Abstract
Parathyroid hormone (PTH) is central to calcium homeostasis and bone maintenance in vertebrates, and as such it has been used for treating osteoporosis. It acts primarily by binding to its receptor, PTH1R, a member of the class B G protein-coupled receptor (GPCR) family that also includes receptors for glucagon, calcitonin, and other therapeutically important peptide hormones. Despite considerable interest and much research, determining the structure of the receptor–hormone complex has been hindered by difficulties in purifying the receptor and obtaining diffraction-quality crystals. Here, we present a method for expression and purification of the extracellular domain (ECD) of human PTH1R engineered as a maltose-binding protein (MBP) fusion that readily crystallizes. The 1.95-Å structure of PTH bound to the MBP-PTH1R-ECD fusion reveals that PTH docks as an amphipathic helix into a central hydrophobic groove formed by a three-layer α–β–βα fold of the PTH1R ECD, resembling a hot dog in a bun. Conservation in the ECD scaffold and the helical structure of peptide hormones emphasizes this hot dog model as a general mechanism of hormone recognition common to class B GPCRs. Our findings reveal critical insights into PTH actions and provide a rational template for drug design that targets this hormone signaling pathway.
Keywords: calcitonin, crystal structure, glucagon, maltose-binding protein, osteoporosis
Parathyroid hormone (PTH) is a classic endocrine hormone that was first identified more than 80 years ago as a key regulator of blood calcium levels (1–3). Decades of research have established PTH as a major systemic hormone that controls mineral ion homeostasis and bone maintenance (4, 5). In response to either low calcium or high phosphate levels in the circulation, PTH is synthesized and secreted into the bloodstream as an 84-amino acid polypeptide from the chief cells of the parathyroid glands (6, 7). The primary actions of PTH occur in kidney and bone tissues. In kidney, PTH promotes renal tubular calcium resorption and synthesis of 1,25-dihydroxyvitamin D3 but prevents resorption of phosphate (8). In bone, PTH increases osteoclastic bone resorption as part of calcium homeostasis, and the stimulatory effects of PTH on osteoblasts increase bone mass (9). Daily administration of PTH, or its N-terminal 34-residue fragment PTH-(1–34), enhances bone formation and increases bone density in patients with osteoporosis (10, 11). Both versions of PTH have been used in clinics as a treatment for osteoporosis in recent years, which has provided a major push for research programs on PTH as related to bone and mineral ion homeostasis. Notably, the activation of PTH signaling in osteoblast cells is also critical in providing an environmental niche for the maintenance of hematopoietic stem cells in bone marrow (12).
The pleiotropic effects of PTH are mediated primarily through the binding and activation of the PTH receptor (PTH1R), which is highly expressed in PTH target tissues (13). PTH also binds to a second receptor (PTH2R) with a distinct pharmacology (for review, see ref. 14). Both PTH receptors belong to the class B G protein-coupled receptors (GPCRs). As a class B GPCR, PTH1R contains an N-terminal extracellular domain (ECD) with three conserved disulfide bonds and a C-terminal domain with seven transmembrane helices. In addition to PTH, PTH1R also recognizes PTH-related peptide (PTHrP), a paracrine/autocrine factor originally discovered in many tumors that causes the syndrome of malignancy-associated hypercalcemia (15–18). PTHrP is developmentally regulated and expressed, and it has been shown to play a vital role in bone formation during embryogenesis (19).
There exists a large body of data regarding the structure–function relationships of PTH and PTHrP and their binding to PTH1R, a consequence of the pervasive role of these molecules in bone maintenance and their therapeutic applications for osteoporosis (for review, see ref. 20). The N-terminal 34-residue fragments of PTH and PTHrP [PTH-(1–34) and PTHrP-(1–34)] are capable of fully activating the receptors to the same degree as the full-length PTH and PTHrP-(1–141) (21, 22). NMR studies reveal that PTH-(1–34) and PTHrP-(1–34) share a similar U-shaped tertiary structure (23, 24), whereas the x-ray crystal structure of PTH-(1–34) shows a single continuous helix (25). Studies with ligand variants and receptor/ligand photo cross-linking indicate that the N-terminal fragment of PTH (residues 1–14) binds to the transmembrane domain, albeit with low affinity, and that it is able of activating the receptor (26, 27). The C-terminal fragment of PTH (residues 15–34) binds to the N-terminal ECD of the receptor to confer high affinity and specificity to the receptor (28, 29). This “two-domain” model of PTH binding and activation is further supported by studies with chimeric ligands and receptors (30–32) and has since been demonstrated for other class B GPCR molecules (33–35).
In addition to PTH receptors, the family of class B GPCRs also includes receptors for glucagon, glucagon-like peptides (GLPs), glucose-dependent insulinotropic polypeptide (GIP), calcitonin, growth hormone-releasing hormone, corticotropin-releasing factor (CRF), and other therapeutically important peptides hormones (36). Given the pharmaceutical importance of these receptors, there has been intensive interest in elucidating the structure of their hormone-bound complexes. However, these efforts have been hampered by technical difficulties in purifying soluble and functional proteins in the large quantities required for structural studies. Very recently, the structures of the receptor ECD–ligand complexes have been reported for the murine CRF receptor 2β (CRFR2β) and the human GIPR (37, 38). These structures reveal a similar scaffold of two antiparallel β-sheets, resembling a short consensus repeat (SCR) fold found in immunoglobins (37, 38). Except for a few core residues that form the conserved SCR fold, the ECD of PTH1R shares little sequence homology with that of CRFR2β and GIPR. Thus, a detailed understanding of PTH binding and activation of PTH1R requires a high-resolution structure of a PTH1R receptor–ligand complex.
In this work, we present a general methodology that we developed for expression and purification of the ECD of class B GPCRs, which allowed us to crystallize and solve the PTH–PTH1R ECD structure at 1.95-Å resolution. The structure reveals a “hot dog” model of PTH binding, in which PTH is docked as a straight helix into the central groove of a three-layer fold adopted by the receptor. The detailed interactions from this structure provide an explanation for the binding specificity of PTH to PTH1R and a rational template for optimizing PTH derivatives for therapeutic applications.
Results and Discussion
General Method for Expression, Purification, and Crystallization of the ECD of Class B GPCRs.
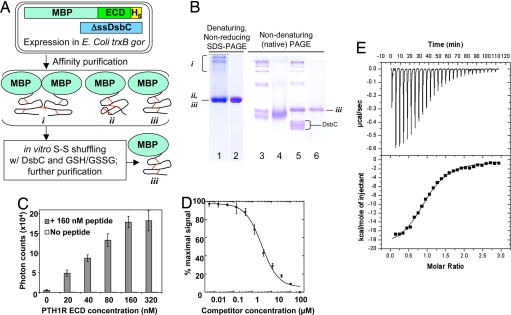
For structural studies of the peptide hormone–receptor ECD complex, we aimed to develop a general method for bacterial expression and purification of the PTH1R ECD that is applicable to all class B GPCRs. This method has five key features (Fig. 1A). First, the PTH1R ECD was fused with the bacterial maltose-binding protein (MBP) at the N terminus to increase protein solubility because MBP is able to improve the stability of the fusion proteins (39). Second, the MBP-PTH1R-ECD fusion contains a C-terminal hexahistidine tag. The inclusion of affinity tags at both ends of the PTH1R ECD ensures purification of the full-length product. Third, the fusion protein was produced in a bacterial strain with mutations in the gor and trxB genes (40), which creates an oxidized cytoplasm to facilitate disulfide bond formation within the PTH1R ECD. We used the same bacterial strain to produce a correctly folded hepatocyte growth factor NK1 fragment, which contains five disulfide bonds (41). Fourth, the fusion protein was coexpressed with DsbC (40), a bacterial disulfide isomerase, which has chaperone activity to further promote correct folding and disulfide bond formation. Despite coexpression with DsbC, much of the ECD was misfolded, as evidenced by the presence of high-molecular-weight species on a nonreducing denaturing gel (lane 1 in Fig. 1B) or multiple species on a native gel (lane 3 in Fig. 1B). To overcome this problem, and as the fifth key feature, the affinity-purified fusion protein was incubated with purified DsbC in the presence of reduced and oxidized glutathione to promote shuffling of the disulfide bonds within the ECD. Overnight incubation of this reaction dramatically reduced the amount of misfolded species (Fig. 1B, lane 5), allowing further purification of the MBP-PTH1R-ECD to a single band in a native gel, indicating its chemical and conformational homogeneity (lane 6 in Fig. 1B).
Fig. 1.
Purification and function of the MBP-PTH1R-ECD fusion. (A) Overview of the method, in which an MBP-ECD fusion protein is coexpressed with DsbC in the oxidizing cytoplasm of an E. coli trxB gor host. Affinity purification of the fusion protein yields multiple protein species, e.g.: (i) multimers linked by intermolecular disulfide bonds; (ii) misfolded monomer; (iii) monomeric, correctly folded protein. The mixture is subjected to in vitro disulfide shuffling in a redox buffer in the presence of DsbC to increase the percentage of the total protein present as species iii, which can be purified to homogeneity. (B) Gel analysis of MBP-PTH1R-ECD-H6 at various stages of purification. Lanes 1, 3, and 4 show samples from affinity purification via the His6 and MBP tags, with lane 4 containing 20 mM DTT. Lane 5 shows a disulfide shuffling reaction incubated overnight at 20°C in the presence of 1 mM each GSH and GSSG, and DsbC. Lanes 2 and 6 show the final purified sample after removing DsbC and remaining misfolded fusion protein. (C) AlphaScreen assay for the association of biotin-PTH-(7–34)-NH2 with the purified MBP-PTH1R-ECD at 20°C. The reaction mixtures contained 15 μg/ml each streptavidin-coated donor beads and Ni-chelate-coated acceptor beads, and protein and peptide as indicated. The data represent the average of duplicate samples. (D) AlphaScreen assay for the ability of PTH(15–34)-NH2 to compete with the interaction of biotin-PTH(7–34)-NH2 and MBP-PTH1R-ECD-His6. The data represent the average of three independent experiments, and the curve yielded an IC50 value of 3.0 μM. (E) Isothermal titration calorimetry analysis of PTH-(15–34)-NH2 binding to the MBP-PTH1R-ECD at 27°C. (Upper) Raw data after subtraction of the peptide heats of dilution. (Lower) Enthalphy vs. the PTH to MBP-PTH1R-ECD-His6 ratio. The best fit of the data yielded a Kd value of 0.98 μM; number of binding sites, 0.985; ΔH = −20,190 calories per mol; and ΔS = −39.8 calories per mol per degree.
The functional activity of the purified protein was assessed by its direct binding to PTH as determined by AlphaScreen assays and isothermal titration calorimetry (ITC). In the AlphaScreen assays, biotinylated PTH was attached to streptavidin-coated donor beads, and the MBP-PTH1R-ECD was attached to nickel-chelate-coated acceptor beads via the C-terminal His6 tag. Incubation of PTH and the purified MBP-PTH1R ECD produced a dose-dependent binding signal, suggesting their direct interaction (Fig. 1C). The affinity of PTH binding to the PTH1R ECD was 1–3 μM, as determined by competition with an unlabeled PTH (Fig. 1D) or by isothermal titration calorimetry (Fig. 1E). This affinity is comparable with that of other peptide hormones for their cognate class B GPCR ECDs (37, 38). The purified MBP-PTH1R-ECD readily produced diffraction-quality crystals in complex with a synthetic PTH fragment (residues 15–34), as described in Methods. A similar protocol was also used successfully to produce protein and crystals of the ECD of the human CRFR1, suggesting that the methodology we derived above is applicable to other class B GPCRs (unpublished results).
The key features of our method are that the targeted protein is affinity-tagged at both ends to ensure the purification of the intact protein and a disulfide-bond shuffling system that does not involve any denaturing/refolding steps. Furthermore, the MBP tag is particularly suitable for crystallization, as successfully demonstrated by a number of fusion proteins for which MBP is critical for crystal packing (42). In our crystals, the MBP tag is also essential for the high quality of the crystals by mediating extensive contacts with the PTH1R ECD or with MBP itself [supporting information (SI) Fig. S1]. The key components of this expression system are flexible, so that our method should be generally applicable to protein production and crystallization of other cell surface receptors or their ligands that contain disulfide bonds.
Overall Structure of the PTH1R ECD Bound to PTH.
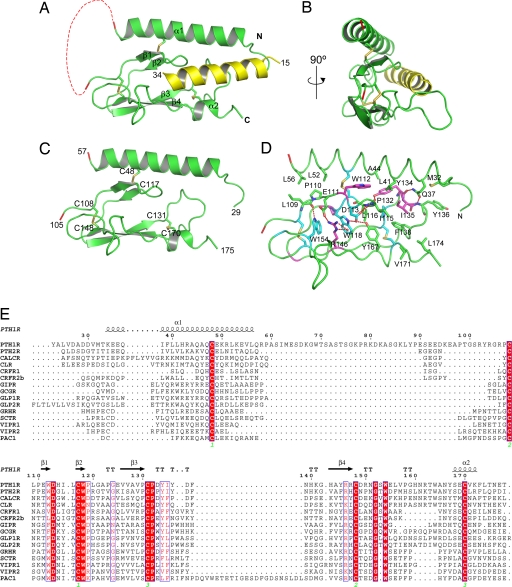
The crystals containing the MBP-PTH1R-ECD fusion protein bound to PTH-(15–34) formed in the P21 space group with two complexes in each asymmetry unit. The structure was solved by molecular replacement using the MBP structure and refined to an R factor of 18.6% at 1.95-Å resolution (43, 44) (Table S1). The PTH1R ECD residues 56–105 and 175–187 of molecule A and residues 58–104 and 176–187 of molecule B were excluded from the final model because of disorder. The two ECD–PTH complexes are very similar as indicated by the rmsd of their Cα atom positions of 0.275 Å; thus, we confine our description of the structure to the complex with molecule B. In the structure, the PTH1R ECD contains two α-helices and four β-strands, and PTH adopts a single continuous helix (Fig. 2A). From the front view, the secondary structure elements of the PTH1R ECD are arranged into a three-layer α–β–βα fold with approximate dimensions of 40 Å × 25 Å × 10 Å (Fig. 2C). The top layer is an extended α-helix formed by the N-terminal portion of the PTH1R ECD (residues 33–57). The middle layer is divided into two halves: the left half is formed by a β-hairpin of the first two β-strands (residues 110–120), and the right half is formed by the extended turn of β-strands 3 and 4 (residues 130–140). The bottom layer is formed by β-strands 3 and 4, α2, and the loop connecting β4 to α2. From a side view, the thickness of the PTH1R ECD is only ≈10 Å (Fig. 2B). The thinness of the PTH1R ECD suggests that it cannot form a large hydrophobic core sufficient to stabilize the ECD structure. Thus, the overall structure of the PTH1R ECD is primarily held together by three interlayer disulfide bonds (Fig. 2C): the first (C48–C117) links the top N-terminal helix to the middle layer β-sheet, the second (C108–C148) links the middle and the bottom β-sheets, and the third (C131–C170) links the middle layer β-turn of strands 3 and 4 with the C-terminal α-helix at the bottom.
Fig. 2.
Structure of the PTH–PTH1R ECD complex. (A and B) Two views of the complex with the ECD in green, PTH in yellow, and the disulfide bonds depicted as sticks. The dashed red line designates the break in the chain caused by the disordered loop between residues 57 and 105. For clarity, MBP is not shown in this and all subsequent figures. All structural figures were prepared with PyMol (55). (C) Overview of the three-layer α–β–βα structure of the PTH1R ECD with PTH removed. (D) Residues that stabilize the ECD fold. The carbon atoms are colored based on amino acid sequence conservation among the 15 human class B GPCR ECDs. Cyan indicates residues that are invariant, magenta indicates residues that have conservative substitutions, and green indicates residues that are not conserved. Hydrogen bonds are depicted as red dashes, and the red sphere is a water molecule. (E) Sequence alignment of human class B GPCR ECDs with the secondary structural elements noted over the sequences. Invariant residues are shown in white lettering on a red background, and conservative substitutions are shown in red lettering.
The topological arrangement of the PTH1R ECD is further supported by interlayer hydrophobic packing interactions centering on the middle β-hairpin and the extended turn of β-strands 3 and 4 (Fig. 2D). The most prominent interactions consist of a series of stepwise packing interactions, starting with the C108–C148 disulfide bond at the left, followed by W154, R146, W118, Y167, the C131–C170 disulfide bond, L116, P132, F138, and I135 to the right (Fig. 2D). These interactions help to hold the structure of the middle and the bottom layers, which are further strengthened by a series of interlayer H-bonds involving E111, D113, W118, R46, and W154. Residues involved in these interactions are mostly conserved among class B GPCRs, including D113, W118, P132, W154, and the six invariant cysteine residues (Fig. 2E). These residues constrain the bottom two β-sheets into a fold that resembles the short consensus repeat, a small protein-binding module found in the immunoglobin superfamily and in the ECDs of class B GPCRs (37, 45). Interestingly, mutation of P132 to a leucine in human PTH1R causes Blomstrand chondrodysplasia, a lethal genetic disorder (46), suggesting that the integrity of these interactions is important for PTH–PTH1R signaling.
Structural Basis for the Binding and Specificity of PTH to PTH1R.
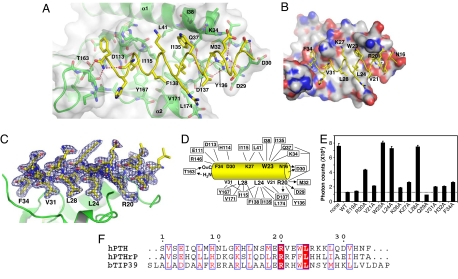
PTH binds to the PTH1R ECD as a single amphipathic α-helix that docks into an extended hydrophobic groove formed by the center-right half of the ECD domain (Fig. 3 A and B). In this mode of PTH binding, the PTH helix resembles a hot dog sitting between the two outer layers (the bun) of the three-layer ECD (Fig. 2 A and B). The binding of PTH to PTH1R ECD buries a solvent-accessible area of ≈1,900 Å2. The center of the PTH-binding groove is formed by the tip of the β-hairpin and the extended β-turn in the middle layer, and the top and the bottom ridges of the groove are formed by the N- and C-terminal helices, respectively. The PTH helix is approximately antiparallel to the C-terminal helix of the ECD such that the N terminus of PTH is predicted to orient toward the transmembrane domain of the full-length receptor, consistent with the two-domain model in which the PTH N-terminal residues activate the receptor.
Fig. 3.
Structural and biochemical analysis of the PTH–ECD interaction. (A) Detail of the PTH–ECD interface. The PTH backbone is shown as a yellow coil and selected side chains as sticks. The ECD is shown as a green ribbon diagram covered by a transparent molecular surface. ECD atoms within 8 Å of PTH are shown in stick representation. Hydrogen bonds are indicated by red dashes, and the red sphere is a water molecule. (B) Molecular surface of the ECD showing the hydrophobic patch that contacts W23, L24, L28, and V31 of PTH. Carbon atoms are colored gray, nitrogen atoms blue, and oxygen atoms red. (C) The σA weighted 2Fo − Fc electron density map for PTH shown as a blue mesh contoured at 1 σ. (D) Interactions between PTH and the ECD where ECD residues are boxed. The lines indicate hydrophobic or van der Wall contacts, and the arrows indicate hydrogen bonds from donors to acceptors. (E) AlphaScreen assay for the ability of alanine scan mutants of PTH-(15–34)-NH2 (100 μM) to compete with the binding of the biotin-PTH-(7–34)-NH2 (40 nM) to the MBP-PTH1R-ECD (40 nM). The data represent the average of duplicate samples. (F) Sequence comparison of human PTH, PTHrP, and bovine TIP39.
Because of the exceptional quality of the electron density map, detailed PTH–PTH1R interactions can be clearly defined in the crystal structure (Fig. 3 C and D). The binding of PTH to the PTH1R ECD appears to be dominated by hydrophobic interactions. Residues V21, W23, L24, L28, V31, and F34, which form an extended hydrophobic face of the PTH helix, pack closely against the central hydrophobic groove formed by the three-layer ECD fold (Fig. 3 A and B). Notably, none of the residues that form the PTH-binding groove is conserved in class B GPCRs, suggesting a unique surface topology for each individual receptor. We examined the surface complementarity between PTH and the PTH1R ECD by using the shape correlation statistic, Sc (47). The Sc value for the complex of 0.782 indicates a high degree of surface topology matching at the interface. Thus, the binding specificity of PTH to PTH1R can be determined in part by complementary shape-matching between the hormone and the receptor. In addition, most of the binding groove residues are conserved between PTH1R and PTH2R, thus providing a basis to account for their similar ligand-binding specificities.
The binding of PTH to the PTH1R ECD is further strengthened by hydrogen bonds at both ends of the PTH helix (Fig. 3D). The amidated C-terminal group forms two H-bonds with the backbone amide and carbonyl of T163. The amidated group also caps the backbone carbonyl of V31 of PTH, thus helping to stabilize the helical conformation. The C-terminal amidation is a general feature of many peptide hormones, and the hydrogen bonds described here help to explain its importance in receptor binding. The N-terminal backbone amide of PTH is capped by the PTH1R residue D30, which also forms a direct H-bond with the side chain of N16 of PTH. Most importantly, R20 of PTH forms a pair of charged interactions with D137 and two H-bonds with the backbone carbonyls of D29 and M32 of PTH1R (Fig. 3 A and D). These polar interactions are particularly important to the binding affinity and specificity of PTH to PTH1R because R20 is completely buried within the receptor–hormone interface (Fig. 3A). Furthermore, the residues that contact R20 are conserved in PTH1R and PTH2R but not in most other class B GPCRs, suggesting that R20 is a key determinant of ligand specificity for PTH and PTH-related ligands.
To validate the structural observations above, we examined the PTH–PTH1R interactions by alanine-scanning mutations of all PTH residues that contact PTH1R. We also chose two residues (E19 and Q29) that do not contact PTH1R as controls. As shown in Fig. 3E, addition of the wild-type unlabeled PTH inhibited >80% of the binding of biotinylated PTH to the MBP-PTH1R ECD. Replacement of any residues in the structure that contact PTH1R affected PTH binding, whereas replacement of the two control residues had little effect. In particular, replacement of the four core residues (R20, W23, L24, and L28) with alanine markedly reduced PTH binding, with mutations in the three hydrophobic residues nearly abolishing PTH binding to the ECD. Our data are in good agreement with the recent study of alanine-scanning of PTH-(17–31) (29). These results thus further support the detailed molecular interactions observed in our crystal structure. The four core residues are well conserved in PTH, PTHrP, and TIP39 (48), a brain ligand for PTH2R, suggesting that the structure presented here can serve as a prototype for their hormone-receptor interactions (Fig. 3F).
A Hot Dog Model for Hormone Recognition by Class B GPCRs.
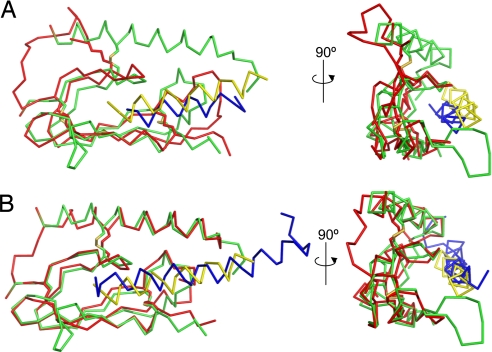
A common structural mechanism of hormone recognition appears to emerge from the structure comparison of the PTH–PTH1R ECD complex with two recently published structures of CRFR2β and GIPR (37, 38) (Fig. 4). The bottom β–β arrangement of the SCR fold is similar in all three ECD structures. Structural alignment of the CRFR2β and GIPR ECDs with the PTH1R ECD yields rmsd values for their Cα atom positions of 2.082 Å and 1.164 Å, respectively. The arrangement of the three disulfide bonds and the packing of the hydrophobic core by key residues (D113, W118, P132, R146, and W154) are also conserved among these three hormone-receptor structures (Fig. 2E). Although the NMR structure of the mouse CRFR2β ECD does not contain the N-terminal helix, our recent crystal structure of the human CRFR1 ECD reveals that it contains an N-terminal helix and a C-terminal short α-helix (unpublished data). Thus, the α–β–βα fold may represent a common structure fold adopted by the ECDs of all class B GPCRs.
Fig. 4.
Comparison of the PTH1R ECD–PTH complex with the mouse CRFR2β ECD–astressin and human GIPR ECD–GIP complexes. (A) Structural alignment between the PTH1R ECD–PTH complex and the NMR solution structure of the mouse CRFR2β ECD bound to astressin (PDB code 2JND). Cα backbone traces are shown. The structures were aligned with PyMol based on the ECD structures. The PTH1R ECD is green and PTH is yellow; the CRFR2β ECD is colored red and astressin blue. (B) Alignment with the crystal structure of the GIPR ECD–GIP complex (PDB code 2QKH). The color scheme is as in A.
Besides the common fold of the receptor ECD, the helical structure of PTH is also observed for the GIP peptide and the CRFR antagonist astressin (37, 38). In each of these structures, the peptide helix binds the middle of the three layers of the ECD structure, resembling a hot dog, with the N-terminal helix and the C-terminal β-sheet serving as the top and bottom bun halves. Because the helical nature has been extensively characterized and documented for many peptide hormones (36), we propose that this hot dog model is generally applicable for ligand–receptor interactions of all class B GPCRs.
Concluding Remarks.
As a classic endocrine hormone, PTH and its receptors have been the subject of intensive study over the last few decades. The high-resolution structure of PTH bound to the PTH1R ECD reveals a hot dog model of hormone recognition, in which the PTH1R ECD assumes a three-layer α–β–βα fold, and PTH adopts an amphipathic helix that fits into the central hydrophobic groove of the receptor ECD. The α–β–βα fold is conserved among class B GPCRs but is distinct from the ECD structures of class A or class C GPCRs (49, 50), illustrating that these three families have evolved different mechanisms of hormone recognition. Because of the helical nature of peptide hormones and the conserved α–β–βα fold, the hot dog model of ligand recognition represents a general mechanism common to all class B GPCRs. Given its pharmaceutical importance, the PTH-PTH1R ECD structure should provide a rational template for designing better PTH mimics for the treatment of osteoporosis and hypercalcemia. Finally, the methodology that we developed is applicable for structural studies of other class B GPCRs, which will expedite our pursuit of the mechanisms of hormone recognition by these therapeutically important receptors.
Methods
Protein Production.
The human PTH1R ECD [residues 29–187, which excludes the native signal peptide (residues 1–22) (51)] was expressed as a fusion protein with MBP and His6 tags at its N and C termini, respectively, from the expression vector pET-Duet1 in the strain Escherichia coli Origami (DE3) (Novagen), which contains trxB gor mutations that promote disulfide bond formation. Details of the plasmid construction are described in SI Methods. Cells harboring this expression plasmid were grown in LB broth to midlog phase at 37°C, cooled to 16°C, and induced with 0.4 mM IPTG for ≈18 h. The cells were harvested and resuspended in buffer C [50 mM Tris·HCl (pH 7.5), 10% (vol/vol) glycerol, 150 mM NaCl, 25 mM imidazole] and then were lysed by homogenization at 10,000 p.s.i. with an Invensys APV homogenizer (Albertslund). The lysate was centrifuged, and the supernatant was loaded on a 50-ml Ni2+-chelating Sepharose column (GE Healthcare). The column was washed with 400 ml of buffer C and eluted with buffer D, containing 50 mM Tris·HCl (pH 7.5), 10% (vol/vol) glycerol, 150 mM NaCl, and 250 mM imidazole.
The peak fractions were pooled and loaded on a 50-ml Amylose column (New England Biolabs). The column was washed with 100 ml of buffer E [50 mM Tris·HCl (pH 7.5), 5% (vol/vol) glycerol, 150 mM NaCl, and 0.5 mM EDTA] and eluted with a linear gradient of 0–10 mM maltose in buffer E. The peak fractions were pooled and subjected to disulfide shuffling in the presence of oxidized (GSSG) and reduced (GSH) glutathione (Sigma) and DsbC purified as described in SI Methods. The shuffling reaction contained 1 mg/ml MBP-PTH1R-ECD-His6 in 50 mM Tris·HCl (pH 7.5), 5% (vol/vol) glycerol, 150 mM NaCl, 0.5 mM EDTA, ≈3 mM maltose, 1 mM GSH, 1 mM GSSG, and DsbC at a 1:1 molar ratio between MBP-PTH1R-ECD-His6 and DsbC. After incubation at 20°C for ≈20 h, the disulfide shuffling reaction mixture was applied to a Ni2+-chelating Sepharose column to remove the DsbC.
The peak fractions eluted from the column were applied to a gel filtration column followed by a QFF anion exchange column, to remove remaining misfolded MBP-PTH1R-ECD-His6. The functional protein eluted from the gel filtration column at a volume corresponding to the monomer. At this point, the protein was >95% pure as judged by SDS/PAGE and native/PAGE. The final protein was dialyzed to a storage buffer containing 50 mM Tris·HCl (pH 7.5), 50% (vol/vol) glycerol, 150 mM NaCl, and 1 mM EDTA and was stored as aliquots at −80°C. A yield of ≈25 mg was obtained from a 6-liter culture.
Binding Assays.
The binding between PTH and the PTH1R ECD was determined by an AlphaScreen assay using a hexahistidine detection kit from PerkinElmer (52). The binding mixtures, containing 40 nM N-terminally biotinylated PTH-(7–34)-NH2, 40 nM of MBP-PTH1R-ECD-His6, 15 μg/ml of streptavidin-coated “donor” beads, and Ni-chelate-coated “acceptor” beads, were incubated in a buffer of 50 mM Mops (pH 7.4), 100 mM NaCl, and 0.1 mg/ml BSA for 1 h. For competition assays, 100 μM unlabeled peptides were added and incubated for 1 additional h before signal recording. Details of peptide synthesis and ITC experiments are described in SI Methods.
Crystallization, Data Collection, and Structure Determination.
Crystals of the purified MBP-PTH1R-ECD bound to a synthetic PTH fragment (residues 15–34) were grown in hanging drops containing a reservoir solution of 100 mM sodium cacodylate (pH 6.5) and 30% (vol/vol) polypropylene glycol P400 (PPG P400). Diffraction data were collected at 21-ID-D (LS-CAT) of the Advanced Photon Source at Argonne National Laboratory (Argonne, IL). The structure was solved by molecular replacement using Phaser (53) with the Protein Data Bank (PDB) coordinates 1ANF (44) and was refined with Refmac5 (54). The statistics of data collection and the refined structure are summarized in Table S1. Details of crystallization, data collection, and structure determination are described in SI Methods.
Acknowledgments.
We thank D. Nadziejka for professional editing of the manuscript; Z. Wawrzak and S. Anderson from LS-CAT for assistance in data collection at sector 21 of the Advanced Photon Source. Use of the Advanced Photon Source was supported by the Office of Science of the U.S. Department of Energy. This work was supported in part by the Jay and Betty Van Andel Foundation, Department of Defense Grant W81XWH0510043, Michigan Economic Development Corporation Grant 085P1000817, and National Institutes of Health Grants DK071662, DK066202, and HL089301.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3C4M).
This article contains supporting information online at www.pnas.org/cgi/content/full/0801027105/DCSupplemental.
References
- 1.Collip JB. The extraction of a parathyroid hormone that will prevent or control parathyroid tetany and that regulates the level of blood calcium. J Biol Chem. 1925;63:395–438. [Google Scholar]
- 2.Greenwald I, Gross J. The effect of the administration of a potent parathyoid extract upon the excretion of nitrogen, phosphorus, calcium, and magnesium, with some remarks on the solubility of calcium phosphate in serum and on the pathogenesis of tetany. J Biol Chem. 1925;66:217–227. [Google Scholar]
- 3.Murray TM, Rao LG, Divieti P, Bringhurst FR. Parathyroid hormone secretion and action: Evidence for discrete receptors for the carboxyl-terminal region and related biological actions of carboxyl-terminal ligands. Endocr Rev. 2005;26:78–113. doi: 10.1210/er.2003-0024. [DOI] [PubMed] [Google Scholar]
- 4.Agus ZS, Gardner LB, Beck LH, Goldberg M. Effects of parathyroid hormone on renal tubular reabsorption of calcium, sodium, and phosphate. Am J Physiol. 1973;224:1143–1148. doi: 10.1152/ajplegacy.1973.224.5.1143. [DOI] [PubMed] [Google Scholar]
- 5.Raisz LG. Stimulation of bone resorption by parathyroid hormone in tissue culture. Nature. 1963;197:1015–1016. doi: 10.1038/1971015a0. [DOI] [PubMed] [Google Scholar]
- 6.Niall HD, et al. The amino acid sequence of bovine parathyroid hormone. I. Hoppe-Seyler's Zeitschr Physiolog Chemie. 1970;351:1586–1588. [PubMed] [Google Scholar]
- 7.Brewer HB, Jr, Ronan R. Bovine parathyroid hormone: Amino acid sequence. Proc Natl Acad Sci USA. 1970;67:1862–1869. doi: 10.1073/pnas.67.4.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pullman TN, Lavender AR, Aho I, Rasmussen H. Direct renal action of a purified parathyroid extract. Endocrinology. 1960;67:570–582. doi: 10.1210/endo-67-5-570. [DOI] [PubMed] [Google Scholar]
- 9.Barnicot NA. The local action of the parathyroid and other tissues on bone in intracerebral grafts. J Anat. 1948;82:232–248. [PMC free article] [PubMed] [Google Scholar]
- 10.Moen MD, Scott LJ. Recombinant full-length parathyroid hormone (1–84) Drugs. 2006;66:2371–2381. doi: 10.2165/00003495-200666180-00008. [DOI] [PubMed] [Google Scholar]
- 11.Neer RM, et al. Effect of parathyroid hormone (1–34) on fractures and bone mineral density in postmenopausal women with osteoporosis. N Engl J Med. 2001;344:1434–1441. doi: 10.1056/NEJM200105103441904. [DOI] [PubMed] [Google Scholar]
- 12.Calvi LM, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 13.Juppner H, et al. A G protein-linked receptor for parathyroid hormone and parathyroid hormone-related peptide. Science. 1991;254:1024–1026. doi: 10.1126/science.1658941. [DOI] [PubMed] [Google Scholar]
- 14.Gensure RC, Gardella TJ, Juppner H. Parathyroid hormone and parathyroid hormone-related peptide, and their receptors. Biochem Biophys Res Commun. 2005;328:666–678. doi: 10.1016/j.bbrc.2004.11.069. [DOI] [PubMed] [Google Scholar]
- 15.Moseley JM, et al. Parathyroid hormone-related protein purified from a human lung cancer cell line. Proc Natl Acad Sci USA. 1987;84:5048–5052. doi: 10.1073/pnas.84.14.5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suva LJ, et al. A parathyroid hormone-related protein implicated in malignant hypercalcemia: Cloning and expression. Science. 1987;237:893–896. doi: 10.1126/science.3616618. [DOI] [PubMed] [Google Scholar]
- 17.Burtis WJ, et al. Identification of a novel 17,000-dalton parathyroid hormone-like adenylate cyclase-stimulating protein from a tumor associated with humoral hypercalcemia of malignancy. J Biol Chem. 1987;262:7151–7156. [PubMed] [Google Scholar]
- 18.Klein RF, Strewler GJ, Leung SC, Nissenson RA. Parathyroid hormone-like adenylate cyclase-stimulating activity from a human carcinoma is associated with bone-resorbing activity. Endocrinology. 1987;120:504–511. doi: 10.1210/endo-120-2-504. [DOI] [PubMed] [Google Scholar]
- 19.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 20.Gardella TJ, Juppner H. Interaction of PTH and PTHrP with their receptors. Rev Endocr Metab Dis. 2000;1:317–329. doi: 10.1023/a:1026522619828. [DOI] [PubMed] [Google Scholar]
- 21.Moseley JM, Gillespie MT. Parathyroid hormone-related protein. Crit Rev Clin Lab Sci. 1995;32:299–343. doi: 10.3109/10408369509084687. [DOI] [PubMed] [Google Scholar]
- 22.Blind E, Raue F, Knappe V, Schroth J, Ziegler R. Cyclic AMP formation in rat bone and kidney cells is stimulated equally by parathyroid hormone-related protein (PTHrP) 1–34 and PTH 1–34. Exp Clin Endocrinol. 1993;101:150–155. doi: 10.1055/s-0029-1211222. [DOI] [PubMed] [Google Scholar]
- 23.Marx UC, et al. Structure of human parathyroid hormone 1–37 in solution. J Biol Chem. 1995;270:15194–15202. doi: 10.1074/jbc.270.25.15194. [DOI] [PubMed] [Google Scholar]
- 24.Barden JA, Kemp BE. NMR solution structure of human parathyroid hormone(1–34) Biochemistry. 1993;32:7126–7132. doi: 10.1021/bi00079a008. [DOI] [PubMed] [Google Scholar]
- 25.Jin L, et al. Crystal structure of human parathyroid hormone 1–34 at 0.9-A resolution. J Biol Chem. 2000;275:27238–27244. doi: 10.1074/jbc.M001134200. [DOI] [PubMed] [Google Scholar]
- 26.Gardella TJ, et al. Determinants of [Arg2]PTH-(1–34) binding and signaling in the transmembrane region of the parathyroid hormone receptor. Endocrinology. 1994;135:1186–1194. doi: 10.1210/endo.135.3.8070362. [DOI] [PubMed] [Google Scholar]
- 27.Luck MD, Carter PH, Gardella TJ. The (1–14) fragment of parathyroid hormone (PTH) activates intact and amino-terminally truncated PTH-1 receptors. Mol Endocrinol. 1999;13:670–680. doi: 10.1210/mend.13.5.0277. [DOI] [PubMed] [Google Scholar]
- 28.Potetinova Z, et al. C-terminal analogues of parathyroid hormone: Effect of C terminus function on helical structure, stability, and bioactivity. Biochemistry. 2006;45:11113–11121. doi: 10.1021/bi060500q. [DOI] [PubMed] [Google Scholar]
- 29.Dean T, Khatri A, Potetinova Z, Willick GE, Gardella TJ. Role of amino acid side chains in region 17–31 of parathyroid hormone (PTH) in binding to the PTH receptor. J Biol Chem. 2006;281:32485–32495. doi: 10.1074/jbc.M606179200. [DOI] [PubMed] [Google Scholar]
- 30.Bergwitz C, et al. Full activation of chimeric receptors by hybrids between parathyroid hormone and calcitonin: Evidence for a common pattern of ligand–receptor interaction. J Biol Chem. 1996;271:26469–26472. doi: 10.1074/jbc.271.43.26469. [DOI] [PubMed] [Google Scholar]
- 31.Stroop SD, Kuestner RE, Serwold TF, Chen L, Moore EE. Chimeric human calcitonin and glucagon receptors reveal two dissociable calcitonin interaction sites. Biochemistry. 1995;34:1050–1057. doi: 10.1021/bi00003a040. [DOI] [PubMed] [Google Scholar]
- 32.Holtmann MH, Hadac EM, Miller LJ. Critical contributions of amino-terminal extracellular domains in agonist binding and activation of secretin and vasoactive intestinal polypeptide receptors: Studies of chimeric receptors. J Biol Chem. 1995;270:14394–14398. doi: 10.1074/jbc.270.24.14394. [DOI] [PubMed] [Google Scholar]
- 33.Runge S, et al. Three distinct epitopes on the extracellular face of the glucagon receptor determine specificity for the glucagon amino terminus. J Biol Chem. 2003;278:28005–28010. doi: 10.1074/jbc.M301085200. [DOI] [PubMed] [Google Scholar]
- 34.Runge S, Wulff BS, Madsen K, Brauner-Osborne H, Knudsen LB. Different domains of the glucagon and glucagon-like peptide-1 receptors provide the critical determinants of ligand selectivity. Br J Pharmacol. 2003;138:787–794. doi: 10.1038/sj.bjp.0705120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du K, Couvineau A, Rouyer-Fessard C, Nicole P, Laburthe M. Human VPAC1 receptor selectivity filter: Identification of a critical domain for restricting secretin binding. J Biol Chem. 2002;277:37016–37022. doi: 10.1074/jbc.M203049200. [DOI] [PubMed] [Google Scholar]
- 36.Hoare SR. Mechanisms of peptide and nonpeptide ligand binding to Class B G protein-coupled receptors. Drug Discov today. 2005;10:417–427. doi: 10.1016/S1359-6446(05)03370-2. [DOI] [PubMed] [Google Scholar]
- 37.Parthier C, et al. Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor. Proc Natl Acad Sci USA. 2007;104:13942–13947. doi: 10.1073/pnas.0706404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grace CR, et al. Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand. Proc Natl Acad Sci USA. 2007;104:4858–4863. doi: 10.1073/pnas.0700682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bessette PH, Aslund F, Beckwith J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tolbert WD, et al. A mechanistic basis for converting a receptor tyrosine kinase agonist to an antagonist. Proc Natl Acad Sci USA. 2007;104:14592–14597. doi: 10.1073/pnas.0704290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smyth DR, Mrozkiewicz MK, McGrath WJ, Listwan P, Kobe B. Crystal structures of fusion proteins with large-affinity tags. Protein Sci. 2003;12:1313–1322. doi: 10.1110/ps.0243403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Navaza J. Implementation of molecular replacement in AmoRe. Acta Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 44.Quiocho FA, Spurlino JC, Rodseth LE. Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure. 1997;5:997–1015. doi: 10.1016/s0969-2126(97)00253-0. [DOI] [PubMed] [Google Scholar]
- 45.Grace CR, et al. NMR structure and peptide hormone-binding site of the first extracellular domain of a type B1 G protein-coupled receptor. Proc Natl Acad Sci USA. 2004;101:12836–12841. doi: 10.1073/pnas.0404702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang P, Jobert AS, Couvineau A, Silve CA. homozygous inactivating mutation in the parathyroid hormone/parathyroid hormone-related peptide receptor causing Blomstrand chondrodysplasia. J Clin Endocrinol Metab. 1998;83:3365–3368. doi: 10.1210/jcem.83.9.5243. [DOI] [PubMed] [Google Scholar]
- 47.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 48.Usdin TB. The PTH2 receptor and TIP39: A new peptide-receptor system. Trends Pharmacol Sci. 2000;21:128–130. doi: 10.1016/s0165-6147(00)01455-3. [DOI] [PubMed] [Google Scholar]
- 49.Fan QR, Hendrickson WA. Structure of human follicle-stimulating hormone in complex with its receptor. Nature. 2005;433:269–277. doi: 10.1038/nature03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunishima N, et al. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 51.Grauschopf U, et al. The N-terminal fragment of human parathyroid hormone receptor 1 constitutes a hormone-binding domain and reveals a distinct disulfide pattern. Biochemistry. 2000;39:8878–8887. doi: 10.1021/bi0001426. [DOI] [PubMed] [Google Scholar]
- 52.Suino K, et al. The nuclear xenobiotic receptor CAR: Structural determinants of constitutive activation and heterodimerization. Mol Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 53.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 54.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 55.DeLano WL. San Carlos, CA: DeLano Scientific; 2002. The PyMOL Molecular Graphics System. [Google Scholar]