Abstract
Follistatin (FS) is one of several secreted proteins that modulate the activity of TGF-β family members during development. The structural and functional analysis of Drosophila Follistatin (dFS) reveals important differences between dFS and its vertebrate orthologues: it is larger, more positively charged, and proteolytically processed. dFS primarily inhibits signaling of Drosophila Activin (dACT) but can also inhibit other ligands like Decapentaplegic (DPP). In contrast, the presence of dFS enhances signaling of the Activin-like protein Dawdle (DAW), indicating that dFS exhibits a dual function in facilitating and inhibiting signaling of TGF-β ligands. In addition, FS proteins may also function in facilitating ligand diffusion. We find that mutants of daw are rescued in significant numbers by expression of vertebrate FS proteins. Since two PiggyBac insertions in dfs are not lethal, it appears that the function of dFS is non-essential or functionally redundant.
Keywords: Follistatin, TGF-beta, Drosophila Activin, Dawdle, DPP, Myoglianin, growth control, heparin sulfate
1. Introduction
Polypeptide cytokines of the transforming growth factor β (TGF-β) family control a wide range of developmental and physiological functions in higher eukaryotes (Massague et al., 2000; Yasuo and Lemaire, 2001). This diverse group of signaling molecules provides positional information required for axis formation and tissue specification, controls various processes such as tissue growth, cell death, and pathfinding of axons in the nervous system, and prevents differentiation of embryonic stem cells (Beattie et al., 2005; Colavita et al., 1998; Ho et al., 2000; James et al., 2005). Many components of this pathway have been linked to tumor formation in humans (Rooke and Crosier, 2001). The highest degree of sequence conservation between various family members is found within the C-terminal domains, which are released as dimers by proteolytic processing. Similarities in sequence and biological activities allow these factors to be divided into at least two distinct subgroups: Bone Morphogenetic Proteins (BMPs) and Activins/Inhibins/TGF-βs (Newfeld et al., 1999). The latter group exhibits an additional intramolecular disulfide bond at the N-terminus after processing. In Drosophila, there are four Activins/TGF-βs, Drosophila Activin (dACT), Dawdle (DAW, also known as Activin-like protein at 23, ALP23, and Anti-Activin, AACT), Myoglianin (MYO), and Maverick (MAV), and three BMP-type ligands, Decapentaplegic (DPP), Screw (SCW), and Glass Bottom Boat (GBB). Each ligand dimer forms a complex with two type II and two type I receptor serine/threonine kinases that phosphorylate SMAD transcription factors. BMP-type ligands signal primarily through the type I receptors Thick veins (TKV) and Saxophone (SAX) and activate Mothers against DPP (MAD) (McCabe et al., 2003; Shimmi et al., 2005). Activins/TGF-β-type ligands are believed to signal through the type I receptor Baboon (BABO), which in turn activates primarily dSMAD2 but to a minor extent also MAD (unpublished data).
TGF-β signaling is regulated by various extracellular proteins. Antagonists like Follistatin (FS), Noggin, Chordin/Short Gastrulation, and DAN/Cerberus bind ligands and prevent interactions with receptors and signaling. In some species, they exhibit overlapping and redundant functions. Recently, it was shown that the simultaneous depletion of FS, Noggin, and Chordin in Xenopus tropicalis results in transformation of ventral into dorsal tissue during embryogenesis (Khokha et al., 2005).
Follistatin was first identified as an inhibitor of Activin in vertebrates. Subsequent studies showed that it also binds other ligands with lower affinities including BMP 2, 4, 6, 7, and Myostatin (Abe et al., 2004; Canalis et al., 2003). Knockout mice of fs die shortly after birth. They are smaller and exhibit defects in skeletal and muscle development (Matzuk et al., 1995). Recently, the crystal structure of the human FS:Activin complex was resolved (Thompson et al., 2005). It provides valuable insight into the function of the different FS domains and a basis to explain the mechanism of ligand inhibition.
In this study, we analyze the function of Drosophila Follistatin (dFS). Like vertebrate FS proteins, dFS is subdivided into a N-terminal domain (N) and three FS domains (FS1-3) (Fig. 1). However, dFS is substantially larger than its vertebrate homologues due to a large basic insertion into FS1. Interestingly, dFS is proteolytically processed, and we find that small processed forms of dFS are able to bind to ligands like dACT. This result suggests a possible different inhibitory mechanism: ligands bound to processed dFS can bind to type II receptors but cannot recruit type I receptors. Consequently, processed dFS might not only sequester ligands but also prevent unbound ligands from interacting with receptor complexes. Among the seven Drosophila TGF-β ligands, we show that dFS primarily inhibits dACT but can also inhibit signaling of other ligands like DPP. In contrast, we find that dFS can augment signaling of the TGF-β member DAW. Our results suggest that dFS might exhibit dual functions in facilitating and inhibiting TGF-β signaling. Analysis of two PiggyBac insertions in dFS reveals that they affect dfs expression. Since homozygous animals of these lines are viable and phenotypically wild type, we have to assume that the function of dFS is non-essential or functionally redundant. Taken together, this study reveals interesting differences between the mechanisms of modulating TGF-β signaling by dFS and its vertebrate orthologues.
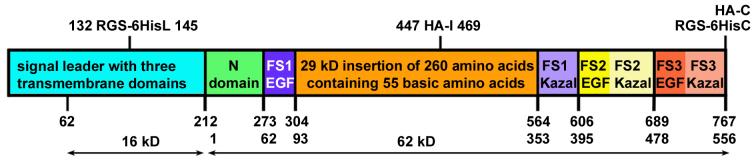
Fig. 1. Structure of Drosophila Follistatin.
Like vertebrate FS proteins, dFS is subdivided into a N-terminal domain (N) and three FS domains (FS1-3). In contrast to its vertebrate orthologues, dFS encodes an unusually long signal peptide trailer, which contains three potential transmembrane domains (84-104, 161-180, 194-211), and a 260 amino acid insertion in FS1, which adds to the overall positive charge of the protein. In this study, we use a truncated cDNA that encodes a shorter signal leader that starts after amino acid 62. The location of the HA and RGS-6His tags in the constructs used in this study are indicated on top. In the absence of protein modifications, the predicted sizes of the signal leader and the processed full-length secreted form are 16 kD and 62 kD respectively.
2. Results
2.1. Structure and function of dFS
The primary structure of dFS shows both similarities and differences compared to its vertebrate orthologues (Haerry and O'Connor, 2002). dFS is divided into a N-terminal domain (N) and three FS domains (FS1-3) that can be further subdivided into EGF-like and Kazal protease inhibitor-like domains (Fig. 1). Compared to its vertebrate orthologues, dFS is substantially larger due to an insertion of 260 amino acids. The insertion contains 55 positively charged amino acids (pI >10) and is located after the heparin binding site in FS1 (Inouye et al., 1992). In contrast to Drosophila, vertebrates produce a second, long isoform of FS by alternative splicing (FS-315). This form contains a C-terminal extension with many negatively charged amino acids that reduce adhesion to sulfates of proteoglycans at the cell surface. It was found that FS-315 is the major circulating form (Schneyer et al., 2004).
To analyze the function of dFS, we cloned our previously isolated dfs cDNA (AF454393) into expression vectors for tissue culture cells and transgenic flies (Haerry and O'Connor, 2002). From comparison to vertebrate orthologues, we assumed the start site to be at nucleotide 577. At this position, the SignalP server website predicts a signal sequence (MALLIGLLLLNFRLTAA-GTCW) that is cleaved equivalently to the vertebrate FS proteins. However, we find expression of a construct that contains this predicted signal sequence but lacks the first 279 nucleotides does not result in a translated protein in culture cells and growth inhibition in transgenic flies. In contrast, the complete cDNA is translated in tissue culture cells and inhibits growth in vivo. There are three potential start codons upstream of the predicted signal peptide sequence. Based on the consensus for Drosophila start sites (Cavener, 1987), the most likely start codon is at nucleotide 181-183 (CAACA-ATG). We note that there is a full-length dfs cDNA that is 671 nucleotides longer, GH04473 (NM_144119). This cDNA extends the coding sequence by 62 additional amino acids (Fig. 1). Although conserved between different Drosophila species, the absence of this additional sequence in our shorter cDNA does not prevent translation and secretion of a growth inhibitory protein. Interestingly, the full-length cDNA does not encode an N-terminal signal peptide sequence either. Instead, several protein analysis programs predict three potential transmembrane domains within the 211 amino acid leader (Fig.1). Thus, in contrast to vertebrate FS proteins, the secretion of dFS might involve an initial membrane-anchoring step.
To investigate where cleavage occurs, we expressed dFS in S2 cells. To monitor dFS fragments, we replace the sequence REQQQHQRQ in the second half of the signal leader with an RGS-6H tag (dFS-HisL). In addition, we introduced HA-epitopes (VYDYPDYASL) in the large insertion domain (dFS-HA-I) or at the C-terminus (dFS-HA-C). When expressing a double-labeled dFS-HisL-HA-I in S2 cells, we detect an 18 kD His-tagged fragment in the cell extract (Fig. 2C). This result is consistent with a single processing event at the equivalent location of vertebrate FS proteins, which is expected to produce a 16 kD fragment (Fig. 1). In the supernatant, we are able to detect a secreted protein via the internal HA-tag but not the N-terminal His tag (Fig. 2A, lane 9 and 13). Thus, the N-terminal leader is not secreted.
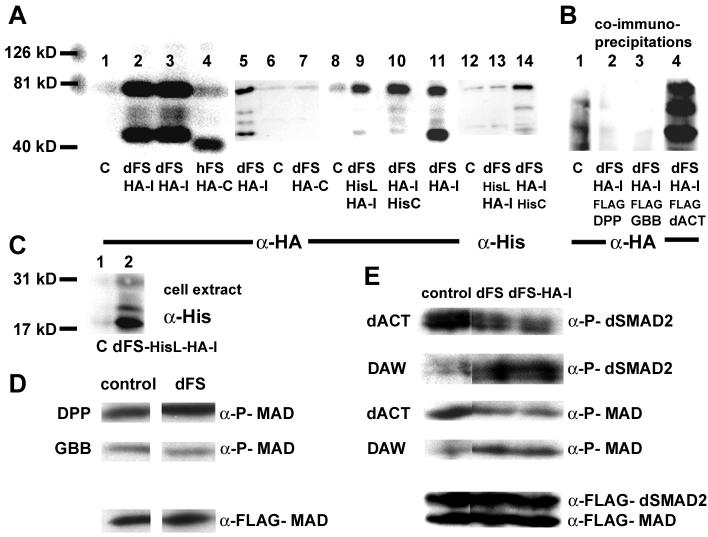
Fig. 2. Functional analysis of dFS in S2 cells.
The molecular markers (126, 81, and 40 kD) are used to determine the size of proteins in (A) and (B). (A) Lanes 1-4, 5-7, 8-11, and 12-14 are independent experiments. (A, lanes 1, 6, 8, and 12) Supernatant from mock-transfected S2 cells. Unlike hFS (A, lane 4), dFS is processed (lanes 2, 3, 5, 11). On western blots, internally tagged dFS-HA-I is detected in the supernatant as two major bands of approximately 75 kD and 45 kD. The upper band is larger than 62 kD, the expected size of the processed form without the signal peptide. Differences in size are likely due to glycosylation. Two minor additional processed forms migrate at about 55 kD. (Lane 4) The C-terminally tagged human FS migrates at approximately 40 kD, about 10 kD higher than expected. (Lane 7) In contrast to hFS-HA-C, a C-terminally tagged dFS cannot be detected on western blots. (Lane 14) Similarly, a C-terminally His/internally HA-double-tagged dFS shows a robust signal with anti-HA but only a weak signal with anti-His antibodies. These results indicate that the C-terminus is removed. (B) In co-immunoprecipitation experiments with FLAG tagged DPP, GBB, and dACT, we can detect all processed forms of dFS-HA-I in the presence of dACT. However, none of the forms co-immunoprecipitates in the presence of DPP or GBB. (C) To study N-terminal processing of dFS, amino acids 132-141 (70-78 in our shorter construct) in dFS-HA-I were replaced by a His-epitope. The His-tag is recognized in cell extracts as an 18 kD protein (lane 2). In the supernatant, this protein is detected by anti-HA but not anti-His antibodies (A, lanes 9 and 13). (D and E) Ligand signaling and phosphorylation of SMADs: Supernatants of dFS or internally tagged dFS-HA-I expressing cells were pooled, divided, and mixed with supernatant of mock or ligand transfected cells and added to cells that over-express FLAG-MAD and dSMAD2. (D) dFS levels that can inhibit dACT do not inhibit DPP and GBB signaling. (E, row 1 and 3) dFS and dFS-HA-I inhibits dACT-mediated activation of dSMAD2 and MAD. (Row 2 and 4) dFS increases rather than decreases phosphorylation mediated by DAW.
On western blots, the internally tagged dFS-HA-I obtained from supernatant migrates as two major bands of approximately 75 kD and 45 kD (Fig. 2A, lanes 2 and 3). The molecular size of both bands differs from the expected 62 kD of a processed form without the signal leader (Fig. 1). Thus, we speculate that the larger form is modified, most likely by glycosylation, while the smaller band represents a proteolytically cleaved protein. Two additional minor bands migrate at about 55 kD. In contrast to dFS, the C-terminally tagged human FS (hFS-HA-C) is not processed and migrates as a single band of approximately 40 kD, about 10 kD larger than expected (Fig. 2A, lane 4). Surprisingly, an identically C-terminally tagged dFS cannot be detected on western blots (Fig. 2A, lane 7). Similarly, the C-terminal RGS-6H tag of a double-tagged dFS (dFS-HA-I-HisC) is barely detectable (Fig. 2A, lane 14). In contrast, the signal of the internal HA tag is very robust (lane 10). Compared to dFS-HA-I (Fig. 2, lane 11), we only detect small amounts of the 45 kD form (lane 10). It appears that the C-terminal His-tag stabilizes the large 75 kD form and prevents further processing. Taken together, our results indicate that dFS is processed at the N-terminus similar to vertebrate proteins. However, unlike vertebrate forms, additional processing steps occur that remove a C-terminal portion of dFS.
It is possible that normal degradation products of dFS can be detected on western blots, since they are more stable compared to those of hFS. To investigate whether processing occurs within the cell or in the supernatant, we separated dFS-HA-I supernatant from cells and incubated samples over five days at 25°C. We do not see any change in the proportions of the processed forms over time (data not shown). The absence of processing in the supernatant suggests that the processed forms are not products of normal degradation.
To assess the function of the processed forms of dFS, we performed co-immunoprecipitation experiments with FLAG-tagged constructs of dACT and two of the other ligands, DPP and GBB. We find that dFS-HA-I precipitates only in the presence of FLAG-tagged dACT (Fig. 2B, lane 4). Interestingly, we can detect all major and minor processing forms, indicating that they all can bind to dACT. We do not see any dFS in the presence of DPP or GBB. These results suggest that the affinity of dFS is higher for dACT than for DPP or GBB.
Next, we performed SMAD phosphorylation signaling assays with the BMP-type ligands DPP and GBB (Fig. 2D) and the Activin/TGF-β-type ligands dACT and DAW and (Fig. 2E). Since SCW, MYO, and MAV do not result in the phosphorylation of SMADs in this assay (data not shown), we cannot test interaction of dFS with these ligands. Using antibodies against phosphorylated SMADs, we find that incubation of supernatant from cells expressing DPP, GBB, dACT, and DAW with cells transfected with MAD and dSMAD2 results in robust SMAD phosphorylation in cell extracts (Fig. 2D and E) (McCabe et al., 2003). We find that the amounts of dFS used in this experiment do not reduce SMAD activation by DPP or GBB (Fig. 2D). However, signaling of dACT is reduced in the presence of dFS or dFS-HA-I (Fig. 2E). With the same amounts of dFS, we note that signaling of DAW is enhanced rather than reduced. Thus, our results show that dFS exhibits opposite effects on dACT and DAW signaling and that processing of dFS does not abolish the binding to dACT.
2.2. dFS inhibits dACT-mediated wing growth
To analyze the function of dFS in vivo, we over-expressed transgenic lines of dFS, dFS-HA-I, and dFS-HA-C with A9-GAL4. We previously used this driver line to analyze signaling of DPP and GBB in wing imaginal discs (Haerry et al., 1998). A9-GAL4 is located on the X chromosome. This line expresses high levels of GAL4 in wing discs but low levels in other tissues as well. We find that animals of most UAS-dFS lines die in combination with A9-GAL4 during pupation. We hypothesize that death most likely occurs due to reduction of signaling by one or several endogenous TGF-β-type ligands. Out of many UAS-dFS lines tested, we identified two that were not lethal when combined with A9-GAL4. Animals from these non-lethal dFS lines exhibit small wings (Fig. 3B). To compare the size, wings of dFS expressing females were mounted on top of A9/+. To determine the size of wings quantitatively, we measured the number of pixels in pictures of five individual female wings. Compared to A9-GAL4 /+, we find that expression of dFS results in a 23% reduction in size (Table 1). This phenotype is similar to that produced by expression of four transgenes encoding dominant negative isoforms of the Activin type I receptor BABO (Fig. 3E). In contrast, expression of dACT with A9-GAL4 results in a 30% increase of wing size (Fig. 3C, Table 1). When two copies of dFS are co-expressed with dACT, the size is reduced by 31% when compared to dACT alone and by 10% when compared to A9/+ (Fig. 3D, Table 1). Again, this result is similar to co-expression of dominant negative BABO isoforms with dACT (Fig. 3F). We would like to note that expression of two copies of dFS is lethal. These animals are only rescued because of co-expression of dACT (Table 2). Taken together, these results show that dFS can inhibit dACT signaling similar to dominant negative dACT type I receptors.
Fig. 3. dFS inhibits dACT mediated growth.
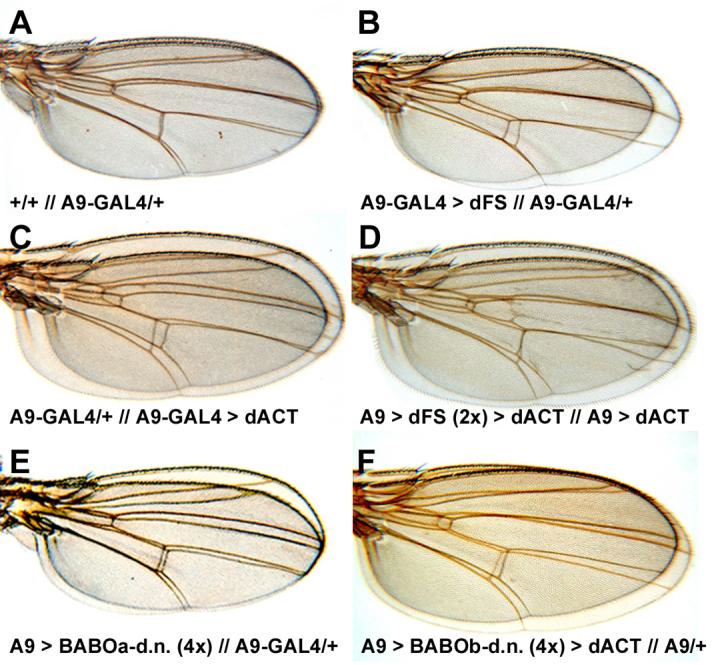
(A-D) Wings of females were mounted on top of each other (top // bottom). The statistical analysis of wing size comparisons is shown in Table 1. (A) Comparison of wild type (y, w) and A9-GAL4/+ wings show no significant difference in size. (B) On average, females of dFS lines that are viable with A9-GAL4 exhibit 23% smaller wings (Table 1). (C) Females that express dACT under the control of A9-GAL4 exhibit 30% larger wings. (D) Co-expression of dACT with two copies of dFS rescues lethality. Wing over-growth is reduced by 31% compared to animals that express dACT alone (Table 1). (E) Expression of dFS is similar to four copies of dominant negative BABO transgenes. (F) Like dFS, co-expression of dominant negative isoforms of BABO with dACT reduces of dACT-induced over-growth. We note that no major patterning defects were obtained in all these experiments.
Table 1. Statistical wing size comparison.
To determine the size of wings, we measured the total pixel number from pictures of five individual female wings using the lasso tool of Adobe Photoshop. The significance of different wing sizes was determined by unpaired t-test analysis assuming equal variances between individual wing samples. Significant differences (<0.05) are shown in bold. Unless indicated otherwise, the samples are compared to wings of heterozygous A9-GAL4 females (1).
| Genotype | Sample Size (N) |
Mean | % | Standard Deviation |
Std. Error |
Significance P-Values |
|---|---|---|---|---|---|---|
| 1. A9/+ | 5 | 1174000 | 100% | 42900 | 19200 | |
| 2. A9> dFS | 5 | 900000 | 77% (1) | 44300 | 19800 | 0.00 (1) |
| 3. A9> dACT | 5 | 1525800 | 130% (1) | 82600 | 36900 | 0.00 (1) |
| 4. A9> dACT > dFS (2x) |
5 | 1055000 | 69% (3) | 65300 | 29200 | 0.00 (3) |
| 5. A9>MYO (2x) | 5 | 1260200 | 107% (1) | 39300 | 17600 | 0.01 (1) |
| 6. A9>MYO (2x) > dFS (1x) |
5 | 1095600 |
122% (2) 87% (5) |
45200 | 20200 |
0.00 (2) 0.00 (5) |
Table 2. Interactions of dFS with TGF-β-type ligands of Drosophila.
Males expressing one of the seven ligands with the X-chromosomal A9-GAL4 were crossed to one copy (1x; columns 2-4) or two copies (2x; columns 5-7) of dFS. The number of males (columns 2 and 5) was used to calculate the expected number of females (columns 4 and 7). Co-expression of four ligands, dACT, MYO, DPP, and SCW, can significantly rescue a lethal line of dFS. While dACT can rescue 38% of females expressing two transgenic copies of dFS, increased levels of DPP and MYO only allow few animals to survive. In contrast, DAW decreases viability of a weak dFS line by 61% from 54% to 33% (bottom rows).
| Ligand | Males without A9-GAL4 |
Females A9>ligand >dFS (1x) |
Rescue females/ males |
Males without A9-GAL4 |
Females A9>ligand >dFS (2x) |
Rescue females/ males |
|
|---|---|---|---|---|---|---|---|
| No ligand | 213 | 0 | 0.0% | 39 | 0 | 0.0% | |
| DPP | 86 | 49 | 57.0% | 69 | 2 | 3.0% | |
| SCW | 105 | 16 | 15.2% | 52 | 0 | 0.0% | |
| GBB | 182 | 0 | 0.0% | 161 | 0 | 0.0% | |
| dACT | 97 | 73 | 75.3% | 171 | 65 | 38.0% | |
| DAW | 112 | 0 | 0.0% | 160 | 0 | 0.0% | |
| MYO | 137 | 28 | 20.4% | 149 | 5 | 3.4% | |
| MAV | 70 | 17 | 24.2% | 77 | 0 | 0.0% | |
| Males without A9-GAL4 |
Females A9>ligand >weak dFS |
Ratio females/ males |
|||||
| No ligand | 66 | 36 | 54.5% | ||||
| DAW | 48 | 16 | 33.3% | ||||
2.3. Interactions between dFS and Drosophila TGF-beta ligands
In vertebrates, FS proteins can inhibit various members of the TGF-β family. We used the lethality of most dFS lines when expressed with A9-GAL4 as a genetic assay to investigate interactions of dFS with individual Drosophila ligands. If A9-GAL4 males are crossed to UAS-dFS, all the daughters die (Table 2; No ligand; 0 offspring). In contrast, the male progeny do not inherit the X-chromosomal A9-GAL4. Males are used to calculate the expected number of female offspring (Table 2; No ligand; 213 expected; 0/213=0%). As a control, we crossed the combination of A9- GAL4 and two copies of UAS-nLacZ to UAS-dFS to analyze whether the number of UAS transgenes affects expression levels. We find that all females die in the presence of the additional UAS transgenes, indicating that GAL4 is not limiting (data not shown). In our assay, co-expression of a ligand that can bind to dFS is expected to reduce the interaction of dFS with endogenous ligands and thus rescue the lethality. Since the amount of secreted protein can vary between insertion lines, we tested several transgenic lines. As shown in Table 2, we find that co-expression of different transgenic lines of GBB and DAW with dFS cannot rescue female lethality. In fact, we find that co-expression of DAW with a non-lethal dFS line reduces the survival from 54.5% to 33.3%. In contrast, lines of dACT can rescue up to 75% of the otherwise lethal dFS expressing females (Table 2). While more than half of the expected females can be rescued by DPP, about one in four is rescued by MAV, one in five by MYO, and one in seven by SCW (Table 2). When individual ligands are co-expressed with two copies of dFS, we find that only dACT significantly rescues the lethality of higher dFS expression (Table 2; 38%). Only about 3% of the expected females survive in the presence of DPP and MYO, and none survive with MAV or SCW. We would like to point out that, aside from different amounts of protein synthesized in each transgenic line, the rescue ability likely depends on different binding affinities between dFS and ligands and not on the signaling strength of the individual ligand. For instance, the dFS lethality can be rescued by co-expression of SCW but not GBB, which is a more potent activator of signaling than SCW (Haerry et al., 1998; Nguyen et al., 1998). Since we use animals of lines that contain both A9-GAL4 and individual ligands in this assay, the lack of rescue is not due to lethality caused by the expression of ligands. In summary, these results indicate that dFS primarily inhibits dACT and to a lesser extent DPP and MYO. Our results are similar to findings in vertebrates showing that FS strongly interacts with Activin (Kd∼45 pM) but binds with a significant lower affinity to other ligands like the DPP orthologues BMP2/4 and the MYO orthologue Myostatin. In contrast to vertebrates, we do not see an interaction between dFS and GBB, the orthologue of BMP 5/6/7. Unlike the other ligands, we find that the presence of DAW reduces the viability of animals that over-express dFS. As discussed later, this result indicates that dFS expression may cause lethality by not only decreasing but also increasing TGF-β signaling.
2.4. dFS inhibits DPP and affects DPP target genes in wing discs
In tissue culture experiments, we can only detect dFS binding to dACT. In rescue assays, however, dFS also interacts with DPP and MYO (Table 2). When over-expressed with A9-GAL4, MYO slightly increases the size of wings by 7% (Table 1). Like dACT, it does not generate any major pattern defects (data not shown). Animals that survive excess dFS by co-expressing two copies of MYO exhibit 13% smaller wings compared to those expressing MYO alone (Table 1). These results indicate that dFS can mitigate over-growth mediated by MYO.
Unlike MYO, expression of DPP in wing imaginal discs results in smaller wings due to cell death and transformation of intervein tissue into dense pigmented vein tissue (Fig. 4C). In contrast, expression of viable dFS lines in wings does not result in pattern defects (Fig. 4B). Animals of lethal dFS lines that are rescued by co-expression of DPP often exhibit almost normal wing patterning (Fig. 4D), suggesting that dFS can inhibit excess DPP signaling. In imaginal discs, DPP signaling via MAD represses the target gene brinker (brk) and results in the activation of the downstream gene spalt (sal) in a stripe close to the source of DPP expression (de Celis et al., 1996; Haerry et al., 1998; Jazwinska et al., 1999; Muller et al., 2003). We find that expression of DPP with A9-GAL4 results in increased growth, repression of brk and expansion of the SAL domain to the entire wing pouch (Fig. 4G and K). In comparison, high-level expression of dFS from two transgenes results in smaller discs, de-repression of brk, and a slight reduction of the area and levels of SAL (Fig. 4F and J). If DPP and dFS are co-expressed, brk and sal expression is restored to almost wild type levels (Fig. 4H and L). These results show that high concentrations of dFS can inhibit DPP and affect the expression of DPP target genes.
Fig. 4. dFS inhibits DPP signaling.
(A-D) Wings of adult females. (A) Wild type wing. (B) Expression of dFS with A9-GAL4 (if not lethal) results in smaller wings without patterning defects. (C) Ectopic expression of DPP with A9-GAL4 results in smaller wings due to apoptosis and vein pattern defects due to transformation of intervein into vein tissue. (D) Co-expression of dFS with DPP restores intervein tissue and the formation of veins. (E-H) brk-LacZ stainings; (I-L) SAL antibody stainings. Compared to wild type discs (E and I), increased levels of dFS results in significantly smaller discs with an expanded area of brk-LacZ staining (F) and reduced levels of SAL (J). Ubiquitous DPP expression results in over-growth, repression of brk (G), and expansion of SAL (K). In comparison to DPP alone, co-expression of dFS with DPP decreases over-growth, partially restores brk-LacZ staining (H), and reduces SAL expression to almost normal levels (L).
2.5. Expression of FS proteins can rescue daw mutants
In tissue culture cells, dFS reduces dACT signaling through dSMAD2 and MAD. However, we observe that dFS increases rather than decreases SMAD phosphorylation in the presence of DAW. On the other hand, DAW exacerbates rather than suppresses the lethal effect of dFS when co-expressed in vivo (Table 2). Taken together, these results suggest that co-expression of DAW with dFS reduces viability by increasing signaling of DAW. This raises the question of whether over-expression of dFS causes lethality by increasing rather than decreasing TGF-β signaling. To answer this question, we examined the effects of dFS expression in daw mutants. Occasionally, small numbers of daw null mutants survive to adulthood (0-6%, Table 3). These animals are fertile, but most of the progeny die and exhibit daw mutant phenotypes. We find that expression of a DAW transgene in most tissues (daugtherless-GAL4, da-GAL4) or in neurosecretory cells (386Y-GAL4) can efficiently rescue daw mutants (Table 3; 66% and 89%). Compared to other ligands like dACT, it appears that DAW can be expressed in any tissue without causing major pattern defects or lethality. Like A9-GAL4, expression of dFS with 386Y-GAL4 is late pupal lethal. Similarly, we find that 85% of larvae that express dFS with 386Y-GAL4 in a daw mutant background pupate but do not survive to adulthood. Since these animals lack DAW, this result indicates that increased signaling of this ligand cannot solely be responsible for lethality caused by over-expression of dFS. However, we noticed that under optimal food conditions, only approximately 32% of daw mutants pupate compared to 85% that also express dFS (Table 2). This finding indicates that expression of dFS can partially compensate for the lack of DAW. To further test this idea, we examined whether human and Xenopus FS (hFS, xFS) proteins exhibit similar effects when expressed in daw mutants. When expressed with the same GAL4 drivers, xFS exhibits weaker growth inhibitory phenotypes than dFS and is not lethal. In comparison, no noticeable growth reduction is observed in animals that express multiple copies of hFS. Interestingly, we find that expression of xFS with 386Y-GAL4 can rescue 23% of daw mutants to adulthood (Table 3). In comparison, 9% of daw mutants are rescued by ubiquitous expression of xFS. We also find that ubiquitous expression of hFS rescues 28% of the expected daw mutants, while no mutants survive with 386Y-GAL4. These results indicate that FS proteins can partially compensate for the lack of DAW. However, since DAW is an active signaling molecule, the mechanism of this interaction is likely indirect and involves other ligands.
Table 3. Rescue of daw mutants.
(Row 1) daw mutants with or without daughterless-GAL4 (da-GAL4) or 386Y-GAL4 GAL4 are crossed to daw lines combined with various UAS-transgenes (Column 1). In these crosses, if all mutants were to survive, they would account for half the number of the heterozygous animals. (Row 2) daw mutants rarely survive to adulthood (2/(77−2)×2=5% of the expected number). Similarly, very few daw animals combined with da-GAL4 and 386Y-GAL4 reach adulthood (6% and 0%). (Row 3) 66% and 89% of daw mutants are rescued by ubiquitous expression of DAW with da-GAL4 or expression in neurosecretory cells with 386Y-GAL4. (Row 4) In the absence of dFS expression, 32% of daw mutants pupate under optimal food condition. Expression of dFS with 386Y-GAL4 increases survival to the pupal stage from 32% to 85%. Expression of dFS with 386Y-GAL4 is late pupal lethal in both wild type and daw mutant background. (Row 5) In contrast to dFS, expression of xFS with da-GAL4 or 386Y-GAL4 is not lethal. While 23% of daw mutants survive to adulthood with 386Y-GAL4, only 9% are rescued by ubiquitously expressed xFS. (Row 6) Ubiquitous expression of hFS rescues 28% of the expected daw mutants. No surviving mutants are observed with 386Y-GAL4.
| * surviving pupae |
daw ; +__ CyO - TM6B |
daw ; da-GAL4 CyO - TM6B |
daw ; 386Y-GAL4 CyO - TM6B |
|---|---|---|---|
|
daw ; +__ CyO - TM6B |
2/77; 5% | 4/129; 6% | 0/109; 0% |
|
daw ; UAS-DAW CyO - TM6B |
0/97; 0% | 28/113; 66% | 37/120; 89% |
|
daw ; UAS-dFS CyO - TM6B |
10*/73; 32%* | NA | 20*/67; 85%* |
|
daw ; UAS-xFS CyO - TM6B |
0/108; 0% | 4/90; 9% | 42/406; 23% |
|
daw ; UAS-hFS CyO - TM6B |
0/80; 0% | 32/262; 28% | 0/121; 0% |
2.6. RNA expression of dfs during development
To investigate possible interactions between dFS and various ligands during development, we performed RNA in situ hybridizations of dfs at various stages and tissues (Fig. 5). We find that dfs is expressed broadly during development. In the embryo, dfs RNA is provided maternally (Fig. 5A) and is present at low levels throughout embryonic development. At the beginning of germ band extension, dfs transcripts accumulate in a stripe at the dorsal side of the embryo that corresponds with the peak levels of DPP signaling (Fig. 5B and C). We note that dfs expression is quite robust along the path of migrating germ cells (Fig. 5B). A possible target of dFS in this area is MYO. As previously reported, myo is expressed in the pole cells (Lo and Frasch, 1999), which exhibit robust staining for phosphorylated MAD (P-MAD, unpublished observation). In contrast to dfs, dact is not transcribed in early embryos, indicating that dACT cannot be the target for early dFS expression (Kutty et al., 1998). In late stage embryos, we notice that dfs mRNA is detected in somatic cells of male but not female gonads (Fig. 5D). At the third instar larval stage, the dfs mRNA accumulates in the brain with little staining in other parts of the CNS (Fig. 5E). Relatively robust staining is seen in all imaginal discs (Fig. 5F-H). In the male germ line, dfs expression is found in primary spermatocytes in larval and adult testes (Fig. 5I and J). In females, low levels of dfs expression are seen at all stages of oogenesis (Fig. 5K). In summary, the expression pattern of dfs overlaps with the reported expression of TGF-beta family members in various tissues indicating that dFS can interact with multiple ligands during development.
Fig. 5. Expression of dfs during development.
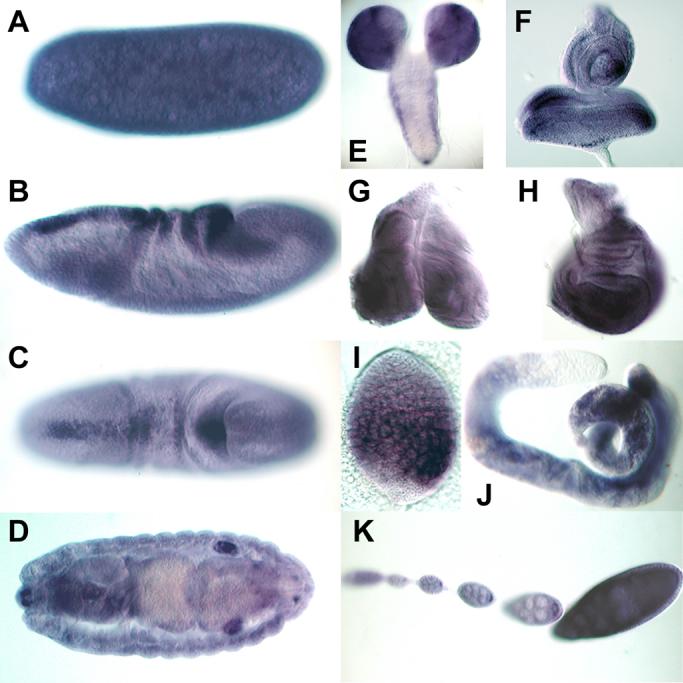
(A) dfs is expressed maternally. (B and C) While low levels of expression are found throughout embryogenesis, first robust zygotic expression is restricted to a dorsal stripe, which surrounds the migrating germ cells. (D) Strong expression is also seen in male but not female gonads. (E-H) In larvae, dfs is broadly expressed in the brain (E) and in imaginal discs (F-H). (I and J) dfs is found in primary spermatocytes of larval and adult testes. (K) It is also ubiquitously expressed in somatic and germ cells during oogenesis.
2.7. dfs loss-of-function phenotype
Since dFS is expressed in many tissues and likely interacts with various ligands during development, we were curious to analyze the dfs mutant phenotype. Database searches for possible transposon insertions in dfs reveal that the closest P-element is located approximately 7 kb upstream of dfs between the two promoters of the neighboring gene scab, which encodes an α-Integrin. They all are semi-lethal, and excision deletions would likely generate double mutants in dfs and scab. However, there are two PiggyBac insertions in the Exelixis collection at Harvard, f00897 and e03941, which were mapped within the dfs transcriptional unit. Using PiggyBac and flanking primers, we confirmed by PCR that f00897 is inserted in the second intron 2.9 kb upstream of the translation start site (data not shown). Similarly, the location of e03941 was corroborated to be 6.2 kb downstream of the transcription start site within intron seven upstream of the last two coding exons. While f00897 is homozygous viable, the original e03941 stock is lethal. However, we found that the lethality can be separated from the transposon. After recombination of heterozygous e03941 females, ten white+ males were selected and isogenized. Out of the ten lines established, we identified five homozygous viable lines. Therefore, we conclude that both f00897 and e03941 are homozygous viable. Both lines are healthy and do not display any noticeable pattern defect or other phenotypes. To examine whether the transposons actually disrupt dfs transcription, we isolated RNA from both lines and performed RT-PCR reactions. As controls, we used the PiggyBac line f04779, which is inserted 6kb downstream of dfs, and heterozygous animals of the Df(2R)Exel7135, which deletes dfs. To avoid amplification from genomic DNA contamination, we used two primers that flank the 8 kb intron where e03941 is inserted. We find that approximately equal amounts of the expected 539 bp cDNA fragments are amplified from the heterozygous deletion and from the f04779 control line (Fig. 6A, lane 1 and 4, arrow). In contrast, the corresponding fragment is approximately four-fold reduced in f00897 flies and missing in e03941 flies (Fig. 6A, lane 2 and 3, arrow). Since the PCR reaction also produced non-specific bands, we digested the reactions with SacII. This restriction digest generates the expected bands of 372bp and 167 bp in the control samples (Fig. 6A, arrows). Again, the amounts of fragments are reduced or missing in the f00897 and e03941 samples. This result indicates that f00897 is a hypomorphic dfs allele. If e03941 prevents transcription past the insertion, it likely generates a mutant dfs mRNA that potentially replaces the conserved third FS domain with a novel C-terminus. To test whether a mutated RNA is actually made, we used two primers upstream of e03941 that span introns 5 and 6 to amplify an expected 1,228 bp cDNA fragment. Again, we obtain reduced levels for f00897 (Fig. 6B, lane 2) compared to the cDNA levels of Df(2R)Exel7135 and f04779 (lane 1 and 4). However, we see normal levels in e03941 flies (lane 3), indicating that transcription is only inhibited downstream of the insertion. As a control, we find that all fly lines produce approximately equal amounts of a 490 bp cg6584 cDNA fragment.
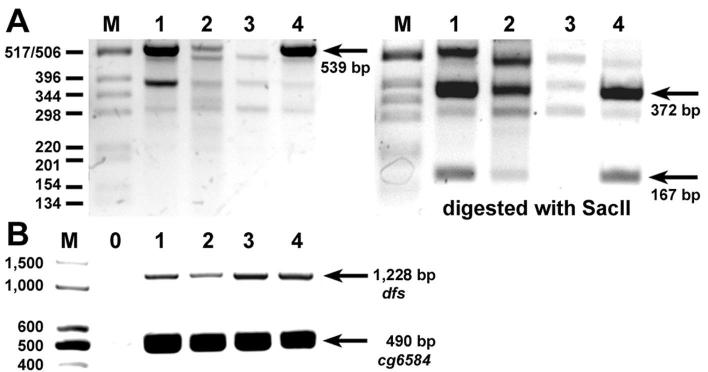
Fig. 6. Expression of dfs in PiggyBac insertion lines.
(Lanes 1-4) RT-PCR reactions were performed from RNA of heterozygous adults of Df(2R)Exel7135 (51E2-11) that deletes dfs (lane 1), homozygous f00897 (lane 2), which is inserted in intron 2, homozygous e03941 (lane 3), which is inserted in intron 7, and as a control f04779, which is inserted 6 kb downstream of dfs. (A) Primers that span intron 7 are used to amplify an expected 539 bp fragment. Density comparison indicates that there is approximately the same amount of the expected cDNA fragment in lane 1 as in lane 4 (arrow), although lane 1 is from heterozygous animals. In contrast, there is approximately four times less cDNA in lane 2 (f00897) and no band in lane 3 (e03941). Digestion of the 539 bp fragment with SacII produces two fragments of 372 and 167 bp (arrows). The corresponding bands are reduced in lane 2 and missing in lane 3. (B) Primers upstream of e03941 that span introns 5 and 6 are expected to produce a 1,228 bp cDNA fragment. Lane 0 is a control without cDNA. Compared to the cDNA levels of Df(2R)Exel7135 and f04779 (lane 1 and 4), reduced levels are seen in f00897 and normal levels for e03941 flies (lane 2 and 3). All fly lines produce approximately equal amounts of a 490 bp cg6584 cDNA fragment.
We also attempted to generate FRT-mediated deletions using two additional PiggyBac insertion lines inserted upstream (f06320) and downstream (f04779) of dfs. Using a Flipase source induced either by heat-shock or under the control of the germ line specific nanos-enhancer, we performed complementation tests of potentially FRT-recombined deletion mutations with the dfs deficiency line Df(2R)Exel7135. Out of several hundred of progeny screened, we only recovered chromosomes that complemented the deficiency and were homozygous viable. Therefore, we have to conclude that similar to many vertebrate species dFS is functionally redundant or not essential and for viability.
3. Discussion
3.1. Structural differences between Drosophila and vertebrate Follistatin
Our analysis shows that there are several differences in the structure of dFS and its vertebrate orthologues. One unique characteristic of dFS is the unusual signal trailer that contains three potential transmembrane domains. Our results suggest that cleavage likely occurs after the third transmembrane domain at the equivalent position of vertebrate FS proteins. We did not obtain any evidence that this feature affects secretion or alters the function of dFS.
A major difference between dFS and its vertebrate homologues is the large basic insertion (Fig. 7). This modification is likely consequential and may alter the activity of dFS. Based on protein folding prediction and the structure of the human FS:Activin complex (Thompson et al., 2005), the insertion is not well structured and is located after the heparin binding site within the EGF sub-domain of FS1 (Innis and Hyvonen, 2003; Inouye et al., 1992). Since this area projects away from Activin, the positively charged amino acids likely increase the affinity of dFS to heparin sulfate proteoglycans on the cell surface and do not contribute to Activin binding. It is conceivable that this feature leads to a reduction of diffusion and an increase in local concentration of dFS. In addition, it may also reduce diffusion of ligands and enhance the stability of ligand-receptor complexes (Fig. 7C). As mentioned earlier, vertebrates have adopted an opposite strategy. They generate a long form of FS by alternative splicing, which contains a negatively charged C-terminal tail that reduces interactions with heparin sulfate. The long form is the major endocrine form in humans. Since it does not efficiently interact with the cell surface, it is not clear whether this form primarily inhibits Activin signaling or actually enhances Activin circulation by preventing unwanted interactions with neighboring cells.
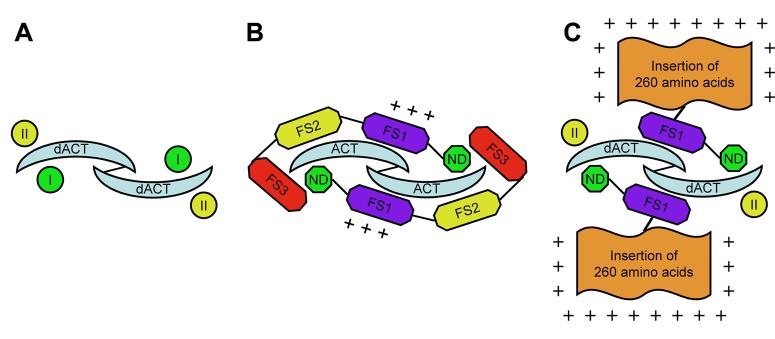
Fig. 7. Interactions of Activins with FS proteins and receptors.
(A) TGF-β-type ligands like dACT form dimers and bind to two type I and two type II receptors (I and II). (B) If two vertebrate FS proteins bind to a ligand dimer, the binding sites of both type I and II receptors are blocked. (C) Proposed model of a complex of processed dFS with ligand and receptor: two processed dFS proteins interact with dACT and block the type I receptor binding sites. Binding of the dFS:dACT complex to type II receptors prevents free ligands from interacting with type II receptors. Thus, shorter forms could be potentially stronger inhibitors than longer forms. The stability of the complex is enhanced through interactions of the additional positively charged amino acids to heparin sulfate proteoglycans at the cell surface.
3.2. Interaction of dFS with ligands
The FS:Activin complex shows that the N domain binds to the wrist region of Activin blocking interaction with the type I receptor (Fig. 7B). The FS2 domain binds to the type II receptor binding site of Activin. In humans, two FS proteins can encircle an Activin dimer preventing interactions with both types of receptors (Fig. 7B). It was found experimentally that Activin bound to FS still could interact with the type II receptor. This result is explained if only one molecule of FS is bound to an Activin dimer leaving the second Activin monomer free to bind to receptors. Unlike vertebrate FS proteins, we found that dFS is proteolytically processed at the C-terminus into two major forms. Based on the migration of epitope tagged forms of dFS on Western blots, the small form contains at least the N and FS1 domains and lacks FS3 and probably part of FS2. Since the small form is immunoprecipitated by dACT, it appears that it can bind to dACT (Fig. 1B). What is the possible role of this small processed form? The following model suggests that the small form could potentially be a stronger inhibitor: if two small processed dFS proteins without FS2 bind to dACT monomers, they prevent interactions with type I but not type II receptors (Fig. 7C). A dFS:dACT:PUT complex is inactive, since dACT cannot recruit the type I receptor BABO. In this scenario, the small dFS forms would not only inactivate bound dACT but also reduce interactions of unbound dACT with type II receptors. If type II receptors were limiting, the small dFS forms would be able to reduce dACT signaling more efficiently than the full-length protein. This hypothesis can be tested by expressing transgenes that encode the small form of dFS. To perform these experiments, we are in the process of determining the structures of the small and large forms of dFS.
In most experiments, we have seen that dFS functions as an inhibitor of TGF-β signaling. As a major exception, dFS enhances rather than inhibits DAW signaling in tissue culture assays. Interestingly, unlike other ligands, DAW does not increase but reduces the viability of animals that over-express dFS. This observation suggests that over-expression of dFS may not only be lethal due to reducing the activities of various TGF-β family members but also due to increased signaling of ligands like DAW. If DAW and dFS physically interact, the question arises how dFS can increase ligand signaling. In the early embryo, it has been shown that Short Gastrulation (SOG), the Drosophila orthologues of the vertebrate Chordin protein, not only inhibits signaling of DPP but also facilitates diffusion, which is necessary for the formation of the peak levels of the DPP gradient (Shimmi et al., 2005). The formation of the DPP gradient depends on the equilibrium between bound and unbound DPP by SOG. Similarly, it is conceivable that ligand binding by the positively charged dFS can reduce ligand diffusion. If the concentration or the affinity of dFS for a ligand is high, the ligand is not released or bound again, and signaling is inhibited. Alternatively, if the concentration of dFS or its affinity for a ligand is low, dFS binding can locally increase ligand concentration, while subsequent ligand release will enhance signal transduction. Consistent with such a model, we see that in tissue culture assays dFS rather increases than decreases the level of MAD activation by DPP (Fig. 2D), while presumably higher levels of dFS inhibit DPP signaling in the wing (Fig. 3D, H, and L). Such a dual function could probably be at work during dorsal-ventral axis formation. While dact is not expressed in the early embryo, dfs is present in a dorsal stripe (Fig. 5B and C). This pattern corresponds with the highest levels of DPP signaling. Expression of dfs is rather late in the formation of the DPP gradient and is potentially regulated by DPP. It is conceivable that initial low levels of dFS could, like SOG, function to increase the local concentration of DPP and contribute to the formation and maintenance of the dorsal peak activity of DPP signaling. In contrast, higher levels of dFS protein that accumulate by the end of the dorsal-ventral axis formation process may inhibit DPP and contribute to terminating the dorsal DPP signal. Recently, a computational model that described ligand distribution and signaling in the presence of a cell surface BMP-binding protein was developed that supports this idea (Umulis et al., 2006). A dual function of FS proteins in facilitating and inhibiting TGF-β signaling is also supported by the finding that fs mutants in mice exhibit overlapping phenotypes with activin knock out animals (Matzuk et al., 1995). Consequently, FS was originally described as enhancer of Activin signaling. Finally, facilitating diffusion and redistribution of dACT and potentially other ligands is probably the best mechanism to explain why dFS and vertebrate FS proteins from frogs and humans can partially compensate for the lack of DAW (Table 3). We can speculate that the vertebrate FS proteins exhibit lower affinities for Drosophila ligands like dACT and diffuse better since they lack the basic insertion of dFS. Most likely, these differences could explain why expression of the vertebrate but not dFS can rescue small but significant numbers of daw mutant animals. Taken together, it is possible that distinct affinities for dFS account for the different interactions seen with various ligands. Further studies are necessary to investigate such a possible dual function of dfs in early embryos and during development.
3.3. dfs loss-of-function mutations
We have shown that the two PiggyBac insertions affect dfs transcription. Interestingly, both lines are homozygous viable and do not show any obvious pattern defects. These results suggest that dFS function not essential for viability. In mice, a mutation in fs results in various defects with lethal consequences. In contrast, FS, Chordin, and Noggin exhibit overlapping functions in Xenopus tropicalis. It is necessary to reduce all three inhibitors to transform ventral into dorsal tissue during embryogenesis of this species (Khokha et al., 2005). Although there is no Noggin-like protein in Drosophila, it is possible that dFS also shares overlapping functions with SOG. sog overlaps with dfs in many tissues, and it is possible that SOG can substitute for dFS in its absence.
The analysis of dfs RNA levels in homozygous f00897 flies shows that they are substantially reduced. Thus, this insertion can be regarded as a hypomorphic allele. It is conceivable that the amount of protein synthesized in this mutant is sufficient for normal development. However, further reduction of dFS by combining f00897 with the deficiency Df(2R)Exel7135 that entirely removes dfs does not affect viability either. In contrast to f00897, the PiggyBac insertion in e03941 clearly disrupts transcription of a full-length mRNA. Based on the location of the insertion, the truncated mRNA likely encodes an altered protein that still contains the N, FS1, and FS2 domains. It lacks the entire FS3 domain and likely contains additional C-terminal amino acids due to altered or absence of splicing. Since the small form is still able to bind dACT (Fig. 2B), a partially functional dFS protein could still be present in e03941 flies, if proper processing would occur. However, it is unlikely that such an altered protein is processed correctly into the small form of dFS. Since we did not obtain homozygous lethal lines in our FRT-mediated deletion screen, it appears that dfs is not lethal. Taken together, the lack of any obvious phenotypes in homozygous f00897 and e03941 lines suggest that dFS is not essential for normal development or is redundant like in some vertebrate species.
4. Materials and methods
4.1. Constructs and generation of transgenic lines
For tissue culture assays, cDNAs were cloned into pAcpA, which contains the Actin 5C promoter and polyA regions (McCabe et al., 2003). For expression in vivo, we cloned cDNAs into pUAST (Brand and Perrimon, 1993) and pUAST2 that contains the unique restriction sites NheI, NaeI, XhoI, Asp718, SacII, NotI, EcoRI, XbaI, and BglII. In this study, we used expression vectors for dACT, DAW (GH14433), DPP, SCW, MYO (BG00784), MAV, dFS, dFS-HA-I, dFS-HA-C, hFS, MAD, FLAG-MAD, dSMAD2, FLAG-dSMAD2, and nuclear LacZ. To obtain higher protein levels in S2 cells, the dACT cDNA was fused with the signal peptide sequence of TKV2. UAS-MAD flies were obtained from S. Newfeld. UAS-xFS lines were generated by P-element mobilization using a line from K. Yu and E. Bier. All other pUAST constructs were injected at a concentration of 1.5 μg/μl and 0.5 μg/μl of Δ2-3 helper plasmid. Some of our constructs have been used in previous studies in collaboration with other groups. Details about individual constructs are available upon request.
4.2. Fly strains and size comparison of wings
daw mutations were obtained by P-element excision deleting 2.5 kb including the second promoter, the start codon, and most of the coding sequence (unpublished results). In this study, we used GAL4 lines under the control of A9 (Haerry et al., 1998), 386Y (Taghert et al., 2001), and daughterless (da) (Wodarz et al., 1995). Flies were raised at 25°C using food prepared according to the Bloomington recipe. To obtain higher expression levels, single transgenes on the same chromosome were recombined.
To compare the size of discs and wings, pictures were taken at identical magnification. We used the lasso and histogram tools in Adobe Photoshop to determine the number of pixels of wings and discs. For statistical analysis, we measured for each sample the size of five female wings or of three male discs (SAL). The significance of different sizes was determined by unpaired t-test analysis assuming equal variances between individual wing and disc samples.
The PiggyBac lines f00897, e03941, f04779, and f06320 were obtained from the Exelixis collection at Harvard. The location of all insertion lines was confirmed by PCR with PiggyBac and location specific primers. f00897 (WH plus) is a homozygous viable insertion 2.9 kb upstream of the translation start site in intron 2. e03941 (RB plus), is inserted 6.2 kb downstream of the start site in intron 7 before the last two coding exons. It was originally homozygous lethal, but we find that the lethality can be segregated away from the e03941 insertion by recombination. f06320 (WH minus) is inserted upstream of dfs between the two promoters of scab (cg8096). It is homozygous viable but semi-lethal over Df(2R)Exel7135. f04779 (WH plus) is located downstream of dfs between cg8079 and cg12954. The FRT sites of f00897 and f04779 are in the same orientation and can be used to generate deletions. Similarly, the FRT sites of e03941 and f06320 are compatible for recombination. To generate deletions, females of the genotypes (1) hs-Flp/+; f00897/f04779, and hs-Flp/+; f06320/e03941 were heat shocked twice for two hours with 24 hours recovery time and crossed to white males. In addition, females of the genotype f00897/f04779; nos-GAL4, UAS-FLP/+, and f06320/e03941; nos-GAL4, UAS-FLP/+ were crossed to white males. Approximately one hundred males with the unique gradient-shaped eye color of f00897 and e03941 were selected from each scheme and mated with Df(2R)Exel7135/CyO females. If a deletion occurs, the weakly expressed white+ transgenes of f04779 and f06320 will be lost but the stronger gradient-shaped eye color phenotypes of f00897 and e03941 should remain. Since the eye color might change if a deletion occurs, we also selected 75 (f04779/f00897) and 40 (f06320/e03941) animals with lighter eye colors from the two schemes for complementation tests. Since all of the chromosomes complemented Df(2R)Exel7135, 40-60 lines from each of the four classes were selected to establish homozygous stocks. We found that all of the lines obtained were homozygous viable and did not show any obvious defects.
4.3. Tissue culture assays
The procedure of the signaling assay was previously described (McCabe et al., 2003). For co-immunoprecipitation experiments, supernatant of 2×108 cells from four dishes transfected with 20 μg of pAcPA-dFS-HA-I DNA was collected after three days and mixed with supernatant from 4×107 cells transfected with 5 μg of pAcpA, pAcpA-FLAG-DPP, FLAG-GBB, or FLAG-dACT. After one hour, the mixture was incubated with 10 μl of Anti-FLAG beads (Sigma A2220). The beads were collected in spin columns, washed six times with TTBS, and the bound material was released from the beads with 1× SDS loading buffer. The samples were run using PAGE and analyzed on Western blots using anti-HA-biotin antibodies from Roche (11666851001) and the ABC kit from Vector (PK-6102).
4.4. RNA in situ hybridizations and immunohistochemistry
Antisense RNA probes for dact, dfs, myo and DAW were prepared using a Digoxigenin-labeling kit from Roche (1175025). In situ hybridizations were performed for one night and washed over three nights according to a previously described protocol (Mason et al., 1994). The labeled RNA was detected with an anti-Digoxigenin-AP-Fab antibody (Roche 1093274). As positive controls, we stained discs of animals expressing transgenes under the control of ptc-GAL4.
LacZ expression was monitored using anti-γ-GAL monoclonal antibodies (1:1000) from Promega (Z3781). Anti-SAL antibodies (rabbit, 1:30) were obtained from R. Schuh. γ-GAL and SAL were detected using AP-coupled secondary antibodies from Promega (S3721 and S3731) and an AP-substrate kit from Vector (SK-5400).
4.5. RT-PCR reactions
RNA from homozygous PiggyBac adult flies and from heterozygous Df(2R)Exel7135 (51E2-11) was isolated using the Trizol reagent from Invitrogen. We generated cDNA using random hexamer primers and M-MLV Reverse Transcripase from Promega. The 539 bp dfs fragment was amplified in 35 cycles using the primers CGGGGCCACTGCAAGAG and CTAGGCCGATGTGGTCG. The two primers GAGAGCTGCAAGGGCTTC and CCACGGCGATGGAACGTC were used to amplify the 1,228 bp dfs fragment. We used two primers specific for the transcript of cg6584, CCAGGAATTCTTATGGATAACAAGAGCGAGAAG and GATCCTCGAGATCACTGCTGGGCAATGGGC, as an internal control.
Acknowledgements
We thank the Basler and the Affolter labs for the brk-LacZ lines. The anti-SAL antibody was a generous gift from R. Schuh. We are grateful to the Taghert lab for 386Y-GAL4, the Newfeld lab for UAS-MAD, and the Bier lab for UAS-xFS. We also would like to thank the Bloomington and Kyoto stock centers for various fly stocks. This work was supported by a Center of Excellence grant from the state of Florida.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe Y, Minegishi T, Leung PC. Activin receptor signaling. Growth Factors. 2004;22:105–110. doi: 10.1080/08977190410001704688. [DOI] [PubMed] [Google Scholar]
- Beattie GM, Lopez AD, Bucay N, Hinton A, Firpo MT, King CC, Hayek A. Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells. 2005;23:489–495. doi: 10.1634/stemcells.2004-0279. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Canalis E, Economides AN, Gazzerro E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr Rev. 2003;24:218–235. doi: 10.1210/er.2002-0023. [DOI] [PubMed] [Google Scholar]
- Cavener DR. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavita A, Krishna S, Zheng H, Padgett RW, Culotti JG. Pioneer axon guidance by UNC-129, a C. elegans TGF-beta. Science. 1998;281:706–709. doi: 10.1126/science.281.5377.706. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Barrio R, Kafatos FC. A gene complex acting downstream of dpp in Drosophila wing morphogenesis. Nature. 1996;381:421–424. doi: 10.1038/381421a0. [DOI] [PubMed] [Google Scholar]
- Haerry TE, Khalsa O, O'Connor MB, Wharton KA. Synergistic signaling by two BMP ligands through the SAX and TKV receptors controls wing growth and patterning in Drosophila. Development. 1998;125:3977–3987. doi: 10.1242/dev.125.20.3977. [DOI] [PubMed] [Google Scholar]
- Haerry TE, O'Connor MB. Isolation of Drosophila Activin and Follistatin cDNAs using novel MACH amplification protocols. Gene. 2002;291:85–93. doi: 10.1016/s0378-1119(02)00463-8. [DOI] [PubMed] [Google Scholar]
- Ho TW, Bristol LA, Coccia C, Li Y, Milbrandt J, Johnson E, Jin L, Bar-Peled O, Griffin JW, Rothstein JD. TGFbeta trophic factors differentially modulate motor axon outgrowth and protection from excitotoxicity. Exp Neurol. 2000;161:664–675. doi: 10.1006/exnr.1999.7290. [DOI] [PubMed] [Google Scholar]
- Innis CA, Hyvonen M. Crystal structures of the heparan sulfate-binding domain of follistatin. Insights into ligand binding. J Biol Chem. 2003;278:39969–39977. doi: 10.1074/jbc.M211284200. [DOI] [PubMed] [Google Scholar]
- Inouye S, Ling N, Shimasaki S. Localization of the heparin binding site of follistatin. Mol Cell Endocrinol. 1992;90:1–6. doi: 10.1016/0303-7207(92)90094-m. [DOI] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- Jazwinska A, Rushlow C, Roth S. The role of brinker in mediating the graded response to Dpp in early Drosophila embryos. Development. 1999;126:3323–3334. doi: 10.1242/dev.126.15.3323. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Yeh J, Grammer TC, Harland RM. Depletion of three BMP antagonists from Spemann's organizer leads to a catastrophic loss of dorsal structures. Dev Cell. 2005;8:401–411. doi: 10.1016/j.devcel.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Kutty G, Kutty RK, Samuel W, Duncan T, Jaworski C, Wiggert B. Identification of a new member of transforming growth factor-beta superfamily in Drosophila: the first invertebrate activin gene. Biochem Biophys Res Commun. 1998;246:644–649. doi: 10.1006/bbrc.1998.8678. [DOI] [PubMed] [Google Scholar]
- Lo PC, Frasch M. Sequence and expression of myoglianin, a novel Drosophila gene of the TGF-beta superfamily. Mech Dev. 1999;86:171–175. doi: 10.1016/s0925-4773(99)00108-2. [DOI] [PubMed] [Google Scholar]
- Mason ED, Konrad KD, Webb CD, Marsh JL. Dorsal midline fate in Drosophila embryos requires twisted gastrulation, a gene encoding a secreted protein related to human connective tissue growth factor. Genes Dev. 1994;8:1489–1501. doi: 10.1101/gad.8.13.1489. [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Matzuk MM, Lu N, Vogel H, Sellheyer K, Roop DR, Bradley A. Multiple defects and perinatal death in mice deficient in follistatin. Nature. 1995;374:360–363. doi: 10.1038/374360a0. [DOI] [PubMed] [Google Scholar]
- McCabe BD, Marques G, Haghighi AP, Fetter RD, Crotty ML, Haerry TE, Goodman CS, O'Connor MB. The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction. Neuron. 2003;39:241–254. doi: 10.1016/s0896-6273(03)00426-4. [DOI] [PubMed] [Google Scholar]
- Muller B, Hartmann B, Pyrowolakis G, Affolter M, Basler K. Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell. 2003;113:221–233. doi: 10.1016/s0092-8674(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Newfeld SJ, Wisotzkey RG, Kumar S. Molecular evolution of a developmental pathway: phylogenetic analyses of transforming growth factor-beta family ligands, receptors and Smad signal transducers. Genetics. 1999;152:783–795. doi: 10.1093/genetics/152.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, Park S, Marques G, Arora K. Interpretation of a BMP activity gradient in Drosophila embryos depends on synergistic signaling by two type I receptors, SAX and TKV. Cell. 1998;95:495–506. doi: 10.1016/s0092-8674(00)81617-7. [DOI] [PubMed] [Google Scholar]
- Rooke HM, Crosier KE. The smad proteins and TGFbeta signalling: uncovering a pathway critical in cancer. Pathology. 2001;33:73–84. [PubMed] [Google Scholar]
- Schneyer AL, Wang Q, Sidis Y, Sluss PM. Differential distribution of follistatin isoforms: application of a new FS315-specific immunoassay. J Clin Endocrinol Metab. 2004;89:5067–5075. doi: 10.1210/jc.2004-0162. [DOI] [PubMed] [Google Scholar]
- Shimmi O, Umulis D, Othmer H, O'Connor MB. Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell. 2005;120:873–886. doi: 10.1016/j.cell.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Hewes RS, Park JH, O'Brien MA, Han M, Peck ME. Multiple amidated neuropeptides are required for normal circadian locomotor rhythms in Drosophila. J Neurosci. 2001;21:6673–6686. doi: 10.1523/JNEUROSCI.21-17-06673.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson TB, Lerch TF, Cook RW, Woodruff TK, Jardetzky TS. The structure of the follistatin:activin complex reveals antagonism of both type I and type II receptor binding. Dev Cell. 2005;9:535–543. doi: 10.1016/j.devcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Umulis DM, Serpe M, O'Connor MB, Othmer HG. Robust, bistable patterning of the dorsal surface of the Drosophila embryo. Proc Natl Acad Sci U S A. 2006;103:11613–11618. doi: 10.1073/pnas.0510398103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Lemaire P. Generation of the germ layers along the animal-vegetal axis in Xenopus laevis. Int J Dev Biol. 2001;45:229–235. [PubMed] [Google Scholar]