuA4 function to the point of inviability. Indeed, the cocrystallization results of Yan et al. (2002) suggest differences in cofactor binding by the mutants. Whereas cocrystals of wild-type Esa1 and Esa1-E338Q were obtained with both CoA and Ac-CoA, Esa1-C304S could be crystallized only with CoA and not Ac-CoA. Consistent with this, in vitro HAT reactions at pH 9.2 bypassed the catalytic defect best with Esa1-E338Q, but very poorly with Esa1-C304S, while the activity of Esa1-C304S,E338Q remained undetectable (Figure 7). A model to incorporate these results is shown in Figure 8 and is formally analogous to the molecular switch cycle of Ras family GTPases (Wittinghofer and Pai 1991). We propose that NuA4 complexes cycle between active and inactive states, with respect to their essential transcription functions, in which CoA-bound Esa1 signals the active and Ac-CoA-bound Esa1 signals the inactive state (Figure 8A). The results of Yan et al. (2002) and Figure 7 imply that Esa1-C304S may be impaired for the binding of Ac-CoA, resulting in a loss of acetylation activity but retention of some ability to bind CoA and activate NuA4 complexes (Figure 8B). Weak binding of Ac-CoA (not illustrated) would permit limited acetylation activity at pH 9.2 in vitro. In contrast, Esa1-E338Q retains the ability to bind both Ac-CoA and CoA but it is unable to carry out protein acetylation without Glu 338. This defect can be effectively bypassed at pH 9.2 in vitro. However, Esa1-E338Q is much poorer than Esa1-C304S at activating NuA4 because Ac-CoA effectively acts as a competitive inhibitor (Figure 8C). Finally, this model predicts that Esa1-C304S,E338Q will be defective in binding CoA and activating NuA4 and thus resulting in cell lethality (Figure 8D). The regulatory role of Esa1 might be coupled to NuA4 and picNuA4 through recruitment to appropriate target sites or by direct modulation of activity. At present, we favor the direct regulatory model since current biochemical evidence suggests that other NuA4 subunits and additional histone domains have major roles in recruitment and nucleosome recognition (Galarneau et al. 2000; Brown et al. 2001; Downs et al. 2004; Berndsen et al. 2007b).
Figure 8.—
An Esa1 cofactor regulatory cycle. (A) The acetylation cycle for NuA4 complexes containing wild-type Esa1. NuA4 complexes inactive for essential functions are shown in red (“EF inactive”) and active complexes are shown in green (“EF active”). Here EF active and EF inactive refer to the essential functions of NuA4 complexes, such as transcriptional coactivation (Reid et al. 2000; Nourani et al. 2004; Durant and Pugh 2007) and not the catalytic activity of Esa1 itself. We propose that Esa1 positively activates NuA4 in the CoA-bound state but not the Ac-CoA-bound state. This cycle is facilitated by acetylation of lysine substrates in the histone tails, depicted in blue. Thus, the acetylation cycle of the wild-type enzyme produces both acetylated histones, or other protein substrates, and CoA-bound Esa1 to activate the essential functions of NuA4. (B) NuA4 complexes containing Esa1-C304S have an abbreviated regulatory cycle. The amino acid replacement S304, the Esa1 catalytic pocket, is indicated in red. We propose that Esa1-C304S is defective for binding Ac-CoA but retains binding of CoA on the basis of the results of acetylation reactions at pH 9.2 in vitro (Figure 7) and the crystallographic results of Yan et al. (2002). Thus, NuA4 complexes containing Esa1-C304S can be activated by CoA binding, conferring cell viability, but this is impaired relative to wild-type Esa1 in the absence of the full acetylation cycle. The binding of histone-tail lysines is unknown in this case. (C) NuA4 complexes containing Esa1-E338Q have a blocked regulatory cycle. The amino acid replacement Q338 is indicated in red. We propose that Esa1-E338Q retains the ability to bind both Ac-CoA and CoA but is unable to acetylate lysine substrates because the target ɛ-amino groups cannot be deprotonated in the absence of E338 (Yan et al. 2000, 2002; Berndsen et al. 2007a). Like Esa1-C304S, Esa1-E338Q is able to partially activate NuA4 complexes by binding CoA. However, this activity is impaired relative to Esa1-C304S since Ac-CoA now becomes a competitive inhibitor of CoA binding. Lysine substrates may bind, but they cannot be acetylated. (D) NuA4 complexes containing Esa1-C304S,E338Q. We propose that Esa1-C304S,E338Q is largely unable to bind either CoA or Ac-CoA, and therefore it fails to activate the essential functions of NuA4 complexes, leading to cell death. The binding status of histone-tail lysines is unknown.
Finally, it is worth noting that the essential function of Esa1 could well involve more than one pathway. For example, both protein acetylation and NuA4 regulation might have partially redundant roles in the control of gene expression. In that case, if E338 has the predominant role in protein acetylation (Berndsen et al. 2007a), and C304 has the predominant role in NuA4 regulation, then the Esa1-C304S,E338Q double mutant would lack both activities and be inviable.
Our results, and those of others, demonstrate that the catalytic HAT activity of Esa1 is important for the normal physiology of budding yeast. By selecting esa1 mutants with a range of sensitivity to genotoxic and cytotoxic stresses, we have defined a strong correlation between the severity of the mutant phenotype and the relative loss of histone H4 acetylation in vivo. However, our genetic, molecular, and biochemical characterization of Esa1 catalytic-site mutants challenges the conventional view that the essential roles of NuA4 and picNuA4 are to regulate the catalytic activity of Esa1 for global and targeted histone H2A, H2A.Z, and H4 acetylation. Instead, we propose that Esa1 is also an essential regulator of NuA4 and picNuA4 activities. Both of these complexes contain other protein subunits essential for viability and, although the activities of these are currently unknown, future experimentation is likely to reveal one or more additional roles vital for cell survival. The evolutionary conservation of Esa1 and NuA4 complexes with human MYST proteins and complexes, such as Tip60, suggests that these mechanisms may have important implications for human chromatin function.
Acknowledgments
We thank David Auble, Patrick Grant, and David Brautigan for discussion, and Bob Kadner for shared equipment. This work was supported by grant GM60444 from the National Institute of General Medical Sciences, National Institutes of Health, to M.S.
References
- Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant et al., 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18 5108–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg, D., D. Burke and J. Strathern, 2005. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Babiarz, J., J. Halley and J. Rine, 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen, C. E., B. Albaugh, S. Tan and J. Denu, 2007. a Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry 46 623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen, C. E., W. Selleck, S. J. McBryant, J. C. Hansen, S. Tan et al., 2007. b Nucleosome recognition by the Piccolo NuA4 histone acetyltransferase complex. Biochemistry 46 2091–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon et al., 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419 411–415. [DOI] [PubMed] [Google Scholar]
- Boudreault, A. A., D. Cronier, W. Selleck, N. Lacoste, R. T. Utley et al., 2003. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 17 1415–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza et al., 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292 2333–2337. [DOI] [PubMed] [Google Scholar]
- Clarke, A., E. Samal and L. Pillus, 2006. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol. Biol. Cell 17 1744–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, A. S., J. E. Lowell, S. J. Jacobson and L. Pillus, 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs, J., S. Allard, O. Jobin-Robitaille, A. Javaheri, A. Auger et al., 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16 979–990. [DOI] [PubMed] [Google Scholar]
- Doyon, Y., and J. Côté, 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 14 147–154. [DOI] [PubMed] [Google Scholar]
- Doyon, Y., W. Selleck, W. S. Lane, S. Tan and J. Côté, 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24 1884–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant, M., and B. Pugh, 2006. Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae. Mol. Cell. Biol. 26 2791–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant, M., and B. Pugh, 2007. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 27 5327–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early, A., L. S. Drury and J. F. Diffley, 2004. Mechanisms involved in regulating DNA replication origins during the cell cycle and in response to DNA damage. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau, L., A. Nourani, A. A. Boudreault, Y. Zhang, L. Heliot et al., 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5 927–937. [DOI] [PubMed] [Google Scholar]
- Iizuka, M., and B. Stillman, 1999. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 274 23027–23034. [DOI] [PubMed] [Google Scholar]
- Keogh, M., T. Mennella, C. Sawa, S. Berthelet, N. Krogan et al., 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor, M., S. Venkatasubrahmanyam, M. Meneghini, J. Gin, J. Jennings et al., 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2 E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N., K. Baetz, M. Keogh, N. Datta, C. Sawa et al., 2004. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA 101 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch, T., L. Florens, W. Macdonald, S. Swanson, R. Glaser et al., 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306 2084–2087. [DOI] [PubMed] [Google Scholar]
- Lafon, A., C. S. Chang, E. M. Scott, S. J. Jacobson and L. Pillus, 2007. MYST opportunities for growth control: yeast genes illuminate human cancer gene functions. Oncogene 26 5373–5384. [DOI] [PubMed] [Google Scholar]
- Le Masson, I., D. Y. Yu, K. Jensen, A. Chevalier, R. Courbeyrette et al., 2003. Yaf9, a novel NuA4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Mol. Cell. Biol. 23 6086–6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M., A. McKenzie, 3rd, D. Demarini, N. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953–961. [DOI] [PubMed] [Google Scholar]
- Megee, P. C., B. A. Morgan and M. M. Smith, 1995. Histone H4 and the maintenance of genome integrity. Genes Dev. 9 1716–1727. [DOI] [PubMed] [Google Scholar]
- Millar, C., F. Xu, K. Zhang and M. Grunstein, 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourani, A., R. T. Utley, S. Allard and J. Côté, 2004. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 23 2597–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiwara, H., A. Ui, S. Kawashima, K. Kugou, F. Onoda et al., 2007. Actin-related protein Arp4 functions in kinetochore assembly. Nucleic Acids Res. 35 3109–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, J. L., V. R. Iyer, P. O. Brown and K. Struhl, 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6 1297–1307. [DOI] [PubMed] [Google Scholar]
- Rohde, J. R., and M. E. Cardenas, 2003. The TOR pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 23 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck, W., I. Fortin, D. Sermwittayawong, J. Côté and S. Tan, 2005. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol. Cell. Biol. 25 5535–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, E. R., A. Eisen, W. Gu, M. Sattah, A. Pannuti et al., 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA 95 3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, K., M. Langer and J. Denu, 2000. Kinetic mechanism of human histone acetyltransferase P/CAF. Biochemistry 39 11961–11969. [DOI] [PubMed] [Google Scholar]
- Tanner, K., R. Trievel, M. Kuo, R. Howard, S. Berger et al., 1999. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem. 274 18157–18160. [DOI] [PubMed] [Google Scholar]
- Wallis, J. W., G. Chrebet, G. Brodsky, M. Rolfe and R. Rothstein, 1989. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58 409–419. [DOI] [PubMed] [Google Scholar]
- Winzeler, E., D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901–906. [DOI] [PubMed] [Google Scholar]
- Wittinghofer, A., and E. F. Pai, 1991. The structure of Ras protein: a model for a universal molecular switch. Trends Biochem. Sci. 16 382–387. [DOI] [PubMed] [Google Scholar]
- Yan, Y., N. A. Barlev, R. H. Haley, S. L. Berger and R. Marmorstein, 2000. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol. Cell 6 1195–1205. [DOI] [PubMed] [Google Scholar]
- Yan, Y., S. Harper, D. W. Speicher and R. Marmorstein, 2002. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat. Struct. Biol. 9 862–869. [DOI] [PubMed] [Google Scholar]