Abstract
MicroRNAs (miRNAs) are short noncoding RNAs regulating gene expression that play roles in human diseases, including cancer. Each miRNA is predicted to regulate hundreds of transcripts, but only few have experimental validation. In chronic lymphocytic leukemia (CLL), the most common adult human leukemia, miR-15a and miR-16-1 are lost or down-regulated in the majority of cases. After our previous work indicating a tumor suppressor function of miR-15a/16-1 by targeting the BCL2 oncogene, here, we produced a high-throughput profiling of genes modulated by miR-15a/16-1 in a leukemic cell line model (MEG-01) and in primary CLL samples. By combining experimental and bioinformatics data, we identified a miR-15a/16-1-gene signature in leukemic cells. Among the components of the miR-15a/16-1 signature, we observed a statistically significant enrichment in AU-rich elements (AREs). By examining the Gene Ontology (GO) database, a significant enrichment in cancer genes (such as MCL1, BCL2, ETS1, or JUN) that directly or indirectly affect apoptosis and cell cycle was found.
Keywords: cancer, microRNA, targets
MicroRNAs (miRNAs) are short noncoding RNAs of ≈19–24 nt, that regulate gene expression by imperfect base-pairing with complementary sequences located mainly, but not exclusively, in the 3′ UTRs of target mRNAs. MiRNAs represent one of the major regulatory family of genes in eukaryotic cells by inducing translational repression and transcript degradation (1–4). Different algorithms such as TargetScan (5), PicTar (6), and Diana microT (7) have been developed to identify miRNA targets, but only few of these predictions have been experimentally validated, supporting the rationale for a combination of bioinformatics and biological strategies to this aim. Two independent studies predicted that 20–30% of human genes could be controlled by miRNAs (8, 9). Deviations from normal miRNA expression patterns play roles in human diseases, including cancer (for reviews see refs. 10–15).
The miR-15a/16-1 cluster resides at chromosome 13q14.3, a genomic region frequently deleted in B cell chronic lymphocytic leukemias (CLLs), and the two members of the cluster are cotranscribed and down-regulated in the majority of CLL patients (16). CLL is a disease with a frequent association in families (10–20% of patients have at least one first-degree relative with CLL) (17). Previously, we identified germ-line or somatic mutations in several miRNAs (including miR-16-1) in ≈15% of CLL patients, with the majority of the patients having a known personal or family history of CLL or other hematopoietic and solid tumors (18). These findings, together with the identification of an abnormal miR-15a/16-1 locus in the NZB strain of mice that naturally develop CLL (19), suggest that this cluster might play also a role in familial CLL.
Among the targets of miR-15a and miR-16, we identified the antiapoptotic protein Bcl2, which is overexpressed in the malignant, mostly nondividing B cells of CLL (20), and in many solid and hematologic malignancies (21). Restoration of miR-15-a/16-1 induces apoptosis in MEG-01, a cell line derived from acute megakaryocytic leukemia (22). These data support a role for miR-15a and miR-16-1 as tumor-suppressor genes (TSGs) in CLLs and perhaps in other malignancies in which these genes are lost or down-regulated.
Here, to investigate the mechanism of action of miR-15a and miR-16-1 as tumor suppressors in leukemias, we analyzed the effects of miR-15a and miR-16-1 on transcriptome and proteome in MEG-01 leukemic cells. This approach allowed us to validate a number of target genes, whose expression was also investigated in cases of CLL.
Results
In Vivo Effects of miR-15a/miR-16-1 Transfection into MEG-01 Leukemic Cells.
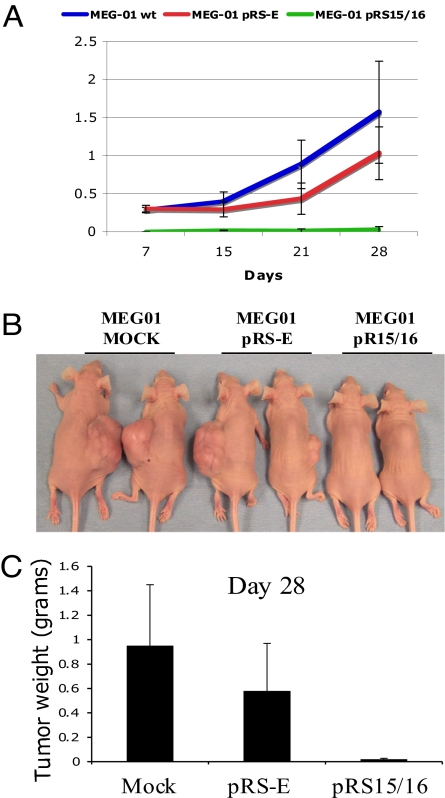
We reported that miR-15a/16-1 cluster induces apoptosis of MEG-01 cells by activating the intrinsic apoptosis pathway as identified by activation of the APAF-1–caspase9–PARP pathway (22). To further investigate the effect of these miRNAs, we tested their tumor-suppression function in vivo. Ten million viable MEG-01 cells, transfected in vitro with pRS15/16, pRS-E, or mock transfected, were inoculated s.c. in the flanks of immunocompromised “nude” mice (5 per group). As shown in Fig. 1A, the miR-15a/16-1 cluster inhibits the growth of MEG-01 tumor engraftments. After 28 days, tumor growth was completely suppressed in three of five (60%) mice inoculated with miR-15a/16-1-transfected MEG-01 (Fig. 1B). At day 28, the average tumor weights for the untreated and empty vector-treated mice were 0.95 ± 0.5 g and 0.58 ± 0.39 g, respectively; in mice inoculated with miR-15a/16-1-treated cells, the average was 0.020 ± 0.01 g (P < 0.003) (Fig. 1C). Thus, the results of these experiments demonstrate the tumor-suppressor function of miR-15a/16-1 cluster in MEG-01 leukemia cells.
Fig. 1.
MiR15a/16-1 cluster inhibits the growth of MEG-01 tumor engraftments in nude mice. (A) Growth curve of engrafted tumors in nude mice injected with MEG-01 cells pretransfected with pRS-E or pRS15/16 or mock transfected. (B) Comparison of tumor engraftment sizes of mock-, pRS-E-, and pRS15/16-transfected MEG-01 cells 28 days after injection in nude mice. (C) Tumor weights ± SD in nude mice.
Transcriptional Effects of Exogenous Expression of miR-15a and mirR-16-1.
To characterize the molecular basis of miR-15a/16-1 tumor suppression in leukemias, we first investigated the effect of miRNAs on genome-wide transcription of protein-coding genes. We transiently transfected the pRS15/16 vector into MEG-01 cells. This vector contains a genomic region encoding for both miRNAs as described (22). Transfection with the empty vector (pRS-E) was used as control. The success of transfection was assessed by measuring the expression levels of miR-15a, and miR-16-1 by quantitative (q)RT-PCR as described in ref. 18 (data not shown). Genome-wide transcriptome was investigated by using Affymetrix microarray. The microarray analysis clearly shows a different pattern of gene expression among pRS15/16- and pRS-E-transfected cells [supporting information (SI) Fig. 3]. After transfection with miR-15a/16-1 cluster, 355 probes (265 genes) were significantly up-regulated and 5,304 probes (3,307 genes) down-regulated (SI Table 5). The cluster analysis, performed with the differentially expressed genes, shows a clearly distinct gene expression profile between pRS15/16- and pRS-E-transfected cells (SI Fig. 3). Among the down-regulated probes, 140 (85 genes) are predicted as targets of miR-15/16 by three of the most used software algorithms (TargetScan, PicTar, and MiRanda), that are built on different prediction criteria and, therefore, used in combination, give the highest probability of target identification. If we consider only one prediction program, we found that 370, 332, and 312 transcripts, respectively, are predicted to be direct targets of these miRNAs (SI Fig. 3, SI Table 6). Among the up-regulated genes, there are no commonly predicted targets. Therefore, the miR-15a/16-1 cluster seems to regulate, directly or indirectly, ≈14% (265 genes up- and 3,307 down-regulated) of the 25,000 total predicted genes in the human genome (23) (SI Fig. 4).
AU-Rich Elements (AREs) Are More Frequently Found Among miR-15a/miR-16-1 Down-Regulated Genes, in MEG-01.
Because for miR-16-1 both a direct interaction in the “seed” region of the target mRNAs (22) and an ARE-mediated mRNA instability (24) have been reported, we investigated the frequency of ARE-containing mRNAs among the miR-15a/16-1-deregulated transcripts. As shown in SI Table 7, the number of genes containing AREs in their 3′ UTR was 36 of 265 (13.6%) up-regulated genes, and 666 of 3,307 (20.1%) among the down-regulated genes. This difference was statistically significant, with a χ2 value of 6.674 (P = 0.0098). Among the 85 genes that are predicted targets of miR-15a/16-1, 28 (32.9%) contain AREs, whereas among the remaining 3,222 down-regulated genes that are not commonly predicted targets, 638 (19.8%) mRNAs contain AREs (χ2 value = 8.89, P = 0.003). According to the number of motifs in the ARE stretch, the ARE-mRNAs can be clustered into five groups, containing five (cluster I), four (cluster II), three (cluster III), and two (cluster IV) pentameric repeats, whereas cluster V contains only one pentamer within the 13-bp ARE pattern as described (25). The ARE-cluster distribution of the miR-15a/16-1 deregulated genes is shown in Table 1. These results indicate that AREs are more frequently found among down-regulated targets of miR-15a/16-1, especially the commonly predicted targets, further confirming the influence of AREs in miR-16 targeting.
Table 1.
Cluster distribution of ARE-mRNAs deregulated in MEG-01 cells after miR-15a/16-1 cluster transfection
| ARE cluster | Up-regulated genes, n (%) | Down-regulated genes, n (%) |
|---|---|---|
| I | 1 (2.8) | 4 (0.6) |
| II | 0 (0) | 8 (1.2) |
| III | 5 (13.9) | 76 (11.4) |
| IV | 4 (11.1) | 84 (12.6) |
| V | 26 (72.2) | 494 (74.2) |
Gene Ontology (GO) of Genes Deregulated by miR-15a/16-1 Cluster.
Genes found to be differentially expressed in MEG-01 cells after transfection with pRS15/16 versus pRS-E were analyzed with the GeneSpring Gene Ontology browser tool to identify the Gene Ontology categories most represented in down-regulated genes (Table 2 and SI Table 8). These results show that the miR-15a/16-1 cluster directly or indirectly affects the expression of many cell cycle-related genes. In particular, many genes involved in the different transition checkpoints of the cell cycle are targeted by the miRNAs. Consistent with our previous finding that BCL2 is a target of miR-15a/16-1, in this GO ontology analysis, the category “antiapoptosis” (GO:6916) is significantly represented among the down-regulated transcripts.
Table 2.
Most significant GO categories after miR-15a/16-1 cluster transfection in MEG-01 cells.
| GO ID | GO description | P* |
|---|---|---|
| GO:7049 | Cell cycle | 2.7E-13 |
| GO:278 | Mitotic cell cycle | 1.7E-12 |
| GO:87 | M phase of mitotic cell cycle | 1.5E-11 |
| GO:7067 | Mitosis | 1.6E-10 |
| GO:51301 | Cell division | 2.7E-10 |
| GO:75 | Cell cycle checkpoint | 1.8E-05 |
| GO:82 | G1/S transition of mitotic cell cycle | 3.2E-04 |
| GO:7095 | Mitotic G2 checkpoint | 2.2E-03 |
| GO:6916 | Antiapoptosis | 4.4E-03 |
| GO:31575 | G1/S transition checkpoint | 4.8E-03 |
| GO:31572 | G2/M transition DNA damage checkpoint | 8.6E-03 |
| GO:31576 | G2/M transition checkpoint | 8.6E-03 |
| GO:43069 | Negative regulation of programmed cell death | 1.3E-02 |
| GO:43066 | Negative regulation of apoptosis | 1.7E-02 |
*P values establish whether there is a significant enrichment of down-regulated genes belonging to that GO category when compared with all genes present on the arrays.
Effect of miR-15a and miR-16-1 on MEG-01 Proteome.
Because both transcriptional and translational levels of miRNA-dependent gene regulation have been described (26), to investigate the effects of miR-15a/16-1 on MEG-01 cells at the protein level, we analyzed the proteins differentially expressed between MEG-01 cells transfected with pSR15/16 or pRS-E vector 48 h after transfection. By proteomics analysis, we identified proteins whose intensity was reduced 4-fold or more in the pRS15/16 group with respect to the pRS-E group. We isolated 27 different proteins (Table 3 and SI Table 9). Interestingly, BCL2, which we had already shown as a target of miR-15a/16-1 (22), and WT1, another predicted target of these miRNAs, were identified. The targeted proteins have a variety of biological functions and can be grouped into four groups. The first group includes proteins that play a role in regulation of cell growth and cell cycle (Ruvbl1, Anxa2, Rcn1, Cct7, Sugt1, Cdc2, Psf1), another category is formed by antiapoptotic proteins (Grp78, Bcl2, Pdia2), and proteins involved in human tumorigenesis, either as oncogenes, or as tumor-suppressor genes (Wt1, MageB3, Rab9B). The remaining 14 proteins have different biological functions, and we identified them as “others.” Among the 27 experimentally identified down-regulated proteins, 8 (29.6%) are predicted targets of miR-15a/16 by at least one of the prediction algorithms. Finally, among this group of eight proteins, two (Bcl2, and Cfl2) were present also in the group of down-regulated mRNAs.
Table 3.
Examples of proteins down-regulated by the miR-15a/16-1 cluster identified by proteomics in MEG-01 cells
| Group | Protein | Gene description | Z-Score | Comments |
|---|---|---|---|---|
| Cell growth & cell cycle | Ruvbl1 | RuvB-like 1; TATA binding protein interacting protein 49 KDa | 2.01 | — |
| Sugt1 | Suppressor of G2 allele of SKP1 | 2.43 | — | |
| Cdc2 | Cell division cycle 2, G1 to S and G2 to M | 2.43 | — | |
| Psf1 | GINS complex subunit 1 (Psf1 homolog) | 2.43 | — | |
| Antiapoptotic | Grp78 | Heat shock 70-kDa protein 5 (glucose-related protein, 78 kDa) | 2.43 | — |
| Bcl2 | B-cell CLL/lymphoma 2 | 2.43 | Predicted and validated target of miR-15a/16 (22) | |
| Pdia2 | Protein disulfide isomerase family A, member 2 | 2.43 | — | |
| Oncogenesis | Wt1 | Wilms tumor 1 | 2.43 | Predicted target of miR-15a/16; Validated qRT-PCR in MEG-01 |
| MageB3 | Melanoma antigen family B, 3 | 2.43 | — | |
| Rab9B | RAB9B member RAS oncogene family | 2.16 | Predicted target of miR-15a/16 | |
| Others | Cdh26 | Cadherin-like 26 | 2.43 | — |
| Crhbp | Corticotropin releasing hormone-binding protein | 2.43 | Predicted target of miR-16 | |
| Actr1A | ARP1 actin-related protein 1 homolog A, centractin alpha | 2.43 | Predicted target of miR-15a/16 | |
| Cshl1 | Chorionic somatomammotropin hormone-like 1 precursor | 2.43 | Predicted target of miR-16 | |
| Hla-B | Major histocompatibility complex, class I, B | 2.43 | — | |
| Tpi1 | Triosephosphate isomerase 1 | 2.43 | Predicted target of miR-15a/16 | |
| Hsp90AB1 | Heat shock protein 90-kD protein 1, β | 2.43 | — | |
| Cfl2 | Cofilin 2 | 1.72 | Predicted target of miR-16 | |
| AldoA | Aldolase A, fructose-bisphosphate | 2.43 | — |
For complete version, see SI Table 9. Z-score, probability of identification of the protein (2.43 = 99%, 1.28 = 90%).
Validation of the Results in the MEG-01 Cell Line.
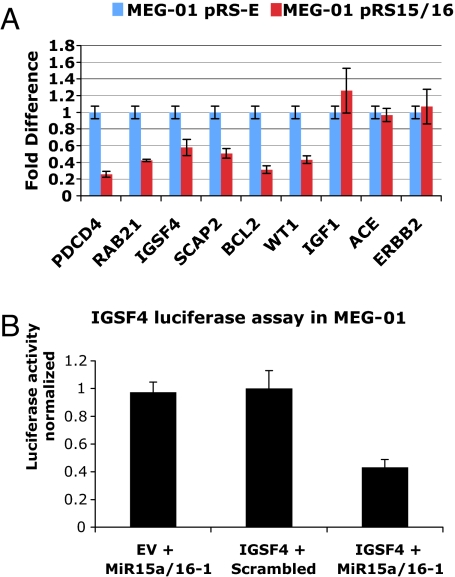
To validate the results obtained by transcriptomic or proteomic analyses, we assayed the expression of nine genes (four identified by the EST microarray, two by proteomics, and three identified by neither of the techniques and therefore considered as negative controls), by qRT-PCR in MEG-01 cells transfected with pRS15/16 or pRS-E (control). As shown in Fig. 2A, the transfection with miR-15a/16-1 reduces the expression of both microarray identified mRNAs (PDCD4, RAB21, IGSF4, SCAP2) and proteomics identified proteins (Bcl2, Wt1). MiR-15a/16-1 transfection does not affect the expression of any of the control genes (IGF1, ACE, and ERBB2).
Fig. 2.
Validation of some of the targets of miR-15a/16-1 identified by microarray or proteomics in MEG-01. (A) qRT-PCR validation of PDCD4, RAB21, IGSF4, SCAP2 (down-regulated in the microarray), BCL2, and WT1 (down-regulated in proteomics). IFG1, ACE, and ERBB2 are negative controls. The results were normalized to pRS-E-transfected cells. Samples were normalized with β-tubulin. (B) Luciferase assay of IGSF4 in MEG-01 cells, showing that the miR-15a/16-1 cluster directly targets this gene.
We also performed the luciferase assay on one of the validated genes (IGSF4) and demonstrated that the miR-15a/16-1 cluster directly targets IGSF4 (Fig. 2B). The direct interactions with BCL2 and DMTF1 were proved by us and others (7, 22). Therefore, we were able to consistently confirm the MEG-01 profile of down-regulated genes and identified another direct target of miR-15a/16-1 in this leukemic model.
Variation of Expression of miR-15a/mir 16-1 Targets in Primary CLLs.
Because MEG-01 is a leukemia cell model with abnormal 13q14 and loss of the miR15a/16-1 cluster (similar to CLL) but is a megakaryocytic established leukemic cell line, we decided to investigate the effects of the different expression of miR-15a/16-1 cluster also in primary CLLs. Therefore, to verify whether some of the targets of miR-15a/16-1 identified in MEG-01 cells were inversely correlated to the expression of these two miRNAs in CLL patients, we selected a group of 16 CLL samples in whom the expression of miR-15a/16-1 had already been determined by miRNA microarray analysis in our previous studies (18, 27). We have shown that a signature of 13 miRNAs distinguished between indolent and aggressive CLL and that loss of the miR-15a/16-1 cluster is a characteristic of indolent CLLs (18). First, we validated the expression of miR-15a/16-1 by qRT-PCR and confirmed the microarray data by qRT-PCR (data not shown). Among the considered 16 patients, 8 have higher expression of miR-15a/16-1, with respect to the other 8 patients (P = 7.7 × 10−6 at microarray analysis, P = 0.019 at qRT-PCR analysis). The comparison between eight CLLs with high and low miR-15a/16-1 expression by EST oligonucleotide microarray analysis showed 678 Affymetrix probes (539 genes) significantly differentially expressed among the two groups (SI Table 10). Overall, 82 of 539 genes (15.2%) are ARE mRNAs, and 4 are predicted as targets by all three bioinformatics algorithms.
A Signature of miR-15a/16-1 Down-Regulated Transcripts.
We selected genes that were low in miR-15/16 high-expressor CLLs and high in miR-15/16 low-expressor CLLs, which were intersected with genes down-regulated in MEG-01 cells after transfection with pRS15/16. A signature of 60 genes (70 probes) emerged (Table 4 and SI Table 11). Thirteen of these genes (21.7%) are ARE-mRNAs, distributed in cluster III (7.8%), IV (7.8%), and V (84.6%). No statistically significant enrichment in ARE-mRNAs was observed in this signature with respect to both the total of down-regulated mRNAs in MEG-01 (P = 0.76) and the total of repressed transcripts in patients with high expression of miR-15a/16-1 (P = 0.14). We performed the GO analysis of these 70 transcripts and found, among the significantly represented categories, some of those previously identified in transfected MEG-01 and involved in regulation of cell cycle and apoptosis, such as “antiapoptosis” (GO:6916), “negative regulation of apoptosis” (GO:43066), and “negative regulation of programmed cell death” (GO:43069) (SI Table 12). The consistency of the results in MEG-01 and in CLL patients confirms the validity of our in vitro model and identifies GO categories and a panel of protein coding genes, whose expression is consistently controlled by the cluster.
Table 4.
Examples of the CLL signature of miR-15a/16-1 down-regulated genes by microarray
| Gene symbol | Map | Gene name |
P |
|
|---|---|---|---|---|
| CLL | MEG-01 | |||
| HSDL2 | 9q32 | Hydroxysteroid dehydrogenase like 2 | 0.00178 | 0.00254 |
| SLC35A1 | 6q15 | Solute carrier family 35 (CMP-sialic acid transporter), member A1 | 0.00178 | 0.00341 |
| ECHDC1 | 6q22.33 | Enoyl coenzyme A hydratase domain containing 1 | 0.00323 | 0.00582 |
| CARD8 | 19q13.32 | Caspase recruitment domain family, member 8 | 0.00446 | 0.00078 |
| OMA1 | 1p32.2-p32.1 | OMA1 homolog, zinc metallopeptidase (S. cerevisiae) | 0.00446 | 0.0158 |
| UGP2 | 2p14-p13 | UDP-glucose pyrophosphorylase 2 | 0.00446 | 0.000629 |
| CREBL2 | 12p13 | cAMP responsive element binding protein-like 2 | 0.00457 | 0.0199 |
| Cep63 | 3q22.1 | Centrosome protein Cep63 | 0.0049 | 0.0137 |
| PNN | 14q21.1 | Pinin, desmosome-associated protein | 0.0049 | 0.00359 |
| TRA1 | 12q24.2-q24.3 | Tumor rejection antigen (gp96) 1 | 0.00496 | 0.0211 |
| SLC35B3 | 6p24.3 | Solute carrier family 35, member B3 | 0.00601 | 0.0208 |
| RHOT1 | 17q11.2 | Ras homolog gene family, member T1 | 0.00695 | 0.0197 |
| LARS | 5q32 | Leucyl-tRNA synthetase | 0.00696 | 0.00239 |
| RAD51C | 17q22-q23 | RAD51 homolog C (S. cerevisiae) | 0.0075 | 0.00334 |
| WASPIP | 2q31.1 | Wiskott–Aldrich syndrome protein interacting protein | 0.00783 | 0.0108 |
| MCL1 | 1q21 | Myeloid cell leukemia sequence 1 (BCL2-related) | 0.00863 | 0.011 |
| ASXL2 | 2p24.1 | Additional sex combs like 2 (Drosophila) | 0.00875 | 0.000503 |
| ARFIP1 | 4q31.3 | ADP-ribosylation factor interacting protein 1 (arfaptin 1) | 0.0114 | 0.0108 |
| HERC6 | 4q22.1 | Hect domain and RLD 6 | 0.0116 | 0.00107 |
| TIA1 | 2p13 | TIA1 cytotoxic granule-associated RNA-binding protein | 0.0116 | 0.0116 |
| VPS45A | 1q21-q22 | Vacuolar protein sorting 45a (yeast) | 0.0117 | 0.000788 |
| HLC-8 | 17q25.1 | Lung cancer-related protein 8 | 0.0124 | 0.0164 |
| HACE1 | 6q21 | HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 | 0.0125 | 0.0115 |
| ARV1 | 1q42.2 | ARV1 homolog (yeast) | 0.0156 | 0.000825 |
| NT5C2L1 | 6q22.1 | 5′ nucleotidase, cytosolic II-like 1 | 0.0172 | 0.0139 |
| PDCD6IP | 3p23 | Programmed cell death 6 interacting protein | 0.0214 | 0.00276 |
| GTF2H1 | 11p15.1-p14 | General transcription factor IIH, polypeptide 1, 62 kda | 0.0217 | 0.00125 |
| MSH2 | 2p22-p21 | mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) | 0.0242 | 0.00192 |
| JUN | 1p32-p31 | v-jun sarcoma virus 17 oncogene homolog (avian) | 0.0281 | 0.00059 |
| ALDH6A1 | 14q24.3 | Aldehyde dehydrogenase 6 family, member A1 | 0.0297 | 0.00798 |
| SCAP2 | 7p21-p15 | Src family-associated phosphoprotein 2 | 0.0298 | 0.0108 |
For complete version, see SI Table 12. In bold: ARE-mRNAs; P, difference between groups with high and low miR-15a/16-1 expression.
Discussion
In this study, we show that miR-15a/16-1 exert a tumor suppressor function in vivo by inhibiting the growth of tumor engraftments of leukemic cells in nude mice. To investigate the molecular bases of miR-15a/16-1 tumor-suppressor function, we performed an extensive microarray analysis of the deregulated genes after transfection of MEG-01 cells with pRS15/16, a vector expressing miR-15a/16-1, and using the same empty vector (pRS-E) as a control. Interestingly, we confirmed some of the targets observed by other groups in different models, such as CDK6, CDC27, and RAB11FIP2 (28) in solid tumor cell lines and ACVR2A in Xenopus laevis (29). We matched our experimentally identified down-regulated genes with the targets of miR-15a/16-1 commonly predicted by three of the most widely used algorithms for the identification of miRNA-targets (PicTar, TargetScan, MiRanda), and found 85 genes (2.6%) in common. Interestingly, by matching our results with a computational method that identifies miRNA targets by predicting miRNA regulatory modules (MRMs) or groups of miRNAs and target genes that are believed to participate cooperatively in posttranscriptional gene regulation (30), we found 5 of 13 (38.5%) miR-15/16 MRM predicted genes (ATP2B1, FBXW7, PPM1D, SON, and WT1) among our differentially expressed genes. This percentage represents the highest among all of the considered prediction algorithms. As expected, among the 265 experimentally up-regulated mRNAs, none is predicted as target of miR-15a/16. This finding could be explained by indirect effects, for example by the regulation of transcription factor(s) targeted by these two miRNAs. The effects of the exogenous expression of miR-15a/16-1 in MEG-01 cells was also investigated by proteomics 48 h after the transfection. We also studied different time-from-transfection intervals to analyze the effects of miR-15a/16-1 at a transcriptional (24 h) or translational (48 h) level, because after 24 h, mRNA silencing is maximal, but secondary transcriptional effects due to protein depletion are minimal (31). Our proteomic approach was able to detect 27 targets of miR-15a/16-1, approximately one-third of which are also predicted targets. Interestingly, 25% (two of eight) of the predicted targets were down-regulated both in the transcriptome and in the proteome. Among the miR-15a/16-1 down-regulated genes, we demonstrated that IGSF4 is a direct target of the cluster. IGSF4 was originally identified as a tumor-suppressor gene in lung cancer and is involved in cell adhesion (32, 33). Sasaki et al. (34) have demonstrated that TSLC1/IGSF4 acts as an oncoprotein involved in the development and progression of adult T cell leukemia (ATL). It can be hypothesized that by directly silencing IGSF4, miR-15a/16-1 could exert a more general antileukemic effect.
We also studied by microarray the down-regulated mRNAs in eight CLL patients with high levels of miR-15a/16-1 with respect to eight CLL patients with low levels of these two miRNAs and identified a signature of 60 genes in common between CLLs and MEG-01 transfected with miR-15a/16-1. This signature (which includes ≈2% of the down-regulated genes in MEG-01 and ≈11% of those repressed in patients) contains oncogenes such as MCL1, JUN, SCAP2, TRA1, PDCD6IP, RAD51C, and HSPA1A/1B, which could explain the oncosuppressor effect of miR-15a/16-1 observed in MEG-01 both in vitro (22),and in vivo (present work). MCL1 is an antiapoptotic BCL-2 family member that contributes to B cell survival in CLL and has been associated with resistance to chemotherapy (35, 36). Despite the fact that MCL-1 expression is not different in ZAP 70-positive (aggressive) vs. ZAP 70-negative (indolent) B-CLL cells (37), it represents a relevant therapeutic target in both acute and chronic lymphoid malignancies, because its silencing is sufficient to promote apoptosis in ALL and CLL cells and increase sensitivity to rituximab-mediated apoptosis (38). Interestingly, miR-29b has also been identified to target Mcl1 in a cholangiocarcinoma model (39), and many pieces of evidence converge in defining a role of the miR-29 family as TSGs in both solid (40) and hematologic malignancies (41). Our findings give a rationale to an association of miR-15a/16-1 and miR-29s in the treatment of CLL. Moreover, a sustained signaling through the B cell receptor promotes survival of B-CLL cells both by induction of MCL1 and, to a less extent, by activation of c-JUN NH2-terminal kinase (JNK) (42). Therefore, by targeting both MCL1, and c-JUN transcripts, the impact of the miR-15a/16-1 cluster on the survival of B-CLL cells could be even more robust. The presence of BCL2 in the proteomics list confirms our previous statement of a posttranscriptional regulation of this target (22). Moreover the repression of LARS (leucyl-tRNA synthetase), involved in the same pathway of RARS (arginyl-tRNA synthetase), and the presence of RARS among the down-regulated genes in MEG-01 confirms our previous hypothesis that this pathway could be targeted by miR-15a/16-1 (16). Interestingly, the signature includes also many important tumor-suppressor genes (RNASEL, HACE1, CEP63, CREBL2, MSH2, TIA1, and PMS1) and reveals an intriguingly possible explanation for the link between miR-15a/16-1 expression and CLL prognosis. We described that in CLL patients with unmutated IgVH, and high expression of ZAP-70 (poor prognosis), the levels of miR-15a/16-1 are higher than in CLL patients with a better prognosis (18). The observed coexistence of oncogenes and TSGs in miR-15a/16-1 CLL signature could give a molecular explanation as to why high levels of these two miRNAs are associated with CLLs with a worse prognosis (18). High miR-15a/16-1 levels could down-regulate many TSGs and consequently negatively affect many oncosuppressor pathways, therefore leading to a more oncogenic phenotype.
Recently, it has been demonstrated that miR-16 is critically involved in ARE-mediated mRNA instability (24). In MEG-01 cells, we found that ARE-mRNAs are significantly more represented among the down-regulated genes (20.1%) than among the up-regulated (13.6%, P = 0.0098). Although the identified signature is not enriched with ARE-mRNAs, it shows a predominance (84.6%) of cluster V ARE-mRNAs (which reflects the higher number of members of this cluster in both MEG-01 and patients), indicating that a higher number of pentameric AU-repeat does not correspond to a higher silencing effect by miR-15a/16-1. Finally the GO analysis of the deregulated genes indicates that miR-15a/16-1 impacts strongly on metabolic pathways, on nucleic acid-binding pathways, and the activities of translation factors. It has been shown that in solid tumor cell lines miR-16-down-regulated transcripts are enriched with genes whose silencing causes an accumulation of cells in G0/G1 and that this function does not depend on AU-rich elements (28). Accordingly, we found that some of the described miR-16 targets whose disruption triggered G0/G1-cell accumulation were down-regulated also in our cell model (CDK6, CDC27, RAB11FIP2) and that some of the previously described GO categories [namely “mitotic cell cycle” (GO:278), and “cell cycle” (GO:7049)] are represented also in our data. In contrast with the previous report, we found a statistically significantly higher number of ARE-mRNAs among the down-regulated targets with respect to the up-regulated. These differences may reflect cell-specific functions of miR-15a/16-1, whereas the common finding that miR-15a/16-1 targets “cell cycle”-involved genes, both in solid and in hematologic tumor models, suggests a more general and robust effect of the cluster on this group of genes. In conclusion, our work describes miR-15a/16-1 deregulated genes in both a leukemic cell model and in primary CLLs, and identifies a signature of common genes whose silencing characterizes the miR-15a/16-1-induced phenotype in CLL. These findings could have important significance for the development of therapeutic approaches for CLLs.
Materials and Methods
Cell Culture and Patient Samples.
The human megakaryocytic MEG-01 cell line was purchased from the American Type Culture Collection and grown in 10% FBS RPMI medium 1640, supplemented with 1× nonessential amino acids and 1 mmol of sodium pyruvate at 37°C and 5% CO2. For the patient study, we used 16 CLL samples obtained after informed consent from patients diagnosed with CLL at the CLL Research Consortium institutions. Briefly, blood was obtained from CLL patients and mononuclear cells were isolated through Ficoll/Hypaque gradient centrifugation (Amersham Pharmacia Biotech) and processed for RNA extraction according to the described protocols (18). For all of the samples, the microarray expression data were known as reported in ref. 18, and we further performed confirmation with qRT-PCR.
In Vivo Studies.
Animal studies were performed according to institutional guidelines. MEG-01 cell lines were transfected in vitro with p-Retrosuper vector (43) expressing miR-15a/miR-16-1 (pRS15/16). Untransfected (mock) or cells transfected with the same empty plasmid (pRS-E) served as tumorigenic controls. At 24 h after the transfection, 107 viable cells were injected s.c. into the left flanks of 5-week-old female nude mice (Charles River Breeding Laboratories), five mice per transfected or control cell line. Tumor diameters were measured on days 7, 15, 21, and 28. After 28 days, the mice were killed, necropsies were performed, and tumors were weighted. Tumor volumes were calculated by using the equation V (in mm3) = A × B2/2, where A is the largest diameter, and B is the perpendicular diameter.
In Vitro Transfection.
MEG-01 cells were transiently transfected with 1 μg/ml (final concentration) pRS-15/16 or pRS-E vector by using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. After 24 h, total RNA was extracted by using TRIzol reagent (Invitrogen) according to the manufacturer's instructions.
Microarray Hybridization and Data Analysis.
Two samples obtained from the MEG01 cell line transfected with pRS-15/16 and pRS-E vector, each one in triplicate, and 16 CLL samples were analyzed by microarray using Human Genome U133A Plus 2.0 GeneChip arrays (Affymetrix). The .CEL files generated by the GeneChip scanner were imported in GeneSpring GX 7.3 software (Agilent Technologies) and further processed. Details about the microarray experiment are described in SI Text.
MiRNA Target Prediction.
The analysis of miRNA predicted targets was determined by using the algorithms TargetScan (http://genes.mit.edu/targetscan/), PicTar (http://pictar.bio.nyu.edu/), and miRanda (http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl).
Adenylate Uridylate-Rich Elements (ARE)-Containing Genes Identification.
The ARE-mRNA database version 3.0 (ARED; http://rc.kfshrc.edu.sa/ared/), as described (44), was used (for details see SI Text).
Two-Dimensional PAGE and Protein Identification by Matrix Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) and Mass Spectrometry (MS).
MEG-01 cells were transiently transfected for 48 hr with 1 μg/ml (final concentration) pRS15/16 or pRS-E vector by using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions and the details of the two-dimensional PAGE, and protein identification by MALDI-TOF and MS are described in SI Text.
qRT-PCR.
qRT-PCR analysis for miRNAs was performed in triplicate with the TaqMan MicroRNA assays kit (Applied Biosystems) according to the manufacturer's instructions and as described (45). For normalization, 18S RNA was used; qRT-PCR analyses for other genes of interest were performed by reverse transcription of RNA to cDNA with gene-specific primers and IQ SYBR green Supermix (Bio-Rad) according to the manufacturer's instructions. β-Tubulin was used for normalization.
Luciferase Reporter Assay.
For luciferase reporter experiments, a IGSF4 3′ UTR segment of 237 bp was amplified by PCR from human cDNA and inserted into the pGL3-control vector with SV40 promoter (Promega) by using the XbaI site immediately downstream from the stop codon of luciferase. Details about the microarray experiment are described in SI Text. The experiments were performed in triplicate.
Supplementary Material
Acknowledgments.
We thank Dr. D. Vandre for performing the proteomics analysis and Dr. S. Yoon for the prediction data. This work was supported by National Cancer Institute grants (to C.M.C., T.J.K., and T.D.S.), by a CLL Global Research Foundation grant, by funding from the University of Texas System Regents Research Scholar Award and the University of Texas M. D. Anderson Research Trust (G.A.C.), by grants from the Italian Ministry of Health (to C.M.C.), and by the Italian Ministry of University Research and the Italian Association for Cancer Research (to M.N.). A.C. was supported by an American–Italian Cancer Foundation Fellowship. M. Ferracin was a recipient of a fellowship from the Fondazione Italiana per la Ricerca sul Cancro.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data were submitted to the Minimum Information About a Microarray Experiment (MIAME) Database (accession no. E-MEXP-1482).
This article contains supporting information online at www.pnas.org/cgi/content/full/0800121105/DC1.
References
- 1.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagga S, Pasquinelli AE. Identification and analysis of microRNAs. Genet Eng. 2006;27:1–20. doi: 10.1007/0-387-25856-6_1. [DOI] [PubMed] [Google Scholar]
- 3.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 4.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 6.Lall S, et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 7.Kiriakidou M, et al. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Croce CM. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin Oncol. 2006;33:167–173. doi: 10.1053/j.seminoncol.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. Investigation of MicroRNA alterations in leukemias and lymphomas. Methods Enzymol. 2007;427:191–213. doi: 10.1016/S0076-6879(07)27011-9. [DOI] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 13.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: A developing story. Curr Opin Genet Dev. 2005;15:200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Fabbri M, Ivan M, Cimmino A, Negrini M, Calin GA. Regulatory mechanisms of microRNAs involvement in cancer. Expert Opin Biol Ther. 2007;7:1009–1019. doi: 10.1517/14712598.7.7.1009. [DOI] [PubMed] [Google Scholar]
- 16.Calin GA, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catovsky D. Definition and diagnosis of sporadic and familial chronic lymphocytic leukemia. Hematol Oncol Clin North Am. 2004;18:783–794. doi: 10.1016/j.hoc.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 18.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 19.Raveche ES, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitada S, et al. Expression of apoptosis-regulating proteins in chronic lymphocytic leukemia: Correlations with in vitro and in vivo chemoresponses. Blood. 1998;91:3379–3389. [PubMed] [Google Scholar]
- 21.Sanchez-Beato M, Sanchez-Aguilera A, Piris MA. Cell cycle deregulation in B-cell lymphomas. Blood. 2003;101:1220–1235. doi: 10.1182/blood-2002-07-2009. [DOI] [PubMed] [Google Scholar]
- 22.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein LD. Human genome: End of the beginning. Nature. 2004;431:915–916. doi: 10.1038/431915a. [DOI] [PubMed] [Google Scholar]
- 24.Jing Q, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 25.Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. ARED: Human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 27.Calin GA, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linsley PS, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martello G, et al. MicroRNA control of Nodal signalling. Nature. 2007;449:183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 30.Yoon S, De Micheli G. Prediction and analysis of human microRNA regulatory modules. Conf Proc IEEE Eng Med Biol Soc. 2005;5:4799–4802. doi: 10.1109/IEMBS.2005.1615545. [DOI] [PubMed] [Google Scholar]
- 31.Jackson AL, et al. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 32.Murakami Y. Involvement of a cell adhesion molecule, TSLC1/IGSF4, in human oncogenesis. Cancer Sci. 2005;96:543–552. doi: 10.1111/j.1349-7006.2005.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuramochi M, et al. TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat Genet. 2001;27:427–430. doi: 10.1038/86934. [DOI] [PubMed] [Google Scholar]
- 34.Sasaki H, et al. Overexpression of a cell adhesion molecule, TSLC1, as a possible molecular marker for acute-type adult T-cell leukemia. Blood. 2005;105:1204–1213. doi: 10.1182/blood-2004-03-1222. [DOI] [PubMed] [Google Scholar]
- 35.Moshynska O, Sankaran K, Pahwa P, Saxena A. Prognostic significance of a short sequence insertion in the MCL-1 promoter in chronic lymphocytic leukemia. J Natl Cancer Inst. 2004;96:673–682. doi: 10.1093/jnci/djh122. [DOI] [PubMed] [Google Scholar]
- 36.Johnston JB, et al. Role of myeloid cell factor-1 (Mcl-1) in chronic lymphocytic leukemia. Leuk Lymphoma. 2004;45:2017–2027. doi: 10.1080/10428190410001723317. [DOI] [PubMed] [Google Scholar]
- 37.Bogner C, et al. Cyclin E but not bcl-2, bax or mcl-1 is differentially expressed in ZAP 70-positive and ZAP 70-negative B-CLL cells. Ann Hematol. 2006;85:458–462. doi: 10.1007/s00277-005-0076-y. [DOI] [PubMed] [Google Scholar]
- 38.Hussain SR, et al. Mcl-1 is a relevant therapeutic target in acute and chronic lymphoid malignancies: Down-regulation enhances rituximab-mediated apoptosis and complement-dependent cytotoxicity. Clin Cancer Res. 2007;13:2144–2150. doi: 10.1158/1078-0432.CCR-06-2294. [DOI] [PubMed] [Google Scholar]
- 39.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pekarsky Y, et al. Tcl1 expression in CLL is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 42.Petlickovski A, et al. Sustained signaling through the B-cell receptor induces Mcl-1 and promotes survival of chronic lymphocytic leukemia B cells. Blood. 2005;105:4820–4827. doi: 10.1182/blood-2004-07-2669. [DOI] [PubMed] [Google Scholar]
- 43.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 44.Bakheet T, Williams BR, Khabar KS. ARED 3.0: The large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.