Abstract
The 85-kb breast cancer-associated gene BRCA1 is an established tumor suppressor gene, but its regulation is poorly understood. We demonstrate by gene conformation analysis in both human cell lines and mouse mammary tissue that gene loops are imposed on BRCA1 between the promoter, introns, and terminator region. Significantly, association between the BRCA1 promoter and terminator regions change upon estrogen stimulation and during lactational development. Loop formation is transcription-dependent, suggesting that transcriptional elongation plays an active role in BRCA1 loop formation. We show that the BRCA1 terminator region can suppress estrogen-induced transcription and so may regulate BRCA1 expression. Significantly, BRCA1 promoter and terminator interactions vary in different breast cancer cell lines, indicating that defects in BRCA1 chromatin structure may contribute to dysregulated expression of BRCA1 seen in breast tumors.
Keywords: transcriptional regulation, chromatin conformation, gene repression, mammary gland, breast cancer
Expression of the tumor suppressor gene BRCA1 is reduced in a significant proportion of human breast tumors (1–3). Although up to one-third of these cases can be explained by promoter hypermethylation (4, 5) for most cases the cause is unknown. Understanding the underlying mechanisms of BRCA1 gene repression is critical for generating effective strategies for re-establishing BRCA1 expression and thus restoring its tumor suppressor function.
Transcriptional initiation of protein-coding genes depends on a coordinated interplay of protein–DNA and protein–protein interactions (6). In addition to the assembly of RNA polymerase II (Pol II) with basal transcription machinery on the gene promoter, numerous transcription factors are recruited to either activate or repress transcription. As many of these factors associate with DNA sequences distant to the promoter, transcriptional regulation often involves long-range DNA associations, possibly mediated by the formation of chromatin loops (7). Chromatin loops can be detected by the chromosome conformation capture (3C) technique (8), which involves formaldehyde cross-linking of chromatin in live cells, digesting DNA with restriction enzymes, and then religating DNA in dilute solution to favor intramolecular ligation. PCR is then used to detect the presence of such ligation products. 3C has been used to study the normal regulation of genes in multiple eukaryotic species and supports a looping model for gene activation and repression. For example, transcriptional activation of the β-globin gene in mouse is associated with interactions between multiple hypersensitive sites spanning >50 kb of DNA (9), whereas repression of the maternal IGF2 gene is linked to a long-range association between IGF2 and H19 loci, restricting access to an IGF2 enhancer (10).
Several human diseases are associated with mutations in long-range control elements (11). Examples include Campomelic dysplasia, which can be caused by deletion of critical regulatory elements ≈50 kb upstream of the SOX9 gene (12), Aniridia, which is associated with mutations up to 75 kb 3′ of the Aniridia gene PAX6 (13), and Blepharophimosis syndrome, where deletion of conserved sequences 230 kb upstream of the FOXL2 gene has been detected in some patients (14).
BRCA1 transcription is controlled at least in part by a bidirectional promoter (15, 16), the activity of which can be modulated by positive and negative regulatory sequences within BRCA1 introns (17). A 140-kb P1 artificial chromosome containing human BRCA1 plus 60 kb of flanking sequence can rescue the embryonic lethal phenotype of Brca1 null mice (18), suggesting that all of the sequences required for correct temporal and spatial expression are contained within this sequence. The identity of these elements, how they associate with one another, and whether they contribute to breast tumorigenesis is unknown.
We describe the analysis of potential long-distance interactions associated with BRCA1 and demonstrate the existence of BRCA1 gene loop structures between the promoter and sequences including the introns and the termination region. Significantly, this latter gene loop structure is altered in response to estrogen stimulation and in several breast cancer cell lines.
Results and Discussion
Long-Range Associations Involving the BRCA1 Promoter and Regulatory Regions.
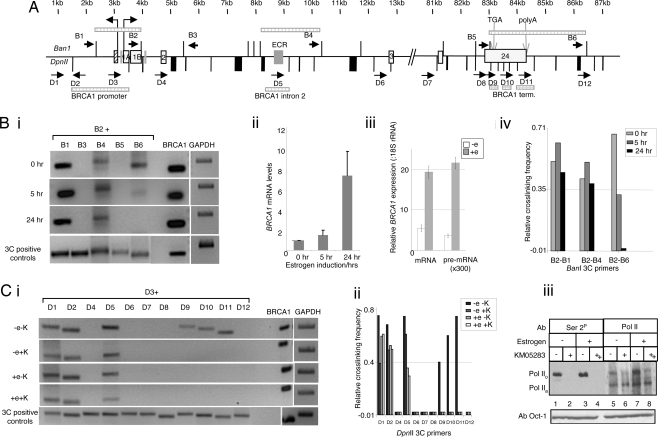
We performed 3C analysis on the human BRCA1 gene, using primers flanking either BanI and DpnII restriction sites. Initially our studies focused on previously characterized regulatory regions of BRCA1: the promoter (16) including 3 kb upstream of transcription; an evolutionarily conserved region in intron 2 (17), and the 3′ end of the gene, including the 3′UTR (19) and 2 kb downstream of exon 24 (Fig. 1A). Critical controls are essential for correct interpretation of 3C data (20). We therefore confirmed that all 3C primers amplified in vitro-generated 3C products (Fig. 1 Bi and Ci, labeled 3C positive controls), there was a nonlinear relationship between detection of 3C products and distance between associated restriction fragments (Fig. 1 Bi and Ci Top), 3C primers did not amplify undigested and ligated, or digested but not ligated, chromatin (data not shown), and the sequence of all 3C products was correct (data not shown).
Fig. 1.
Dynamic long-range association between regulatory regions of BRCA1. (A) Diagram of BRCA1, exons 1–24 (bold lines) flanked by 3 kb upstream of exon 1 and 2 kb downstream of exon 24 (thin lines) with BanI (above sequence) and DpnII (below sequence) sites indicated. Upright tall arrows indicate divergent transcription initiation positions for NBR2 and BRCA1. BRCA1 exons are indicated by open rectangles, and an NBR2 exon is indicated by a striped rectangle. The gray rectangles indicate regions known to possess regulatory (promoter, enhancer, or repressor) activity. Vertical lines represent restriction sites. Black labeled arrows indicate primer direction. Checked boxes represent restriction fragments involved in gene looping based on 3C analysis. (B) (i) BanI 3C analysis of BRCA1 in the MCF7 breast adenocarcinoma cell line before and after 5 or 24 h of 100-nM estrogen stimulation and artificial 3C products (see Materials and Methods) using primer B2 in combination with one of the other BanI primer as indicated. BRCA1 load represents the product of PCR analysis of the same BanI 3C using BRCA1-specific internal primers in intron 2. (ii) qRT-PCR analysis of BRCA1 mRNA, relative to 18S rRNA, after induction with 100 nM estrogen showing a slight increase after 5 h and a significant increase after 24 h. (iii) qRT-PCR analysis of BRCA1 mRNA and pre-mRNA, relative to 18S rRNA, after induction with 100 nM estrogen showing a significant increase in both after 24 h of stimulation. (iv) Quantitation of 3C band intensity relative to intensity of 3C control bands (see Materials and Methods). (C) 3C and Western analysis of MCF7 cells that are either unstimulated or stimulated with 100 nM E2 for 24 h and then untreated or treated with the transcription inhibitor KM05283 for 1 h. (i) 3C analysis of MCF7 cells grown in serum-free phenol red free media (−e) or stimulated with 100 nM estrodiol for 24 h (+e), and either untreated (−K) or treated with KM05283 for 1 h (+K). 3C-positive controls and BRCA1 loading control were as for Bi. (ii) Quantitation of 3C band intensity relative to intensity of 3C control bands (see Materials and Methods). (iii) Western analysis with antibodies against Pol II Ser-2 phosphoylated CTD (Ser 2P) or the N terminus of the large subunit of Pol II showing that the transcription inhibitor has worked. Anti-Oct antibody provides loading control.
Using these validated 3C primers, we initially analyzed the conformation of BRCA1 in the breast cancer cell line MCF7. 3C analysis of chromatin from cells grown in defined media (serum-free phenol-red free) showed that the BRCA1 5′ region (primers B2 and D3) associates with sequences in BRCA1 intron 2 (primers B4 and D5) and sequences at the 3′ end of the gene (primers B6 and D9 and primers D10 and D11; Fig. 1 B and C). No evidence for interactions between the BRCA1 promoter and sequences elsewhere in intron 2 (primers D4 and D6), intron 22 (primer D7), intron 23 (primer B5), or 2 kb downstream of exon 24 (primer D12) was found.
The Association Between 5′ and 3′ Ends of BRCA1 Is Lost upon Estrogen Stimulation.
To investigate whether induction of BRCA1 expression was associated with changes in the 3C profile, we examined the effect of stimulating MCF7 cells with estrogen (β-estradiol; E2). This treatment induces BRCA1 mRNA levels (21, 22), indirectly through associated changes in cell proliferation (23). We therefore analyzed BRCA1 transcription levels in MCF7 cells either without E2 (defined media as above) or after 5 and 24 h of E2 stimulation. Quantitative RT-PCR (qRT-PCR) showed that BRCA1 mRNA levels increased slightly after 5 h and 5- to 7-fold after 24 h (Fig. 1Bii and ref. 24). Using RT-PCR primers that discriminate between pre-mRNA and mature mRNA (19), we also showed that increased BRCA1 expression occurred largely at the pre-mRNA level, indicating that E2 treatment activates transcription rather than increases mRNA stability (Fig. 1Biii). 3C analysis was performed by using the BanI 3C primers described above and showed a significant decrease in the 3C product after 5 h of estrogen stimulation between the 5′ and 3′ ends of BRCA1 (Fig. 1 Bi and Biv). Significantly, after 24 h BRCA1 5′ to 3′ end association was no longer detectable. Consistent with the results of BanI 3C, the association between the BRCA1 promoter and terminator region detected by DpnII 3C was also lost after estrogen stimulation (Fig. 1 Ci and Cii). In contrast, association between the 5′ end and intron 2 of BRCA1 was unchanged upon E2 stimulation. Overall, these data indicate that the 5′ and 3′ ends of BRCA1 are juxtaposed when gene expression is repressed and released upon transcriptional induction. The timing of the change in BRCA1 chromatin structure further suggests that it is likely to be an indirect consequence of E2 stimulation, as is E2-mediated induction of BRCA1 (23). Interestingly, for FMP27 and SEN1 in yeast, a similar promoter and terminator association was detected. This gene loop conformation was associated with the promoter in a transcriptionally poised state but not under conditions of complete repression (24, 25).
Long-Range Associations Between 5′ and 3′ Ends of BRCA1 Depend on Basal Transcription.
To further investigate the mechanism of promoter and terminator interaction in BRCA1, we examined the effect of transiently inhibiting transcription after E2 induction. We used KM05283, which specifically inhibits transcriptional elongation by blocking phosphorylation of Pol II on the carboxyl-terminal domain (CTD) Ser-2 (26) (Fig. 1Ciii). DpnII 3C analysis of cells treated with KM05283 revealed that association between BRCA1 promoter and terminator does not occur if transcription is inhibited. This finding suggests that although the observed promoter-terminator loop is associated with gene repression, transcription is required for this loop to form, which raises the possibility that either transcription of a regulatory molecule is required for loop formation, or transcriptional elongation plays an active role. It is also possible that BRCA1 is autoregulated so that when threshold levels of BRCA1 mRNA are reached, loop formation is initiated to repress future transcription and so maintain low levels required for certain physiological situations (see Fig. 4). Precedents exist for negative transcriptional autoregulatory mechanisms in genes encoding Mashi, a neural-specific basic helix–loop–helix transcription factor (27) and Hairy-related transcription factors (28).
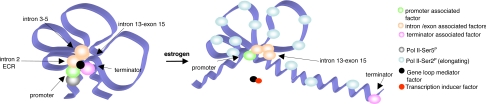
Fig. 4.
Proposed model of BRCA1 gene looping. Diagram shows two possible states of the human BRCA1 gene during transcriptional regulation. (Left) A factor (black) mediates a long-range association between the BRCA1 promoter and terminator, which in turn drives formation of a chromatin loop preventing further transcription. We refer to this as regulated repression of BRCA1, based on our finding that loop formation is transcription-dependent. (Right) When transcription of BRCA1 is induced (e.g., upon estrogen stimulation), we propose that expression of another factor (red) is induced that interferes with the function of the black factor, causing chromatin to become relaxed and promote transcription.
Other Long-Range Associations Across the BRCA1 Gene Exist, but Do Not Change in Response to Estrogen.
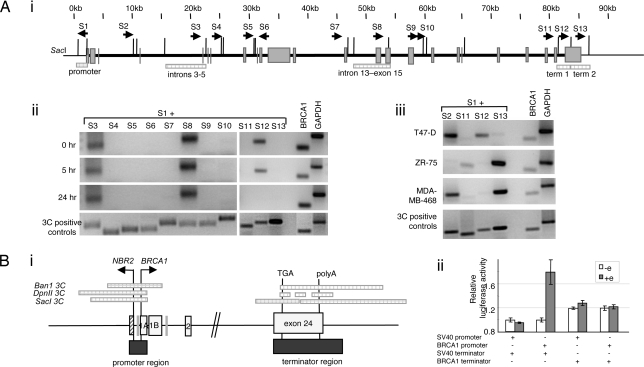
To address the possibility that other long-range associations occur between the 5′ end of BRCA1 and other regions of the gene, we performed 3C analysis with primers mapping to 10 SacI restriction fragments spanning ≈100 kb, including BRCA1 and ≈2 kb of 5′ and 3′ flanking region (Fig. 2Ai). This analysis revealed two additional gene loops, one between sequences in the 5′ end of BRCA1 (primer S1) and sequences in a region spanning introns 3–5 (primer S3) and another region spanning intron 13 to exon 15 (primer S8; Fig. 2Aii). No 3C products were detected in MCF7 cells by using validated combinations of primers mapping to the 5′ end and sequences intron 7 (primer S4), intron 8 (primers S5 and S6), intron 13 (primer S7), intron 16 (primer S9), intron 23 (primer S11), or sequences 2 kb downstream of exon 24 (primer S13). In contrast to the association between BRCA1 5′ end and 3′ end, associations between the 5′ end of BRCA1 and sequences detected by primers S3 and S8 were unchanged upon E2 stimulation (Fig. 2Aii).
Fig. 2.
Estrogen-responsive promoter-terminator gene loop of BRCA1. (A) SacI 3C analysis across BRCA1, before and after estrogen stimulation. (i) Map of BRCA1 showing SacI restriction sites with relevant exons and primers. 3C-positive fragments are indicated by checked boxes. (ii) SacI 3C analysis of MCF7 cells grown in serum-free phenol-red free media (0 h) or treated with 100 nM E2 for 5 or 24 h. 3C-positive controls and loading controls are as for Fig. 1. (iii) SacI 3C analysis of three different human breast cancer cell lines grown in complete media (T47-D, ZR-75, MDA-MB-468) show differential chromatin loops. (B) (i) Map showing the overlap between interacting 3C fragments identified by using three different restriction fragments and the corresponding sequences used to examine the regulatory activity of BRCA1 promoter and terminator sequences in reporter assays (dark shaded boxes). (ii) Luciferase activity in cells transfected with reporter constructs containing combinations of the SV40 or BRCA1 promoter and the SV40 and BRCA1 terminator region and either grown in serum-free phenol-red free media (empty bars) or stimulated with 100 nM E2 (filled bars). Data shows that the significant induction of the BRCA1 promoter by E2 (P = 0.008) is suppressed in the presence of the BRCA1 terminator (P = 0.67).
Altered BRCA1 Chromatin Loops in Breast Cancer Cell Lines.
BRCA1 mRNA levels are lower in most breast tumors than in normal tissue (1). To begin to address the possibility that this observation may be caused by differential gene loop associations across the BRCA1 gene, resulting in abnormal regulation of BRCA1 gene expression, we performed 3C analysis on three additional cell lines derived from breast tumors: T47-D, ZR-75, and MDA-MB-468. 3C products were detected in the three cell lines (Fig. 2Aiii). In T47-D cells, we observed the same association between BRCA1 promoter and terminator (primers S1 and S12), as seen with MCF7 cells (Fig. 2Aii). However, this association was not observed in ZR-75 and MDA-MB-468 cells where instead another promoter–terminator association was observed involving the adjacent 3′ SacI fragment (primers S1 and S13). This interesting difference between promoter–terminator region associations in the different breast tumor cell lines may relate to their different properties. Significantly, BRCA1 has been shown to have altered promoter DNA–protein interactions in a cell line in which BRCA1 expression is undetectable (30). Although BRCA1 repression may be caused by the associated promoter hypermethylation, an alternate explanation could be the formation of an aberrant chromatin loop between the 5′ and 3′ ends of the gene.
Estrogen-Mediated Induction of the BRCA1 Promoter in Vitro Is Repressed by Sequences in the 3′ End of BRCA1.
Our results suggest that sequences at the 5′ and 3′ ends of BRCA1 colocalize and that this event is associated with transcriptional repression. To investigate the possibility that this association contributes to the regulation of BRCA1, we generated luciferase reporter constructs driven by either the simian virus 40 (SV40) or the BRCA1 promoters and followed by sequences from the 3′ end of either SV40 or BRCA1, including both polyadenylation and terminator sequences. A map showing how the fragments used in these assays correspond to the 3C-positive restriction fragments is shown in Fig. 2Bi. We then examined the luciferase activity of cells transfected with each of these four constructs, before and after E2 induction. The BRCA1, but not the SV40 promoter, responds to E2 stimulation when followed by 3′ sequences from SV40 (Fig. 2Bii). This finding is consistent with previous reports (16) and with our data showing that induction of BRCA1 occurs at a transcriptional level (Fig. 1Biii). Interestingly, when sequences from the 3′ end of BRCA1 replace the SV40 3′ sequences, this induction is no longer evident. Possibly, in the context of this reporter system, the 3′ end of BRCA1 represses estrogen-mediated induction of the BRCA1 promoter. Transcriptional repressor elements mapping to the 3′ end of other genes have been described, including one in the 3′ UTR of the cyclin-dependent kinase inhibitor p21 (29) and three in the 3′ UTR of the serine protease inhibitor 2.3 gene (26). The existence of a transcriptional repressor element in the 3′ region of BRCA1 raises the possibility that the promoter–terminator interaction participates in this transcriptional repression mechanism.
BRCA1 Chromatin Loops in Mouse Mammary Tissue.
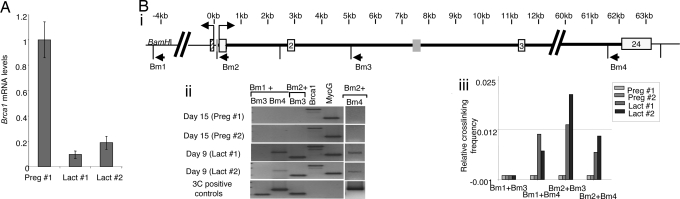
If chromatin looping is an important means of regulating BRCA1 expression, we predict that it may be an evolutionarily conserved process. We therefore performed 3C analysis on chromatin-extracted epithelial cells isolated from mouse mammary glands during lactational development. Brca1 mRNA levels are known to be induced during pregnancy and decrease during lactation (refs. 31 and 32 and confirmed by qRT-PCR in Fig. 3A).
Fig. 3.
Repression of mouse Brca1 is also associated with long-range associations. (A) qRT-PCR of Brca1 mRNA levels relative to 18S rRNA in mammary glands from mice at day 15 of pregnancy (Preg #1) or day 9 of lactation (Lact #1 and Lact #2). Quantitation was performed by using the comparative ΔCt method as described in Materials and Methods. (B) BamHI 3C analysis of Brca1 in pregnant and lactating mammary glands. (i) Map of mouse Brca1 showing BamHI restriction sites and relevant exons and primers as above. (ii) 3C analysis of chromatin extracted from pregnant and lactating mammary glands from two independent mice (nos. 1 and 2). 3C-positive controls and Brca1 and MyoG loading controls are as described above and in Materials and Methods. (iii) Quantitation of 3C band intensity relative to intensity of 3C control bands (see Materials and Methods).
A map of the mouse Brca1 gene and the primers used in 3C experiments is shown in Fig. 3Bi. Again, primers were selected for their suitability in 3C analysis, and the 3C products obtained were verified by sequence analysis. 3C analysis indicated the presence of Brca1 chromatin loops in mammary epithelial cells during lactation (Fig. 3Bii), when Brca1 expression was repressed. As with our findings for human BRCA1 (Figs. 1 and 2), we observed associations between the 5′ region of Brca1 contained within a 4.5-kb BamH1 fragment detected by primer Bm1 or the adjacent 2-kb nucleotide BamHI fragment (detected by primer Bm2) and 3′ region of Brca1 (1.9-kb BamHI fragment containing exon 24, detected by primer Bm4). Significantly, this association was not detectable in mammary epithelia from pregnant mice, when Brca1 expression is induced (Fig. 3A). This result supports the hypothesis that the BRCA1 promoter and terminator regions are juxtaposed when BRCA1 expression is repressed and that this association is released when gene expression is induced.
A 3C product consistent with an association between the promoter region and intron 2 of Brca1 was also observed (detected by primers Bm2 and Bm3). Interestingly, the intron 2 fragment contains the mouse orthologue of the conserved transcriptional regulatory sequence identified in human BRCA1 (17). If this represents the same BRCA1 promoter to intron 2 association observed in human MCF7 cells, it is interesting that, whereas the 3C pattern is unchanged upon estrogen induction in human cells, it is only present in mouse cells when Brca1 is repressed. This temporal difference may reflect the more complex regulatory pathways controlling BRCA1 expression in vivo in mouse mammary glands versus effects of estrogen on cultured human breast cancer cell lines.
A Possible Model for BRCA1 Gene Looping.
We propose a model for transcriptional regulation of the human BRCA1 by gene looping (Fig. 4). We depict the uninduced BRCA1 in a “four-leaf clover” conformation, where the promoter associates with sequences in the BRCA1 introns and with the terminator region (Figs. 1 and 2). We propose that a “gene loop mediator” factor, which could be protein or RNA, recruits proteins and/or RNAs that associate with the promoter and terminator regions of BRCA1, juxtaposing them and the DNA sequences to which they interact. In this looped conformation transcription of BRCA1 is disabled, maintaining BRCA1 expression in a “regulated repressed” state. We suggest that the repression is regulated because the associated gene loop is transcription-dependent (Fig. 1C). Possibly new transcription is required to synthesize the mediator factor. The stimulus for such transcription could be any physiological situation necessitating down-regulation of BRCA1 and could even be regulated by BRCA1 itself, potentially representing an autoregulatory loop. It is known that overexpression of BRCA1 can be toxic, so the existence of such a regulatory mechanism is a reasonable, albeit unproven, possibility.
When BRCA1 transcription is induced, for example as a consequence of estrogen stimulation, we predict that the expression of another regulatory factor is induced. We hypothesize that this factor is able to sequester the mediator protein proposed above, thus allowing the chromatin conformation to relax into a “three-leaf clover” conformation. In this conformation the BRCA1 promoter is sufficiently exposed to allow RNA Pol II to perform its role in transcriptional elongation.
Conclusions
These studies demonstrate an interaction between the promoter and terminator region of BRCA1 that is associated with gene silencing. Further investigations into the molecular mechanisms and consequences of such BRCA1 gene loops and whether they contribute to breast tumorigenesis are clear priorities for the future.
Materials and Methods
Cell Culture.
MCF7, T47-D, ZR-75, and MDA-MB-468 cells were obtained from Cancer Research UK and grown according to their recommendations. MCF7 cells were grown in serum and phenol red free RPMI medium 1640 (Invitrogen) for 24 or 48 h, then washed twice and grown in the same media, with or without 10−8 M estrogen (Sigma) for 5 or 24 h.
Animals.
Female C57BL/6 mice were obtained from the University of Queensland Central Animal Breeding House at 6 weeks of age. At 10 weeks they were estrous-matched and mated, with conception being diagnosed by vaginal plugs. Two independent pools of at least two pregnant mice were killed 15 days later, and gestation date was confirmed by morphological examination of fetuses. Two independent pools of two lactating mice were killed 9 days after parturition. Inguinal, abdominal, and thoracic mammary glands were collected, microdissected from any associated lymph nodes, fat, tendons, or musculature, and pooled. Mammary epithelial cells were isolated (33) by mincing tissue with scalpel blades, digesting with 12 mg/g collagenase (Worthington), filtering through 100-μm mesh and separating from stromal cells by differential adhesion to tissue culture plastic after 1 h. Unattached cells were then divided and used immediately for either 3C or expression analysis.
3C Analysis.
3C analysis was performed as described (34). Briefly, chromatin was cross-linked with 1% formaldehyde and nuclei were isolated by using Nonidet P-40. DNA was digested with 800 units of restriction enzyme and ligated in 8 ml of 1× ligation buffer. 3C products were phenol/chloroform-extracted, ethanol-precipitated, and quantitated by spectroscopy before PCR analysis as follows: initial denaturation of 95°C/5 min, 35 cycles of 95°C/45s, 60°C/45s, 72°C/60s, final extension of 72°C/5 min. Restriction enzymes were chosen based on their proximity to regions of interest, size of restriction sites flanking these regions, and their lack of colocalization with repetitive elements in BRCA1 using the sequence information available from the National Center for Biotechnology Information database [human BRCA1 accession no. L78833; mouse BRCA1 accession nos. NT_033681 (exons 1–24 and intervening introns) and AL590996 (flanking sequences)]. 3C primers were designed by using the Primer 3 program (http://frodo.wi.mit.edu) under the following criteria: optimal length, 26 nt (range 24–28); optimal melting temperature, 62°C (range 60–64); optimal size, 200 nt with restriction site midway between the two primers. Primers for human and mouse BRCA1 were obtained from Sigma-Aldrich and Geneworks. Artificial 3C products used in controls were generated by digesting and ligating PCR products spanning 100 nt 5′ and 3′ of the restriction sites. Two types of loading controls were also included to control for the efficiency of the 3C method and for the total amount of DNA used in the 3C analysis. First, a nonestrogen responsive housekeeping gene (hGAPDH and mMyoG) was analyzed by SacI, BanI, and DpnII 3C PCR in human-derived samples and BamHI 3C PCR in the case of mouse. Second, a BRCA1 loading control was generated by amplifying the same 3C chromatin with primers mapping to BRCA1 intron 2. Primer sequences are provided in supporting information (SI) Table S1 and Table S2.
BRCA1 mRNA Analysis (qRT-PCR).
Total RNA was extracted from either cultured cell lines or primary mammary epithelial cells by using TRIzol (Invitrogen) and DNase-treated with DNA-free (Ambion). cDNA was synthesized from 5 μg of RNA by using BRCA1 and 18S-specific reverse transcriptase primers and SuperScript III (Invitrogen). BRCA1 pre-mRNA, BRCA1 mRNA, and 18S rRNA transcripts were quantitated by using qPCR with SYBR green reaction mix (Qiagen) or SYBR green PCR master mix (Applied Biosystems), primer mix (200 nM for BRCA1 mRNA; 50 nM for BRCA1 pre-mRNA and 18S rRNA). All RT-PCR primers flanked large intronic sequences, thus preventing amplification of genomic DNA. Cycling conditions were: 10 min (95°C), followed by 45 cycles of 15 s (95°C) and 1 min (58°C). PCR products were quantitated by using the ABI Prism 7900HT Sequence Detection System with ABI Prism 7900HT SDS software (v2.2.2; Applied Biosystems). No reverse transcriptase controls were conducted for genomic DNA contamination. The comparative ΔCt method (Applied Biosystems) was used to determine relative BRCA1 mRNA and pre-mRNA expression, taking into account the primer set efficiencies (E values): target/18S = (E18S)Ct18S/(Etarget)Cttarget (35). Mean E values calculated from two independent experiments were: BRCA1 pre-mRNA, 1.9544; BRCA1 mRNA, 1.7364, 18S rRNA, 1.9998.
Luciferase Assays.
Luciferase reporter plasmids were generated by using sequences from either promoter or terminator regions of SV40 (from pGL3; Promega) or the BRCA1 promoter (499 bp) fragment containing NBR2 exon 1, BRCA1 exon 1A, and the intervening promoter sequences (17) or the BRCA1 terminator region (2,222 bp) fragment containing the BRCA1 exon 24 plus 500 bp of sequence after the polyadenylation signal. MCF7s were plated on 24-well plates in phenol red free RPMI medium 1640 containing 10% FBS. The next day, the serum was removed, and the cells were serum-starved for 24 h before transfection. Cells were transfected under serum-free conditions with 0.8 μg of the luciferase reporter plasmid and 20 ng of pRL-TK by using Lipofectamine 2000 (Invitrogen) for 24 h. Cells were induced with 100 nM E2, and relative luciferase activity was determined 24 h after induction by using a dual luciferase reporter assay kit (Promega) and a Microbeta Trilux Luminometer counter (Wallac) according to the manufacturer's instructions. To correct for any differences in transfection efficiency or cell lysate preparation, Firefly luciferase activity was normalized to Renilla luciferase. The activity of each test construct was calculated relative to no E2 treatment, the activity of which was arbitrarily defined as 1.
Transient Inhibition of Transcriptional Elongation.
MCF7 was grown in 10-cm2 dishes in phenol red free RPMI medium 1640 serum-free media for 24 h. Cells were either stimulated with 100 nM E2 for 24 h or left unstimulated. Transcription was inhibited in one of the E2-stimulated and one of the unstimulated dishes by treating cells with 100 nM KM05283 (a kind gift from S. Murphy, University of Oxford, Oxford) for 1 h. Dishes proceeded to either 3C analysis or expression analysis. For the latter, cells were washed and scraped in ice-cold PBS and extracted with either Trizol (for RNA) or Laemmli buffer (for protein). RNA was then analyzed by qRT-PCR. Protein was analyzed by Western blotting, using standard procedures. Antibodies against the N terminus of the large subunit of Pol II (H-224) (1:500; Santa Cruz biotechnology) or Ser-2 phosphorylated CTD (H5) (1:500; Covance) were used to determine the relative levels of elongating versus total Pol II. Antibody against Oct-1 (Santa Cruz) was used as a loading control.
Acknowledgments.
We thank Kelly Perkins for useful advice; Chanel Smart for expertise and help extracting primary mammary epithelial cells; and John Moore and Adrian Harris (John Radcliffe Hospital, Oxford, United Kingdom) for human cell lines. This work was supported by grants from the Wellcome Trust and the Medical Research Council (to N.J.P.) and a National Health and Medical Research Council grant (to M.A.B.). M.A.B. received a International Union Against Cancer International Cancer Technology Transfer Fellowship for sabbatical leave to Oxford. J.D.F. is supported by a National Breast Cancer Foundation Postdoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801048105/DCSupplemental.
References
- 1.Wilson C, et al. Localization of human BRCA1 and its loss in high-grade, noninherited breast carcinomas. Nat Genet. 1999;21:236–240. doi: 10.1038/6029. [DOI] [PubMed] [Google Scholar]
- 2.Magdinier F, Ribieras S, Lenoir G, Frappart L, Dante R. Down-regulation of BRCA1 in human sporadic breast cancer: Analysis of DNA methylation patterns of the putative promoter region. Oncogene. 1998;17:3169–3176. doi: 10.1038/sj.onc.1202248. [DOI] [PubMed] [Google Scholar]
- 3.Seery L, et al. BRCA1 expression levels predict distant metastasis of sporadic breast cancers. Int J Cancer. 1999;84:258–262. doi: 10.1002/(sici)1097-0215(19990621)84:3<258::aid-ijc10>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 4.Dobrovic A, Simpfendorfer D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 1997;57:3347–3350. [PubMed] [Google Scholar]
- 5.Wei M, et al. BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromosome 17 aneusomy. Cancer Res. 2005;65:10692–10699. doi: 10.1158/0008-5472.CAN-05-1277. [DOI] [PubMed] [Google Scholar]
- 6.Kadonaga J. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 7.Bulger M, Groudine M. Looping versus linking: Toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 8.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 9.Tolhuis B, Palstra R, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 10.Kurukuti S, et al. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleinjan D, van Heyningen V. Long-range control of gene expression: Emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wunderle V, Critcher R, Hastie N, Goodfellow P, Schedl A. Deletion of long-range regulatory elements upstream of SOX9 causes campomelic dysplasia. Proc Natl Acad Sci USA. 1998;95:10649–10654. doi: 10.1073/pnas.95.18.10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crolla J, van Heyningen V. Frequent chromosome aberrations revealed by molecular cytogenetic studies in patients with aniridia. Am J Hum Genet. 2002;71:1138–1149. doi: 10.1086/344396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beysen D, et al. Deletions involving long-range conserved nongenic sequences upstream and downstream of FOXL2 as a novel disease-causing mechanism in blepharophimosis syndrome. Am J Hum Genet. 2005;77:205–218. doi: 10.1086/432083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown M, et al. The 5′ end of the BRCA1 gene lies within a duplicated region of human chromosome 17q21. Oncogene. 1996;12:2507–2513. [PubMed] [Google Scholar]
- 16.Xu C, Chambers J, Solomon E. Complex regulation of the BRCA1 gene. J Biol Chem. 1997;272:20994–20997. doi: 10.1074/jbc.272.34.20994. [DOI] [PubMed] [Google Scholar]
- 17.Wardrop S, kConFab Investigators. Brown M. Identification of two evolutionarily conserved and functional regulatory elements in intron 2 of the human BRCA1 gene. Genomics. 2005;86:316–328. doi: 10.1016/j.ygeno.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Lane T, Lin C, Brown M, Solomon E, Leder P. Gene replacement with the human BRCA1 locus: Tissue-specific expression and rescue of embryonic lethality in mice. Oncogene. 2000;19:4085–4090. doi: 10.1038/sj.onc.1203760. [DOI] [PubMed] [Google Scholar]
- 19.Saunus J, Edwards S, French J, Smart C, Brown M. Regulation of BRCA1 messenger RNA stability in human epithelial cell lines and during cell cycle progression. FEBS Lett. 2007;581:3435–3442. doi: 10.1016/j.febslet.2007.06.046. [DOI] [PubMed] [Google Scholar]
- 20.Dekker J. The three Cs of chromosome conformation capture: Controls, controls, controls. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 21.Spillman M, Bowcock A. BRCA1 and BRCA2 mRNA levels are coordinately elevated in human breast cancer cells in response to estrogen. Oncogene. 1996;13:1639–1645. [PubMed] [Google Scholar]
- 22.Romagnolo D, et al. Estrogen up-regulation of BRCA1 expression with no effect on localization. Mol Carcinog. 1998;22:102–109. doi: 10.1002/(sici)1098-2744(199806)22:2<102::aid-mc5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 23.Gudas J, Nguyen H, Li T, Cowan K. Hormone-dependent regulation of BRCA1 in human breast cancer cells. Cancer Res. 1995;55:4561–4565. [PubMed] [Google Scholar]
- 24.O'Sullivan J. Gene loops juxtapose promoters and terminators in yeast. Nat Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 25.Ansari A, Hampsey M. A role for the CPF 3′ end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paul C, Simar-Blanchet A, Ro H, Le Cam A. Characterization of three transcriptional repressor sites within the 3′ untranslated region of the rat serine protease inhibitor 2.3 gene. Eur J Biochem. 1998;254:538–546. doi: 10.1046/j.1432-1327.1998.2540538.x. [DOI] [PubMed] [Google Scholar]
- 27.Meredith A, Johnson J. Negative autoregulation of Mash1 expression in CNS development. Dev Biol. 2000;222:336–346. doi: 10.1006/dbio.2000.9697. [DOI] [PubMed] [Google Scholar]
- 28.Nakagawa O, et al. Members of the HRT family of basic helix–loop–helix proteins act as transcriptional repressors downstream of Notch signaling. Proc Natl Acad Sci USA. 2000;97:13655–13660. doi: 10.1073/pnas.250485597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rishi A, et al. Transcriptional repression of the cyclin-dependent kinase inhibitor p21WAF1/CIP1 gene mediated by cis elements present in the 3′ untranslated region. Cancer Res. 1997;57:5129–5136. [PubMed] [Google Scholar]
- 30.Rice J, Futscher B. Transcriptional repression of BRCA1 by aberrant cytosine methylation, histone hypoacetylation, and chromatin condensation of the BRCA1 promoter. Nucleic Acids Res. 2000;28:3233–3239. doi: 10.1093/nar/28.17.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marquis S, et al. The developmental pattern of BRCA1 expression implies a role in differentiation of the breast and other tissues. Nat Genet. 1995;11:17–26. doi: 10.1038/ng0995-17. [DOI] [PubMed] [Google Scholar]
- 32.Rajan J, Marquis S, Gardner H, Chodosh L. Developmental expression of Brca2 colocalizes with BRCA1 and is associated with proliferation and differentiation in multiple tissues. Dev Biol. 1997;184:385–401. doi: 10.1006/dbio.1997.8526. [DOI] [PubMed] [Google Scholar]
- 33.Pullan S, et al. Requirement of basement membrane for the suppression of programmed cell death in mammary epithelium. J Cell Sci. 1996;109:631–642. doi: 10.1242/jcs.109.3.631. [DOI] [PubMed] [Google Scholar]
- 34.Vakoc C, et al. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 35.Livak J, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]