Abstract
The DNA end-joining protein Ku70 is one of several proteins that inhibit apoptosis by sequestering the proapoptotic factor Bax from the mitochondria. However, the molecular mechanism underlying Ku70-dependent inhibition of Bax is not fully understood. Here, we show that the absence of Ku70 results in the accumulation of ubiquitylated Bax. Under normal growth conditions, Bax ubiquitylation promotes its degradation. Upon induction of apoptosis in wild-type cells, a significant reduction in the levels of ubiquitylated Bax was observed, whereas in Ku70−/− cells, the ubiquitylated Bax was robustly accumulated. Addition of recombinant Ku70 into a protein extract of Ku70−/− cells resulted in a decrease in the levels of ubiquitylated Bax, even in the presence of proteasome inhibitors. Moreover, an in vitro deubiquitylation assay demonstrated that recombinant Ku70 hydrolyzed polyubiquitin chains into monoubiquitin units. Thus, Ku70 regulates apoptosis by sequestering Bax from the mitochondria and mediating Bax deubiquitylation. These results shed light on the role of proteasome inhibitors as tumor suppressors.
Keywords: apoptosis, ubiquitin
Programmed cell death, or apoptosis, is a natural process of removing unneeded or damaged cells and is required for the proper execution of the organism's life cycle (1, 2). Apoptosis was shown to be involved in numerous processes including embryonic development, response to cellular damage, and aging and as a mechanism of tumor suppression. Thus, understanding the molecular regulation of apoptosis has great therapeutic implications.
Two pathways were shown to induce apoptosis, an extrinsic and an intrinsic one, which differ in the mechanism by which the death signal is transduced (2). Whereas the extrinsic pathway is activated by binding of ligands to a death receptor, the intrinsic pathway is activated by cellular stress, for example DNA damage. The intrinsic pathway involves the release of cytochrome c from the intermembrane space of the mitochondria. Together with apoptotic protease activating factor 1 (APAF1), cytochrome c activates caspase 9, leading to activation of downstream caspases and the induction of the death response.
Key players in the regulation of the intrinsic pathway include the Bcl2 protein family, which can influence the permeability of the outer mitochondrial membrane (3). Members of the Bcl2 protein family are divided into proapoptotic proteins such as Bax, Bak, and Bok and antiapoptotic ones including Bcl2, Bcl-X, Bcl-w, and Mcl-1. Proteins of a third subfamily, known as the BH3-only proteins, are considered to be initiators of apoptosis and probably function by regulating Bcl2-like proteins from the other two subfamilies.
In healthy cells, Bax exists as a monomer, either in the cytosol or weakly bound to the outer mitochondrial membrane. Upon stimulation of apoptosis, Bax undergoes a series of conformational changes and translocates to the mitochondria, where it becomes anchored into the mitochondrial membrane. After its translocation, Bax oligomerizes into large complexes, which are essential for the permeabilization of the mitochondrial membrane (4). Given its central role in mediating apoptosis, Bax is heavily regulated both by binding to other proteins and through posttranslational modifications.
At least four proteins, Bcl2 (5), 14–3-3 (6), humanin (7), and Ku70 (8–11), have been shown to negatively regulate apoptosis by binding to Bax and sequestering it from the mitochondria. Yet, although binding to these proteins suggests an elegant mechanism for the retention of Bax in the cytosol, it does not explain how Bax dissociates from these proteins after apoptotic stimuli. Recently, it was shown that acetylation of two critical lysines on the carboxyl terminus of Ku70 results in the dissociation of Bax from Ku70 and, as a consequence, in an increase in cellular sensitivity to apoptotic stimuli (8, 9). Nevertheless, the observations that the Ku70−/− mutation in mice is not prenatally lethal and that Ku70−/− mice exhibit a significant increase in their basal level of apoptosis only in the brain (12) and in gastrointestinal tissue (13) suggests that additional steps subsequent to the release from Ku70 also regulate Bax activation.
In this article, we show that the absence of Ku70 results in the accumulation of ubiquitylated Bax. Under normal growth conditions, this modification labels Bax for degradation. Induction of apoptosis leads to a reduction in the levels of ubiquitylated Bax, a phenomenon that is significantly delayed in Ku70-deficient cells. Moreover, recombinant Ku70 hydrolyzes polyubiquitin chains into monoubiquitin units, and the addition of recombinant Ku70 into the protein extract of Ku70−/− cells results in a decline in ubiquitylated Bax levels. Thus, Ku70 regulates Bax-mediated apoptosis by sequestering Bax from the mitochondria and by regulating Bax deubiquitylation.
Results
The DNA repair protein Ku70, was recently shown to negatively regulate apoptosis by sequestering Bax away from the mitochondria (Fig. 1) (8, 10). Yet, given the increasing number of proteins that were found to sequester Bax, the unique role of Ku70 in its association with Bax is not fully understood. To increase our understanding of this association, we followed the fate of Bax in the absence of Ku70 under normal growth conditions.
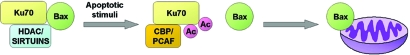
Fig. 1.
Ku70 negatively regulates Bax-mediated apoptosis. The association between Ku70 and the proapoptotic protein, Bax, sequesters Bax away from the mitochondria. Under normal growth conditions, Ku70 is maintained in an unacetylated state by the NAD+-dependent SIRT1 deacetylase and as yet unidentified histone deacetylases (HDACs), which enable its association with Bax. After apoptotic stimuli, the CBP and PCAF acetyltransferases acetylate specific lysines on Ku70, resulting in a conformational change of Ku70 and the liberation of Bax; the released Bax then acts to initiate apoptosis.
Reduction in Ku70 Levels Promotes the Accumulation of High-Molecular-Weight Isoforms of Bax.
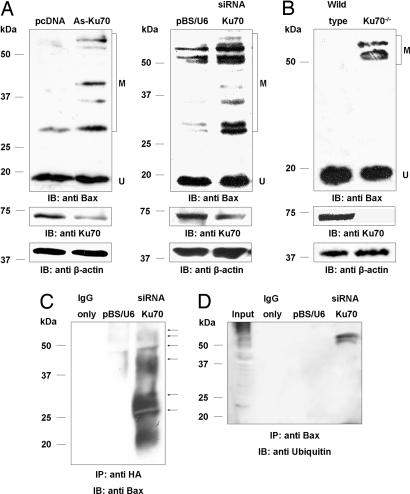
Ku70 levels were knocked down in human osteosarcoma U2OS cells by either overexpressing siRNA or antisense sequences against Ku70 (Fig. 2A). In both cases, the reduction in Ku70 protein levels resulted in the accumulation of high-molecular-weight isoforms of Bax, which were detected by a specific rabbit polyclonal antibody. Similarly, these isoforms were more abundant in Ku70−/− mouse embryonic fibroblasts (MEFs) compared with wild-type MEFs (Fig. 2B).
Fig. 2.
Decrease in Ku70 levels promotes Bax ubiquitylation. (A) Decreasing the level of Ku70 by overexpressing either antisense or siRNA sequences (As-Ku70; siRNA Ku70) against Ku70 in U2OS cells resulted in the accumulation of high-molecular-weight modified forms of Bax. (B) The high-molecular-weight modified forms of Bax were significantly more abundant in Ku70 −/− MEFs compared with wild-type MEFs. (C) HEK293T cells were cotransfected with a HA-tagged ubiquitin-expressing vector together with either control siRNA (center lane) or siRNA against Ku70 (right lane) plasmids. Immunocomplexes were precipitated with the anti-HA antibody, separated by SDS/PAGE, and immunoblotted with an HRP-conjugated anti-Bax antibody. The distinct sizes of ubiquitylated Bax on the immunoblot are marked by small arrows. An additional control of IgG alone was also loaded (left lane) to distinguish between antibodies from the Immunocomplexes and ubiquitylated Bax. (D) Immunocomplexes were precipitated with anti-Bax antibody from HEK293T overexpressing either control siRNA (middle lane) or siRNA against Ku70 (right lane) and immunoblotted with an anti-Ubiquitin antibody. IgG alone was also loaded (left lane). In all experiments, β-actin served as a loading control. U, unmodified Bax; M modified Bax.
Ku70 acetylation results in the dissociation of Bax from Ku70 (8). Consequently, treatment with protein deacetylase inhibitors promotes Ku70 acetylation, and Bax-mediated apoptosis (8). Treatment of human embryonic kidney (HEK)293T cells with deacetylase inhibitors at doses that increase Ku70 acetylation and Bax release without increasing the level of apoptosis, promoted the accumulation of a high-molecular-weight modified forms of Bax (data not shown). Taken together, these results suggest that one reason why Ku70−/− mice do not die prenatally because of excessive apoptosis is that these modifications of the Bax protein negatively regulate Bax-mediated apoptosis.
Association Between Ku70 and Bax Regulates Bax Ubiquitylation.
The increase in the molecular weight of Bax in the absence of Ku70 suggests that, under these conditions, Bax was either heavily modified by numerous low-molecular-weight modifications such as acetylation, methylation, and phosphorylation or by high-molecular-weight modifications including sumoylation or ubiquitylation. Moreover, modified Bax species of similar size to those obtained in cells with reduced levels of Ku70 were previously suggested to represent ubiquitylated Bax (14). To explore the possibility that Bax is ubiquitylated in the absence of Ku70, HEK293T cells were cotransfected with HA-tagged ubiquitin and either an siRNA vector specific for Ku70 or a control siRNA. Immunoprecipitation (IP) of HA tagged proteins, followed by probing of the immunocomplex with an antibody against Bax, demonstrated that Bax is ubiquitylated (Fig. 2C), resulting in the accumulation of specific modified Bax species as in cells with decreased Ku70 levels (Fig. 2 A and C). Reversed IP with antibody against Bax, followed by probing the immunocomplex with an antibody against ubiquitin, gave similar results, further confirming that Bax is ubiquitylated in the absence of Ku70 (Fig. 2D). Notably, the reversed IP showed only the high-molecular-weight forms of ubiquitylated Bax, probably because of intrinsic limitations of the antibodies. Thus, either Ku70 is required for the deubiquitylation of Bax, or the release from Ku70 enables Bax ubiquitylation.
Bax Ubiquitylation Promotes Its Degradation and Decreases upon Apoptotic Stimuli.
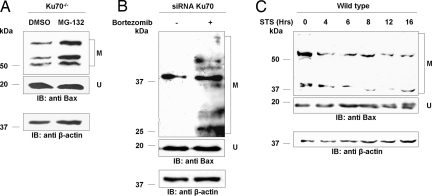
Protein ubiquitylation mediates protein degradation by the proteasome and also serves as a posttranslational modification that regulates protein function. Treatment of Ku70−/− cells with the proteasomal inhibitor MG-132 increased the levels of modified Bax (Fig. 3A). Bortezomib (Velcade or PS-341) is a potent proteasomal inhibitor that was approved as a clinical drug against multiple myeloma (15). Similarly, treatment of HEK293T cells, expressing reduced levels of Ku70, with the proteasomal inhibitor Bortezomib, resulted in the accumulation of modified Bax (Fig. 3B). Thus, Bax ubiquitylation plays an antiapoptotic role by targeting Bax for proteasomal degradation. Staurosporine (STS) was found to induce Bax-mediated apoptosis (16). Time-course analysis of Bax ubiquitylation in wild-type cells after apoptotic stimuli by STS, revealed a significant decrease in the levels of ubiquitylated Bax 4 h after the treatment. (Fig. 3C). These findings suggest that Bax ubiquitylation inhibits Bax function and that Bax must be deubiquitylated to mediate apoptosis.
Fig. 3.
Bax ubiquitylation promotes its degradation and decreases upon apoptotic stimuli. (A) Treatment of Ku70 −/− cells with the proteasomal inhibitor, MG-132, increased the levels of ubiquitylated Bax. (B) Treatment of HEK293 cells overexpressing siRNA against Ku70 with the proteasomal inhibitor Bertozomib (Velcade or PS-341) increased the levels of ubiquitylated Bax. The decrease in Ku70 levels was validated against HEK293 cells overexpressing control siRNA (data not shown) (C) Time-course analysis of ubiquitylated Bax levels by Western blot using polyclonal antibody against Bax before (0) or at various times after the induction of Bax-mediated apoptosis by staurosporine-STS in wild-type cells. In all experiments, β-actin levels served as a loading control. U, unmodified Bax; M, modified Bax.
Ku70-Dependent Bax Deubiquitylation.
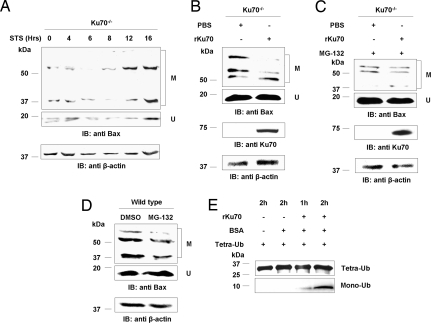
The increase in the levels of ubiquitylated Bax in Ku70-deficient cells suggests that Bax ubiquitylation might protect the cell from Bax-mediated apoptosis when Ku70 dissociates from Bax and no longer sequesters it away from the mitochondria. In such a scenario, Bax would be ubiquitylated after the release from Ku70 as an additional level of protection from uncontrolled apoptosis. Strikingly, time-course analysis of Bax ubiquitylation in Ku70−/− cells (Fig. 4A) or in cells knocked down for Ku70 by siRNA (data not shown) after apoptotic stimulus showed an increase rather than a decrease in the levels of ubiquitylated Bax. Thus, Ku70 may promote deubiquitylation of Bax in general and especially during apoptosis.
Fig. 4.
Ku70 mediates Bax deubiquitylation. (A) Time-course analysis of ubiquitylated Bax levels by Western blot with polyclonal antibody against Bax before (0) or at various times after the induction of Bax-mediated apoptosis by staurosporine-STS in Ku70 −/− cells. (B) In vitro deubiquitylation of Bax in Ku70−/− cell extracts supplemented with PBS or recombinant Ku70-rKU70. (C) In vitro deubiquitylation of Bax in Ku70−/− cells that were grown in the presence of the proteasomal inhibitor MG-132. The extracts were supplemented with PBS or rKu70. (D) Levels of ubiquitylated Bax in wild-type cells that were grown in medium supplemented with DMSO or the proteasomal inhibitor, MG-132. (E) In vitro deubiquitylation assay of homogenous tetraubiquitin chains supplemented with BSA and rKu70 for various time periods, demonstrating that Ku70 possess an intrinsic DUB enzymatic activity. Incubation of tetraubiquitin only or tetraubiquitin together with BSA did not induce hydrolysis of the tetraubiquitin chains into monoubiquitin units. In A–D, β-actin levels served as a loading control. U, unmodified Bax; M, modified Bax.
To test such a possible role for Ku70 in Bax deubiquitylation, we followed the rate of Bax deubiquitylation in vitro in Ku70−/− cell extract supplemented with either recombinant Ku70 or PBS. After 60 min at 37°C, there was a significantly greater extent of Bax deubiquitylation in the extract supplemented with Ku70 (Fig. 4B) even in the presence of the proteasomal inhibitor, MG-132 (Fig. 4C). Moreover, treatment with MG-132 or Bortezomib increased the levels of ubiquitylated Bax in Ku70−/− (Fig. 3 A and B) but not in wild-type cells (Fig. 4D and data not shown). These results suggest that Ku70 might have an intrinsic deubiquitylation (DUB) enzymatic activity. To explore this possibility, recombinant Ku70 was incubated with homogenous hexaubiquitin (K48-Ub6, data not shown) or tetraubiquitin (K48-Ub4) chains for various time periods. Strikingly, the recombinant Ku70 hydrolyzed the polyubiquitin chains into monoubiquitin units (Fig. 4E). The thiol blocking agent, N-ethylmaleimide (NEM), was shown to inhibit the activity of DUB enzymes (17). As expected, Ku70 was sensitive to NEM treatment in vitro (data not shown). Thus, taken together, these results demonstrate that Bax deubiquitylation rather than Bax degradation per se is Ku70-dependent. These findings suggest an attractive model by which Ku70 mediates deubiquitylation of Bax, a process that is enhanced upon apoptotic stimulus to enable cellular death.
Discussion
The DNA repair protein Ku70 was recently shown to inhibit apoptosis by associating with the proapoptotic factor Bax (8, 10, 18, 19). Yet, it was not clear whether Ku70 serves only as a “parking slot” for Bax or whether it mediates other more specific regulatory activities on Bax. Moreover, the observation that Ku70−/− mice do not die prenatally suggested that, in the absence of Ku70, other mechanisms must prevent uncontrolled Bax-mediated apoptosis, but the nature of these pathways was not known. In this study, we explored these questions and followed the fate of Bax in the absence of Ku70 under normal unstressed growth conditions. We showed that in the absence of Ku70, Bax is heavily modified by ubiquitylation, which labels Bax for proteosomal degradation. In wild-type cells, apoptotic stimuli increased the rate of Bax deubiquitylation, whereas, in the absence of Ku70, this process was delayed. Finally, Ku70 possesses an intrinsic DUB activity on polyubiquitin chains and promotes Bax deubiquitylation in vitro.
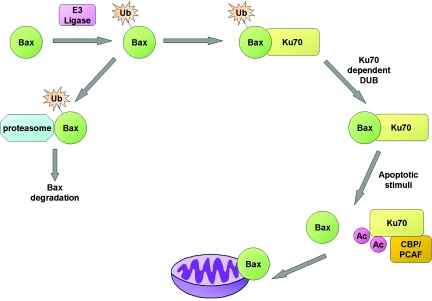
Taken together, our data suggest a mechanism for Bax regulation. Under normal unstressed conditions, after its synthesis, Bax undergoes ubiquitylation, which negatively regulates its proapoptotic function by labeling it for proteasomal degradation. The association with Ku70 mediates and promotes Bax deubiquitylation, a process that is enhanced after apoptotic stimuli. Upon apoptotic stimulus, Ku70 is acetylated by the acetyltransferases CBP or PCAF and releases Bax (8). The free cytosolic unubiquitylated Bax localizes to the mitochondria where it mediates apoptosis (Fig. 5).
Fig. 5.
Proposed model for the regulation of Bax-mediated apoptosis by Ku70. Newly synthesized Bax undergoes ubiquitylation, which negatively regulates its proapoptotic function by labeling it for proteasomal degradation. After its ubiquitylation, Bax associates with Ku70, which mediates Bax deubiquitylation, simultaneously generating and sequestering the active form of Bax away from the mitochondria. After apoptotic stimuli, Ku70 is acetylated by CBP or PCAF and releases Bax. The free cytosolic unubiquitylated Bax can then localize to the mitochondria where it executes programmed cell death.
Our current description of Bax ubiquitylation, combined with the previous findings demonstrating that the acetylation status of Ku70 regulates the association of Bax with Ku70, raises an interesting question regarding the sequence of these regulatory steps under normal and stressed conditions. Under normal growth conditions, the absence of Ku70 leads to the accumulation of ubiquitylated Bax. Time-course analysis of the level of modified Bax in wild type showed that, after staurosporine-mediated apoptotic signaling, ubiquitylated Bax levels decreased, whereas, in the absence of Ku70, this decrease was significantly delayed (Figs. 3C and 4A). Thus, the ubiquitylation of Bax occurs independently of the association between Ku70 and Bax, and Ku70 induces Bax deubiquitylation specifically after an apoptotic signal. The observation that treatment with deacetylase inhibitors increased the level of ubiquitylated Bax demonstrates that the deubiquitylation of Bax occurs before Ku70 acetylation releases Bax from Ku70. Thus, we propose the following order of cellular events: First, newly synthesized Bax is ubiquitylated, and the ubiquitylated Bax then becomes associated with Ku70. This association with Ku70 sequesters Bax from the mitochondria, allowing Ku70 to mediate Bax deubiquitylation, without immediately inducing apoptosis. The deubiquitylation is enhanced upon apoptotic signal, which, later in this process, will lead to the acetylation of K539 and K542 of Ku70 and the dissociation of the Ku70-Bax complex.
Despite the role of Ku70 in mediating Bax deubiquitylation, unmodified Bax was observed in Ku70-deficient cells with or without apoptotic stimuli (this study and refs. 8 and 20). The source of these unmodified Bax proteins could be explained by two possible scenarios. First, it is possible that, in addition to Ku70, other DUB enzymes modify Bax especially after apoptotic stimuli. Yet, this explanation is less probable because our findings do not support this possibility. Even 16 h after the treatment of Ku70-deficient cells with STS, an accumulation rather than a reduction of ubiquitylated Bax was observed. Thus, such other possible DUBs, if they exist, would modify Bax only after the first 16 h after apoptotic stimuli. The second possible explanation and, in our opinion, a more reasonable one, could be that the observed unmodified Bax are newly synthesized Bax before their ubiquitylation or unmodified Bax associated with other proteins than Ku70, such as Humanin or Bcl2.
Ku70−/− cells are more sensitive to Bax-mediated apoptosis (20). Yet, our findings demonstrated that, in the absence of Ku70, the level of ubiquitylated Bax increased. The key for understanding this apparent contradiction lies in the timing of the Ku70–Bax dissociation after the apoptotic signal. After the apoptotic stimuli, Bax translocates to the mitochondria only if it is not associated with Ku70. The release from Ku70 occurs 1 to 4 hours after the apoptotic stimuli when Ku70 becomes acetylated (8). In the absence of Ku70, Bax is not limited by the timing of Ku70 acetylation and can translocate to the mitochondria earlier than in Ku70+/+ cells. Thus, in Ku70-deficient cells, the rate of Bax-mediated apoptosis would be faster than in wild-type cells.
One of the interesting questions that arise is the nature of the biochemical activity of Ku70 during Bax deubiquitylation. Does Ku70 itself catalyze Bax deubiquitylation or does Ku70 only mediate the enzymatic activity of an unknown DUB enzyme on Bax? The findings that Ku70 can hydrolyze various polyubiquitin chains (hexa- or tetra-) in vitro (Fig. 4E), strongly support the direct deubiquitylation activity of Ku70. Interestingly, sequence analysis failed to identify any similarity with the DUB motifs of four known DUB classes (OUT, UCH, USP, and JAMM) (21) in Ku70's protein sequence. However, we have identified in Ku70 a region with similarity to the motif of the UIM/MJD DUB class (21) [supporting information (SI) Fig. S1]. It remains to be determined whether Ku70 expresses DUB enzymatic activity in vivo or promotes the association between a specific DUB and ubiquitylated Bax.
Similar to Bax, another pivotal protein involved in apoptosis that is regulated by ubiquitylation is the transcription factor p53 (22). Ubiquitylation of p53 has been shown to regulate its level and activity. Whereas some forms of ubiquitylation label p53 for degradation, other types lead to the translocation of p53 to the cytosol (23). Nuclear export of p53 might sequester the protein from its main transcriptional targets while maintaining it in reserve in its active form. It would be important to pursue the possibility that Bax might also be regulated by different types of ubiquitylation and that Ku70-dependent deubiquitylation determines the balance between these forms. These different types of ubiquitylation have the potential to control many aspects of Bax biochemistry including its association with other proteins, localization, and dimerization.
Bax was shown to be associated with several proteins in the cytosol, including Bcl2 (5), 14-3-3 (6), humanin (7), and Ku70 (8, 10, 11). It was proposed that the association of each of these proteins with Bax sequesters it away from the mitochondria and prevents apoptosis. Yet, it is not clear why four different proteins are required for sequestering Bax. The finding that, in wild-type cells, apoptotic stimuli results in a decrease in Bax deubiquitylation suggests an antiapoptotic role for Bax ubiquitylation. Thus, the association between Ku70 and Bax serves a dual function, sequestering Bax away from the mitochondria and mediating Bax deubiquitylation. Deubiquitylated Bax in association with Ku70 allows the cells to maintain a reservoir of active Bax for apoptosis. Thus, in the case of exposure to stress, the cells can immediately activate their apoptotic program by Ku70 acetylation and Bax release.
How broad is the DUB activity of Ku70? One could answer this intriguing question from studies of deacetylases. In vitro deacetylation with HDACs as SIRT1 showed no specificity for its deacetylase activity (24). Moreover, we could not find a deacetylation consensus sequence in silico (data not shown). Nevertheless, in vivo SIRT1 and other HDACs deacetylate only a limited range of proteins out of the whole spectrum of acetylated proteins. Thus, the specificity for deacetylases is via the association with their substrates. In an analogy with deacetylases, we suggest that the association between a given DUB and its substrate determines its specificity. Indeed, our results support this mode of operation because Ku70 can deubiquitylate Bax in vivo and in vitro (Fig. 4 B and C) and can deubiquitylate unspecific polyubiquitin chains in vitro (Fig. 4E). It would be of great interest to follow the role of Ku70 as a DUB enzyme in other Ku70-related pathways e.g., DNA double-strand break repair.
Protein ubiquitylation was shown to control many cellular functions including cell cycle, DNA repair, cell proliferation, and apoptosis (25, 26). Given this variety of cellular pathways, it is not surprising that alteration in the ubiquitylation machinery or in ubiquitin signaling was found to occur in many pathologies, including cancer (26). Large bodies of experimental and clinical data have shown associations between defects in the proteolysis of cell cycle components or house keeping genes and various malignancies. In addition, ubiquitylation was shown to regulate several proteins whose activities are important for tumor development, including p53 (27) and NFκB (28). Thus, ubiquitylation can act as a tumor-suppressor mechanism by increasing apoptosis and inhibiting the normal advance through the cell cycle. Recently, an increasing effort was invested in the identification of small molecules that can function as modulators of the ubiquitylation system and thereby inhibit the development of malignancies (15, 29, 30). Similar efforts have been invested in the use of deacetylase inhibitors to promote apoptosis as a cancer therapy (31, 32). Our findings suggest a potential approach to achieve maximal inhibition of a malignancy by induction of apoptosis. Such an approach should consist of the following pharmacological manipulations: First, treatment with proteasomal inhibitors such as MG-132 or Bortezomib to increase the amount of ubiquitylated Bax associated with Ku70. After enough time has elapsed to enable Ku70 to promote the deubiquitylation of Bax, resulting in an increase in the reserve of active Bax, treatment with deacetylase inhibitors should be given to dissociate Ku70 from Bax, thereby inducing Bax-mediated apoptosis. Reversing the order of treatments or giving both treatments simultaneously is expected to cause an accumulation of the less effective, ubiquitylated form of Bax.
Experimental Procedures
Cell Culture and Biological Reagents.
HEK cells, U2OS human osteosarcoma cells, HEK293T cells, and Ku70−/− mouse embryonic fibroblast (MEF) cells were grown in DMEM supplemented with 10% FCS, 100 units/ml penicillin, and 100 mg/ml streptomycin. Staurosporine (STS), Trichostatin A (TSA), Nicotinamide (NAM), and MG-132 were purchased from Sigma.
Expression Vectors, DNA manipulation, and transfections.
pBS/U6 for U6-driven siRNA expression was described (33). The following oligonucleotides were cloned into the pBS/U6 plasmid to produce psiRNA Ku70. 5′-GGAAGAGATAGTTTGATTTAAAATCAAACTATCTCTTCCCTTTTT and 5′-AAAAAGGGAAGAGATAGTTTGATTTTAAATCAAACTATCTCTTCC.
pcDNA expressing an antisense sequence against Ku70 was kindly provided by S. Matzuyama (Case Western Reserve University, Cleveland, OH). All transfections were performed by using Metafectene (Biontex).
Western Blot Analysis, Antibodies, and Immunoprecipitation.
Whole-cell extracts were prepared by using lysis buffer [50 mM Tris (pH 8), 1% Nonidet P-40, 150 mM NaCl, 1 mM MgCl2, 10% glycerol, 1 mM DTT, and 1× EDTA-free protease inhibitor mixture (Roche Diagnostics)]. Protein extraction was performed as described (34). All the experiments were done by using freshly prepared extracts that had not been subjected to freezing and thawing. The following antibodies were used to detect protein expression by Western blot: rabbit polyclonal anti Bax (N-20; Santa Cruz Biotechnology), mouse monoclonal anti actin (A4700; Sigma), mouse monoclonal anti ubiquitin (P4D1; Santa Cruz Biotechnology), and mouse monoclonal anti HA (HA.11; Covance). For immunoprecipitation of ubiquitylated Bax, protein extracts from cells expressing the HA-tagged ubiquitin were precleared by incubating with protein A/G Plus Agarose beads (Santa Cruz Biotechnology) at 4°C for 1 h. The collected supernatant was then incubated for at least 1 h with primary monoclonal anti-HA antibody (Covance), followed by incubation with protein A/G Plus Agarose beads at 4°C for 3 h to overnight. The washed immunoprecipitates were boiled in SDS sample buffer and used for Western blot analysis with an HRP-conjugated polyclonal antibody against Bax (N-20; Santa Cruz Biotechnology). The reversed IP was performed by using the ExactaCruz kit (cat. no. sc-45039; Santa Cruz Biotechnology) according to the manufacturer's protocol. Bax was immunoprecipitated with rabbit polyclonal anti-Bax and probed with monoclonal anti-ubiquitin antibody.
In Vitro Deubiquitylation Assays.
Ku70−/− MEFs cells were lysed in lysis buffer without protease inhibitors. The lysates were incubated with purified Ku70/80 complex (35) or PBS at 37°C for 1 h. Reaction products were separated by SDS/PAGE and detected by immunoblotting with antibodies against Bax, Ku70, and β-actin. For direct in vitro deubiquitylation activity assay, hexaubiquitin or tetraubiquitin chains (Boston Biochem) were incubated for 0, 60, and 120 min with BSA (negative control) or purified Ku70/80 complex as described (36). The reaction products were separated by SDS/PAGE, and the hydrolysis of ubiquitin chains into monoubiquitin units was detected by silver staining.
Apoptosis Induction, Proteosomal Inhibition, and HDAC Inhibition.
To induce apoptosis, cells were grown in medium supplemented with 200 nM staurosporine (STS) for 16 h. Proteasomes were inhibited by adding MG-132 (40 μM) or Bortezomib (10 nM/ml) to the cell growth medium 4 h before protein extracts were prepared.
Histone deacetylases (HDACs) were inhibited by treating cells with 1 mM nicotinamide (NAM) for 6 h and 0.4 μM trichostatin A (TSA) for 16 h.
Acknowledgments.
We thank Doron Ginsberg, Shelley Schwarzbaum (Bar-Ilan University), and members of the H.Y.C. laboratory for helpful comments on the manuscript and Mali Salmon (Bar-Ilan University) for the bioinformatic analysis of Ku70. Bortezomib (Velcade) was kindly provided by Dr. Izhar Harden (Sheba Medical Center, Tel Hashomer, Israel). This work was supported by grants from the Israeli Academy of Sciences, the German–Israeli Foundation, the Binational U.S.–Israel Foundation, the Israel Cancer Association, the Koret Foundation, and the Israel Cancer Research Fund. H.C. was supported by the Alon Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706700105/DCSupplemental.
References
- 1.Zimmermann KC, Bonzon C, Green DR. The machinery of programmed cell death. Pharmacol Ther. 2001;92:57–70. doi: 10.1016/s0163-7258(01)00159-0. [DOI] [PubMed] [Google Scholar]
- 2.Chowdhury I, Tharakan B, Bhat GK. Current concepts in apoptosis: The physiological suicide program revisited. Cell Mol Biol Lett. 2006;11:506–525. doi: 10.2478/s11658-006-0041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reed JC. Proapoptotic multidomain Bcl-2/Bax-family proteins: mechanisms, physiological roles, and therapeutic opportunities. Cell Death Differ. 2006;13:1378–1386. doi: 10.1038/sj.cdd.4401975. [DOI] [PubMed] [Google Scholar]
- 4.Antignani A, Youle RJ. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr Opin Cell Biol. 2006;18:685–689. doi: 10.1016/j.ceb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 6.Nomura M, et al. 14–3-3 Interacts directly with and negatively regulates pro-apoptotic Bax. J Biol Chem. 2003;278:2058–2065. doi: 10.1074/jbc.M207880200. [DOI] [PubMed] [Google Scholar]
- 7.Fiorillo C, et al. Beneficial effects of poly (ADP-ribose) polymerase inhibition against the reperfusion injury in heart transplantation. Free Radic Res. 2003;37:331–339. doi: 10.1080/1071576021000055262. [DOI] [PubMed] [Google Scholar]
- 8.Cohen HY, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 9.Subramanian C, Opipari AW, Jr, Bian X, Castle VP, Kwok RP. Ku70 acetylation mediates neuroblastoma cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:4842–4847. doi: 10.1073/pnas.0408351102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazumder S, Plesca D, Kinter M, Almasan A. Interaction of a cyclin e fragment with ku70 regulates bax-mediated apoptosis. Mol Cell Biol. 2007;27:3511–3520. doi: 10.1128/MCB.01448-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SM, et al. Bcr-Abl-independent imatinib-resistant K562 cells show aberrant protein acetylation and increased sensitivity to histone deacetylase inhibitors. J Pharmacol Exp Ther. 2007;322:132–137. doi: 10.1124/jpet.107.124461. [DOI] [PubMed] [Google Scholar]
- 12.Gu Y, et al. Defective embryonic neurogenesis in Ku-deficient but not DNA-dependent protein kinase catalytic subunit-deficient mice. Proc Natl Acad Sci USA. 2000;97:2668–2673. doi: 10.1073/pnas.97.6.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li GC, et al. Ku70: a candidate tumor suppressor gene for murine T cell lymphoma. Mol Cell. 1998;2:1–8. doi: 10.1016/s1097-2765(00)80108-2. [DOI] [PubMed] [Google Scholar]
- 14.Li B, Dou QP. Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proc Natl Acad Sci USA. 2000;97:3850–3855. doi: 10.1073/pnas.070047997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajkumar SV, Richardson PG, Hideshima T, Anderson KC. Proteasome inhibition as a novel therapeutic target in human cancer. J Clin Oncol. 2005;23:630–639. doi: 10.1200/JCO.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 16.Chechlacz M, Vemuri MC, Naegele JR. Role of DNA-dependent protein kinase in neuronal survival. J Neurochem. 2001;78:141–154. doi: 10.1046/j.1471-4159.2001.00380.x. [DOI] [PubMed] [Google Scholar]
- 17.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, et al. Bax-inhibiting peptide protects cells from polyglutamine toxicity caused by Ku70 acetylation. Cell Death Differ. 2007;14:2058–2067. doi: 10.1038/sj.cdd.4402219. [DOI] [PubMed] [Google Scholar]
- 19.Gomez JA, et al. Bax-inhibiting peptides derived from Ku70 and cell-penetrating pentapeptides. Biochem Soc Trans. 2007;35:797–801. doi: 10.1042/BST0350797. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, et al. Bax-inhibiting peptide protects cells from polyglutamine toxicity caused by Ku70 acetylation. Cell Death Differ. 2007;14:2058–2067. doi: 10.1038/sj.cdd.4402219. [DOI] [PubMed] [Google Scholar]
- 21.Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 23.Toledo F, Wahl GM. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 24.Blander G, et al. SIRT1 shows no substrate specificity in vitro. J Biol Chem. 2005;280:9780–9785. doi: 10.1074/jbc.M414080200. [DOI] [PubMed] [Google Scholar]
- 25.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 26.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 27.Watson IR, Irwin MS. Ubiquitin and ubiquitin-like modifications of the p53 family. Neoplasia. 2006;8:655–666. doi: 10.1593/neo.06439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitsiades CS, Mitsiades N, Hideshima T, Richardson PG, Anderson KC. Proteasome inhibition as a therapeutic strategy for hematologic malignancies. Expert Rev Anticancer Ther. 2005;5:465–476. doi: 10.1586/14737140.5.3.465. [DOI] [PubMed] [Google Scholar]
- 30.Dikic I, Crosetto N, Calatroni S, Bernasconi P. Targeting ubiquitin in cancers. Eur J Cancer. 2006;42:3095–3102. doi: 10.1016/j.ejca.2006.05.041. [DOI] [PubMed] [Google Scholar]
- 31.Kortenhorst MS, Carducci MA, Shabbeer S. Acetylation and histone deacetylase inhibitors in cancer. Cell Oncol. 2006;28:191–222. doi: 10.1155/2006/760183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchion D, Munster P. Development of histone deacetylase inhibitors for cancer treatment. Expert Rev Anticancer Ther. 2007;7:583–598. doi: 10.1586/14737140.7.4.583. [DOI] [PubMed] [Google Scholar]
- 33.Sui G, et al. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen HY, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 35.Affar EB, Shah RG, Poirier GG. Poly(ADP-ribose) turnover in quail myoblast cells: relation between the polymer level and its catabolism by glycohydrolase. Mol Cell Biochem. 1999;193:127–135. [PubMed] [Google Scholar]
- 36.Chen Z, et al. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J Virol. 2007;81:6007–6018. doi: 10.1128/JVI.02747-06. [DOI] [PMC free article] [PubMed] [Google Scholar]