Abstract
Adaptor proteins stimulate the nuclear export of mRNA, but their mechanism of action remains unclear. Here, we show that REF/ALY binds mRNA; but upon formation of a ternary complex with TAP the RNA is transferred from REF to TAP, and overexpression of TAP displaces REF from mRNA in vivo. RNA is also handed over from two other adaptors, 9G8 and SRp20 to TAP upon formation of a ternary complex. Interestingly, the RNA-binding affinity of TAP is enhanced 4-fold in vitro once it is complexed with REF. 9G8 and SRp20 also enhance the TAP RNA-binding activity in vitro. Consistent with a model in which TAP directly binds mRNA handed over from adaptors during export, we show that TAP binds mRNA in vivo by an arginine-rich motif in its N-terminal domain. The importance of direct TAP–mRNA interactions is confirmed by the observation that a mutant form of TAP that fails to bind mRNA but retains the ability to bind REF does not function in mRNA export.
Keywords: gene expression, RNA, SF2/ASF, NXF1 transport
Transport of mRNA from the nucleus to cytoplasm is an essential step in eukaryotic gene expression. Various proteins involved in mRNA export are conserved from yeast to man, including Sub2p, Yra1p, and Mex67p, whose mammalian orthologs are UAP56, REF/ALY, and TAP/NXF1, respectively. Sub2p/UAP56 is an RNA helicase required for spliceosome assembly and mRNA export (1, 2). Sub2p/UAP56 binds to Yra1p/REF, and together they associate with THO proteins to form the TREX complex, which directly couples transcription and export in yeast and indirectly couples transcription and export by splicing in humans (3, 4). UAP56 provides a bridge between THO proteins and REF in TREX (4) and REF is thought to assist recruitment of TREX to the 5′ ends of mRNA by an interaction with the Cap-binding complex (5, 6).
REF/Yra1p interacts with TAP/Mex67p, and in yeast this interaction leads to displacement of Sub2p from Yra1p (7). TAP heterodimerizes with p15 and binds nucleoporins through central and C-terminal domains (8), directing the mRNP to the nuclear pore and promoting transport to the cytoplasm. On the cytoplasmic side of the nuclear pore, Dbp5p triggers displacement of Mex67p from mRNA. Yra1p binds mRNA early during its nuclear maturation but is no longer bound once it reaches the nuclear periphery (9). Consistent with this finding, analysis of Balbiani ring pre-mRNPs shows that UAP56 and REF accompany the mRNP to the nuclear periphery where UAP56 and then REF dissociate during translocation through the pore (10).
Although Yra1p is essential for yeast mRNA export, depletion of REF in higher eukaryotes does not block bulk mRNA export (11), suggesting that other proteins can fulfill this role and that there may be functional redundancy between export adaptors. The shuttling SR proteins 9G8, SRp20, and SF2/ASF directly bind TAP by short arginine-rich peptides (12, 13) and can function as export factors (14). Even in yeast, other proteins can recruit Mex67p to the mRNP, including Yra2p (15) and Npl3p.
The fact that TAP binds RNA weakly in vitro led to the idea that export factors such as REF, which bind RNA avidly, bridge the interaction between TAP and mRNA, leading to the term mRNA export adaptors (15). However, both TAP and Mex67p are readily UV-cross-linked to mRNA in vivo (16–18), suggesting a direct stable interaction at some point during export. Here, we show that mRNA is handed over from export adaptors to TAP and that at least in vitro, export adaptors have the ability to enhance the RNA-binding activity of TAP.
Results
The RNA- and TAP-Binding Sites on REF Overlap.
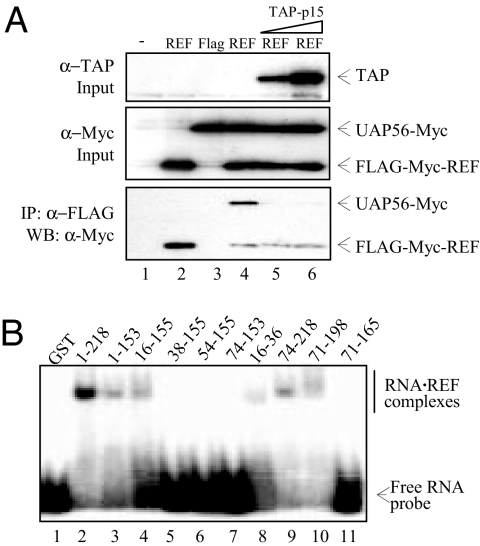
In Saccharomyces cerevisiae, Mex67p binding to Yra1p triggers displacement of Sub2p (7), so we established whether this is the case for the mammalian orthologs. We examined whether UAP56 coimmunoprecipitated (Co-IP) with REF2-I (REF) in the presence of increasing amounts of TAP. This analysis revealed that TAP triggered dissociation of UAP56 from REF (Fig. 1A, lanes 5 and 6).
Fig. 1.
Displacement of UAP56 from REF by TAP. (A) Co-IP assays. Total extracts from 293T cells (Mock, lane 1) transfected with FLAG-Myc-tagged REF (lane 2) or cotransfected with Myc-tagged UAP56 and either control (FLAG) or FLAG-Myc-tagged REF construct (REF) (lanes 3–6) were incubated with increasing amount of purified recombinant GST-TAP-p15 (TAP-p15, lanes 5 and 6) before IP with α-FLAG antibodies. Total extracts (Top and Middle) and purified complexes (Bottom) were analyzed by Western blotting (WB) with the indicated antibodies. (B) EMSA with a 32P-radiolabeled 15-mer RNA in the presence of 1 μM GST control (lane 1) and GST-tagged fusions of REF (lanes 2–11).
We analyzed organization of the resulting REF–TAP–RNA ternary complex by examining how REF binds RNA. NMR techniques had revealed extensive chemical shifts on binding RNA, mapping to the N domain (amino acids 1–73) and RRM (amino acids 74–155) loops 1 and 5 (19). Electrophoretic mobility shift assays (EMSAs) also implicated the N and C variable regions (amino acids 15–71 and 165–198) in RNA binding (20). We used EMSAs to dissect regions of the N variable region involved in RNA binding [Fig. 1B and supporting information (SI) Fig. S1A]. REF and both N and C (amino acids 156–218) domains fused to the RRM bind RNA, and removal of the first 15 or last 20 aa did not alter this activity (Fig. 1B, lanes 3 and 4, and 9 and 10). In contrast, amino acids 38–155 and 54–155 failed to bind RNA, implicating amino acids 16–37 in RNA binding. Indeed, a minimal peptide encompassing amino acids 16–36 fused to GST bound RNA (Fig. 1B, lane 8). These data are consistent with observations that this region displays NMR chemical shifts upon addition of RNA (19). The EMSA data also implicate amino acids 166–198 in RNA binding (Fig. 1B, lanes 10 and 11), and this region together with amino acids 16–36 of REF contain RGG RNA-binding motifs (21). Despite being unable to observe an interaction between the REF RRM and RNA with EMSA (Fig. 1B, lanes 6 and 7 and ref. 20), a weak interaction between the RRM and RNA is detectable by NMR and UV-cross-linking assays (19). Taken together, the data suggest that the N- and C-terminal RGG boxes together with the RRM contribute to RNA binding.
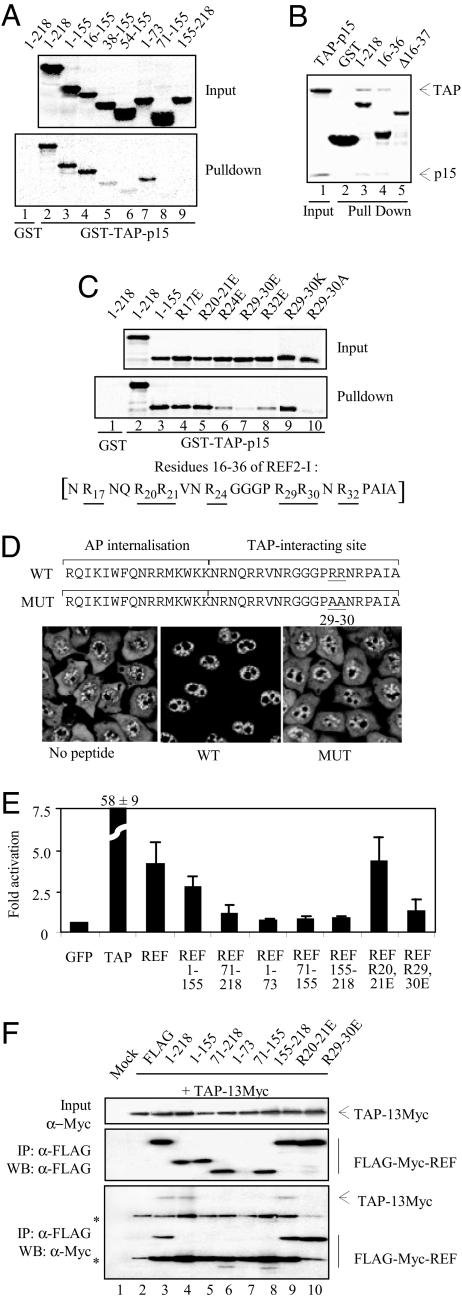
We next examined how REF binds TAP. Full-length REF bound GST-TAP/p15 as did amino acids 1–155 and 16–155 (Fig. 2A), whereas amino acids 38–155 bound GST-TAP/p15 more weakly, suggesting that amino acids 16–37 were important for TAP interaction. The minimal REF RNA-binding peptide, amino acids 16–36, fused to GST was sufficient for TAP interaction by using a pulldown assay (Fig. 2B). Furthermore, when this region was deleted, the remaining weak interaction with the RRM was not detectable by using Coomassie staining (Fig. 2B, lane 5). Weak binding was seen for constructs encompassing the RRM (Fig. 2A, lane 6), which requires amino acids 54–73 for optimal folding, consistent with earlier NMR and biochemical analysis of the TAP–REF interaction (22). The interaction with the TAP-binding site in the N domain (Fig. 2A, lane 7) is weaker than that seen for the N domain + RRM (lane 3), indicating the two domains work together to ensure optimal TAP binding, in contrast to the earlier suggestion that the RRM was unimportant for TAP binding (20). Earlier reports suggested that TAP also interacts with the C domain of REF proteins, yet we only detected this interaction, which mapped to amino acids 155–198, using a less chaotropic buffer (Fig. S1C). Together, these results indicate that TAP binds strongly to amino acids 16–36 and weakly to the RRM and C-terminal binding sites, with the combined action of amino acids 16–36 and the RRM required for optimal interaction. These data are consistent with the observation that the Yra1p N-terminal domain binds Mex67p strongly compared with the C-terminal domain (15).
Fig. 2.
TAP-binding domain in REF. (A) Pulldown assays. GST (control, lane 1) and GST-TAP-p15 (lanes 2–9) were immobilized on glutathione-coated beads. 35S-radiolabeled REF proteins indicated by aa numbers were added to the binding reactions in the presence of RNase. Eluted proteins were analyzed by SDS/PAGE and PhosphorImaging. (B) Pulldown assays. Input corresponds to purified TAP-p15. Recombinant GST (lane 2) and GST-REF (lane 3), GST-REF amino acids 16–36 (lane 4), or GST-REF 1–155 deleted from amino acids 16 to 37 (lane 5) were immobilized on glutathione beads, and TAP-p15 was added. Eluted proteins were analyzed by SDS/PAGE stained with Coomassie blue. (C) Pulldown assays as in A. GST (lane 1) and GST-TAP-p15 (lanes 2–10) were used in pulldown assays with 35S-REF proteins + RNase. Sequence of the TAP-interacting site of REF with underlined arginine positions is shown. (D) (Upper) Peptide sequences of Antennapedia cell-permeable element (AP) fused to REF amino acids 16–36 (WT) or to the same peptide with mutations of R29,R30 (MUT). (Lower) Poly(A)+ RNA localization in HeLa cells (no peptide) and in cells incubated for 72 h with 12.5 μM WT or MUT peptides. (E) Luciferase activity generated by MS2 fusions normalized with a LacZ transfection control in the tethered export assay with the pLUCSALRRE6MS2 reporter. Error bars represent SD values from three datasets, each carried out in triplicate. Fold activations relative to MS2-GFP are shown. (F) Co-IP assays. Total extracts from 293T cells (Mock) cotransfected with Myc-tagged TAP and either control (FLAG) or FLAG-Myc-tagged REF constructs indicated by aa numbers were treated with RNase before IP with α-FLAG antibodies. Total extracts (Top) and purified complexes (Middle and Bottom) were analyzed by Western blotting with the indicated antibodies. Asterisks indicate heavy or light IgG chains. REF amino acids 1–155 and 71–218 comigrate with the light chains of FLAG antibody used for IP.
An arginine-rich motif is essential for TAP interaction with 9G8, SRp20, and SF2/ASF (12, 13). Therefore, we assessed whether arginines present within REF amino acids 16–36 were required for TAP binding. Mutation of R17, R20, and R21 had no effect on the interaction (Fig. 2C, lanes 4 and 5), whereas R24 and R32 mutations reduced binding significantly (Fig. 2C, lanes 6 and 8) and double mutation of R29,R30 drastically reduced TAP binding (Fig. 2C, lanes 7 and 10). Therefore, R29,R30 plays a critical role in binding TAP, with R24,R32 contributing to this interaction. Thus, 9G8, SRp20, and REF all use arginine-rich peptides to bind TAP. Mutation of R29K,R30K (Fig. 2C, lane 9) still supported a strong TAP interaction, indicating that the charge of these amino acids plays a key role in binding.
Analysis of REF Domains in mRNA Export.
To examine whether the N-terminal TAP-binding motif in REF was functional in vivo, we generated fusions of Antennapedia internalization element to amino acids 16–36 of REF (WT) or to a peptide bearing mutations of R29–R30 (MUT) (Fig. 2D). The effects of these peptides on mRNA export in HeLa cells were examined by using fluorescence in situ hybridization (FISH) to detect poly(A)+ RNA. In the absence of peptide, HeLa cells showed a clear mRNA signal in the nucleus and cytoplasm. In contrast, there was a robust nuclear accumulation of mRNA at 72 h in the presence of the WT peptide with little cytoplasmic mRNA staining, indicating that the TAP-binding peptide functions as an inhibitor of mRNA export. Mutations R29A,R30A abolished these effects, highlighting the importance of R29,R30 for interaction with TAP-p15.
There is potentially functional redundancy between mRNA export adaptors, therefore to address specifically the function of REF domains in vivo further we used a tethered assay. In this assay, the activity of an export factor is monitored by tethering it to an inefficiently exported reporter mRNA via bacteriophage MS2 coat protein (MS2) and RNA operator sequences (13) (Fig. 2E). The MS2 fusion protein expression was verified, and all constructs with the exception of MS2-REF (amino acids 71–155) showed good levels (Fig. S2). All MS2 fusions showed nuclear expression by immunofluorescence (data not shown). The tethering of TAP to reporter mRNA led to a 58-fold activation of luciferase activity compared with MS2-GFP, whereas MS2-REF gave rise to ≈4-fold activation. Amino acids 1–155 showed 75% activity compared with full-length REF, whereas the C domain does not function in this assay. Amino acids 71–218, lacking the N-terminal binding site for TAP, showed weak activation, as did the isolated N domain. Thus, although TAP binds the N domain of REF in vitro and a peptide from this domain blocks mRNA export in vivo, the strength of this interaction appears insufficient to promote export of reporter RNA. Given that amino acids 1–155 encoding the N domain and RRM shows a stronger TAP interaction in vitro and good activity in vivo, it may be that the combined action of the N and RRM domains of REF is important for function in vivo providing a stable TAP-binding site. REF with point mutations in R20,R21, which binds TAP well in vitro, functions normally in this assay, whereas R29,R30 mutations that reduces the interaction with TAP in vitro, significantly reduce its mRNA export activity in vivo. To confirm the function of REF domains in vivo correlates with their TAP-binding ability we used a Co-IP assay (Fig. 2F). These data showed that domains of REF that function in the export assay, Co-IP with TAP and vice versa. The Co-IP assay is less sensitive than the in vitro binding assays, which may explain why weaker interactions between TAP and REF domains, e.g., REF (amino acids 1–73), are not detected. We conclude that the combined action of amino acids 16–36 and RRM provides a stable binding site for TAP in vivo, allowing its efficient recruitment to mRNA for export.
TAP Displaces RNA from Export Adaptors, Which in Turn Enhance RNA Binding by TAP.
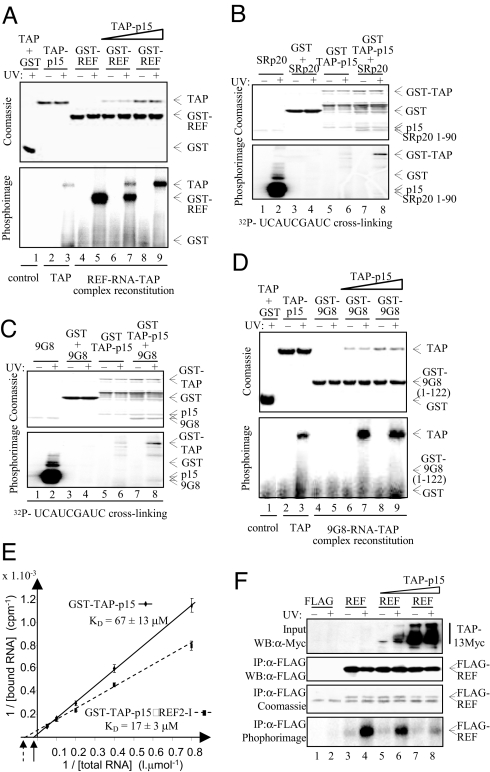
The data presented above together with NMR data (19) show that RNA and TAP bind overlapping regions in REF, suggesting a mutually exclusive interaction, addressed by using RNA UV-cross-linking assays. TAP-p15 showed a weak and GST-REF a strong cross-link with RNA, respectively (Fig. 3A, lanes 3 and 5). When GST-REF was incubated with RNA and then mixed with equimolar TAP-p15 followed by UV cross-linking, there was substoichiometric pull down of TAP-p15 with GST-REF (Fig. 3A, lanes 6 and 7). However, there was a significant reduction in the RNA bound to REF and a concomitant increase in RNA cross-linked to TAP compared with free TAP (Fig. 3A, lanes 3, 7, and 9). When TAP was present in a 5 molar excess (in solution), the interaction between RNA and REF was abolished, whereas there was a ≈5-fold increase in RNA cross-linked to TAP (Fig. 3A, lane 9). We conclude that when TAP binds a REF–RNA complex, RNA is displaced from REF. Even when excess TAP is used in these assays, we fail to see stoichiometric pull down of TAP with GST-REF, yet at the time of UV cross-linking (in solution), REF was clearly saturated with TAP because no RNA was bound to REF, which indicates that during purification after cross-linking, a significant amount of TAP is lost. To examine whether the ability to alter the RNA cross-linking activity of TAP was a general property of adaptors, we carried out assays with 9G8 and SRp20. Using an RNA that cross-linked to SRp20 and 9G8, we showed that RNA is displaced from both proteins on TAP binding and that they enhance the TAP UV cross-linking activity (Fig. 3 B and C, lanes 6 and 8). Similar results were obtained by using SF2/ASF (data not shown). These data show that enhancement of TAP RNA-binding activity is a general property of adaptors.
Fig. 3.
Export adaptors enhance TAP RNA binding. (A) In vitro reconstitution of REF-RNA-TAP-p15 complexes. GST (lane 1), TAP-p15 (lanes 2 and 3), or GST-REF (REF, lanes 4–9) were first incubated with continuously 32P-radiolabeled RNA. Equimolar (lanes 6 and 7) or 5 molar excess (lanes 8 and 9) TAP-p15 was added to the GST and GST-REF RNA-binding reactions. Bound RNA was UV-cross-linked as indicated, treated with RNase, and protein–RNA complexes were purified. Eluted complexes were analyzed on SDS/PAGE by Coomassie blue (Upper) and PhosphorImaging (Lower). (B and C) Cross-linking experiments carried out as described in A except GST-TAP was used to pull down SRp20 (amino acids 1–90) (B) or 9G8 (amino acids 12–98) (C). The indicated end-labeled RNA oligonucleotides were used. Lanes 1 and 2 show free SR proteins; lanes 3–8 represent pulldowns. (D) In vitro reconstitution of 9G8 RNA-TAP-p15 complexes. Experiments were carried out and displayed as in A except GST-9G8 (amino acids 1–122) was used. (E) RNA binding affinities were measured as described in Experimental Procedures for GST-TAP-p15 and GST-TAP-p15–REF2–1 complex. (F) In vivo competition assay. 293T cells transfected with either FLAG control (FLAG) or FLAG-tagged REF (REF) and increasing amounts of Myc-tagged TAP and p15 expression plasmids (TAP-p15) were exposed to UV as indicated. REF complexes were immunopurified with α-FLAG antibodies. (Bottom) Purified complexes were treated with RNase, and the resulting residual mRNAs cross-linked to REF were end-labeled by using [γ32]ATP and polynucleotide kinase before PhosphorImaging. (Top) Increasing expression of TAP was confirmed in total extracts by α-Myc Western blotting (WB). (Top Middle and Bottom Middle) Successful purification of FLAG-REF was confirmed by WB (Top Middle) and Coomassie staining (Bottom Middle). The band found in all lanes in Bottom Middle corresponds to the IgG light chain.
A further cross-linking assay was carried out by using continuously labeled nonspecific RNA that did not bind 9G8; yet in this assay, 9G8 was still capable of enhancing TAP RNA cross-linking (Fig. 3D, lanes 3 and 8). This finding indicates that adaptors do not simply increase the local RNA concentration in this assay upon binding to TAP; rather, they directly alter the TAP RNA UV-cross-linking activity. We confirmed that the increased cross-linking activity seen for TAP corresponded to increased RNA-binding affinity by using a GST pulldown assay with radiolabeled 15-mer RNA followed by quantification of bound and free RNA (Fig. 3E). Under conditions used in this assay, REF efficiently complexed with TAP and in the complex only TAP bound RNA (Fig. S3). Binding analysis (Fig. 3E) showed GST-TAP-p15 bound RNA with a Kd of 67 ± 13 μM, whereas GST-TAP-p15–REF complex bound RNA with a Kd of 17 ± 3 μM, representing a 4-fold higher affinity, which is consistent with the ≈5-fold increase in UV-cross-linking activity.
We next examined whether excess TAP influenced the interaction of REF with RNA in vivo. REF was cross-linked to RNA in vivo and immunoprecipitated under stringent conditions, and the bound RNA was digested to a minimal fragment and end-labeled (Fig. 3F). With normal TAP levels, there was a robust UV-dependent interaction between REF and RNA, consistent with the observation that Yra1p cross-links with mRNA in vivo (9). Overexpression of TAP led to a dose-dependent decrease in the RNA cross-linked to REF (Fig. 3F, lanes 6 and 8). These data are consistent with a model whereby RNA and TAP binding to REF are mutually exclusive in vivo.
The RNA-Binding Domain of TAP Is Required for Its Export Activity in Vivo.
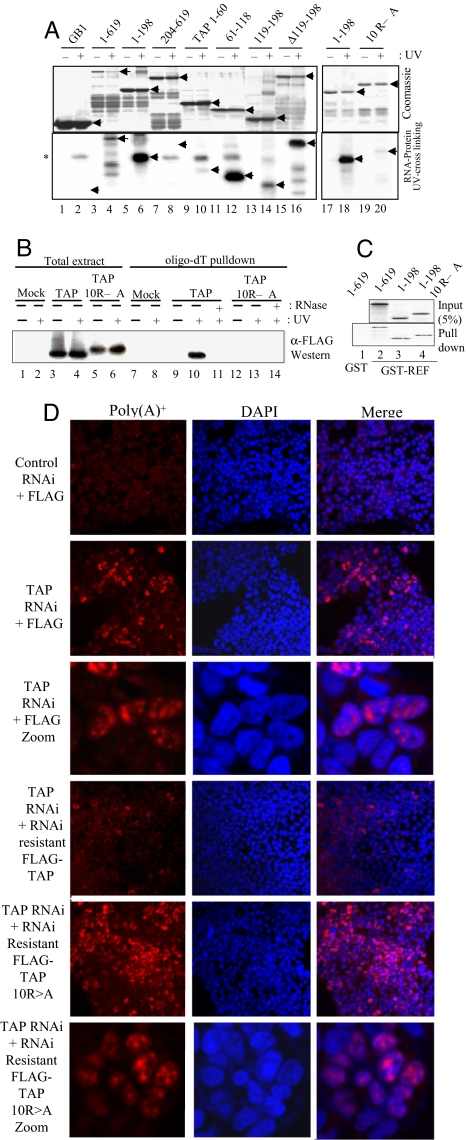
To determine the importance of RNA handover to TAP, we set out to generate mutations in TAP that retained the ability to bind REF but not RNA. To this end, we investigated which aa of TAP were responsible for general RNA binding. Earlier work demonstrated the TAP RNP-type RNA-binding domain (RBD) was dispensable for activity in vivo (23), although it had not been shown that the isolated RBD (amino acids 118–198) bound RNA. Amino Acids 96–198, encompassing the RBD, bind RNA (24), yet a separate study showed that amino acids 61–121 were involved in RNA binding (25). We clarified which region of TAP is responsible for nonspecific RNA binding by using truncations, internal deletions, and mutations (Fig. S1). These results showed that internal RBD deletion, shown to function in vivo, cross-linked with RNA efficiently (Fig. 4A, lanes 4 and 16) and that the isolated RBD cross-linked with RNA inefficiently (Fig. 4A, lane 14). Amino acids 61–118 showed a strong RNA cross-linking activity consistent with earlier work (25) and amino acids 1–60 a weaker activity. Within amino acids 61–118 there are a large number of arginines potentially involved in RNA binding, and despite low overall conservation of this region across organisms, TAP orthologs possess a basic region at the N terminus (Fig. S4). Mutation of 10 arginines within this region (Fig. S4) in TAP amino acids 1–198 drastically reduced RNA binding in vitro (Fig. 4A, lanes 18 and 20), and full-length TAP 10RA did not UV cross-link to mRNA in vivo (Fig. 4B, lanes 10 and 13). However, these mutations did not prevent the interaction with REF (Fig. 4C).
Fig. 4.
Identification of the TAP RNA-binding domain. (A) RNA cross-linking using continuously labeled RNA and GB1-TAP fusions. (B) UV cross-linking mRNP capture assay. 293T cells were transfected with FLAG-TAP-Myc or FLAG-TAP-Myc 10RA vectors and mRNA capture assays carried out after UV irradiation of cells as indicated. TAP proteins cross-linked to the mRNA were detected by Western blotting (WB) with α-Myc Abs. (C) GST pulldowns using GST (lane 1) or GST-REF (lanes 2–4) with GB1-TAP fusions. (D) Rescue of the mRNA export block induced by TAP RNAi. HeLa cells were transfected with pSUPERLUC (control RNAi) or pSUPERTAP (TAP RNAi) together with FLAG, FLAG-TAP, or FLAG-TAP 10RA vectors modified to make the mRNAs resistant to RNAi. Poly(A)+ RNA was detected by using Cy3-oligo(dT)50. The third and sixth sets of panels from the Top show a larger magnification image compared with other panels.
To address whether the 10RA form of TAP was functional in vivo, we assessed its ability to rescue the mRNA export defect seen in mammalian cells after RNA interference (RNAi) knockdown of TAP (26). Rescue cDNA expression vectors were prepared by generating silent mutations in the TAP RNAi vector target sequence for both WT and TAP 10RA. These vectors included an N-terminal nuclear localization signal to counteract potential disruption of the normal TAP nuclear localization signal in the 10RA mutant. Western blot analysis (Fig. S5A) confirmed that these cDNAs were resistant to RNAi. The ability of these cDNAs to rescue TAP knockdown was assessed in HeLa cells treated with actinomycin D for 2 h before detection of poly(A)+ RNA to reduce the nuclear signal from nascent mRNA (27). After TAP knockdown, a robust nuclear accumulation of RNA was observed in many cells at 72 h after transfection. The TAP RNAi-resistant cDNA rescued the TAP knockdown with very few cells showing nuclear accumulation of mRNA. In contrast, the 10RA form of TAP was incapable of rescuing the TAP RNAi phenotype, with many cells showing nuclear accumulation of mRNA. The phenotype seen with the 10RA mutant was more pronounced than that seen with the TAP RNAi vector alone. Consistent with this finding, expression of the 10RA mutant in HeLa cells with a control RNAi vector led to a limited number of cells showing an mRNA export block, indicating that this mutant may act as a weak dominant negative. We conclude that the RNA-binding activity of TAP is required for its mRNA export activity in vivo.
Discussion
Although the importance of adaptors for mRNA export is established, their precise function has remained unresolved. Here, we have shown that at least one function of export adaptors is to hand mRNA over to TAP during export. Therefore, export adaptors play a central role in remodeling the protein–mRNA interactions within the mRNA export complex during transport from the nucleus to the cytoplasm.
Function of REF Domains.
The overlap between DDX39 and TAP-binding sites on REF revealed by previous structural studies (19) explains why recruitment of TAP to REF leads to displacement of UAP56 as demonstrated here. This situation is similar to that found in yeast and is consistent with electron microscopy data from Chironomus tentans, indicating that UAP56 is displaced from the mRNP before REF (10).
Functional studies in Xenopus oocytes had shown that REF can stimulate mRNA export but only when the RRM was present (20). Furthermore, deletion of the RRM from S. cerevisiae Yra1p causes mRNA export defects (15), yet these studies did not identify the weak interaction between the RRM and RNA/TAP. Therefore, the reasons for conservation of the RRM in adaptors and its requirement for activity in vivo were unclear. Structural studies of REF have shown that the N domain binds the RRM in the free state and that interaction with RNA, TAP, or DDX39 triggers a conformational change such that these ligands are embraced by the N domain of REF and the RRM (19). By using REF constructs ± RRM we have shown how the combined interaction with amino acids 16–36 and the RRM provides the stable TAP-binding site required for REF activity in vivo.
The N-terminal TAP-binding peptide in REF is arginine-rich, and the SR proteins 9G8, SRp20, and SF2/ASF also use arginines in a short peptide adjacent to their RRM for interaction with TAP (12, 13), indicating a common mode for TAP binding. Furthermore, the arginine-rich TAP-binding peptide from REF functions as an effective mRNA export inhibitor in vivo. Although the primary interaction with TAP involves arginines, both classes of adaptor show weak interactions with TAP via their RRMs. As well as stabilizing the overall interaction, the RRM may contribute to the specificity of binding, especially given the low complexity of the arginine-rich peptides and their prevalence in other RNA-binding proteins that do not function as export adaptors.
Handover of mRNA from Export Adaptors to TAP.
It has been assumed that export adaptors would remain bound to RNA during export and bridge the interaction between TAP and mRNA. Yet, we have shown that in fact the mRNA is transferred to TAP, which could account for why a TAP–mRNA cross-link is readily detected in vivo (16–18 and this work).
Earlier studies on Yra1p and Mex67p suggested that Mex67p and RNA bound Yra1p simultaneously (28). However, a preformed complex of Mex67p and GST-Yra1p, with Mex67p present in substoichiometric amounts, was used for UV-cross-linking analysis; free GST-Yra1p would have been present and capable of interacting with RNA. We observe that, by using a preformed GST-REF–TAP/p15 complex, where substoichiometric amounts of TAP/p15 are present, RNA cross-links to both TAP and the free GST-REF (data not shown). In light of this observation, Yra1p may also be displaced from mRNA once saturated with Mex67p.
Modifying the TAP RNA-Binding Activity.
When TAP is tethered to pre-mRNA or over expressed it stimulates the direct export of pre-mRNA (23). Simple retroviruses and TAP pre-mRNA have taken advantage of this property by evolving specific high-affinity RNA elements that directly bind TAP, recruiting it and promoting export of RNAs that would normally be exported inefficiently (29, 30). Were TAP to directly bind all cellular pre-mRNAs efficiently, it might lead to premature export, a disastrous situation for the cell, so it is likely that this process is regulated. Recruitment of TAP to mRNA by export adaptors, whose own recruitment is likely to be regulated by RNA-processing events, provides one means to control the activity of TAP.
The in vitro observation that adaptors, including REF, SRp20 and 9G8, can enhance the RNA-binding affinity of TAP up to 4-fold is intriguing. The major nonspecific RNA-binding activity of TAP involves an arginine-rich motif between amino acids 61 and 118, a region overlapping the minimal REF-binding domain in TAP (amino acids 1–202) (14). Therefore, adaptors bind a region that could influence the structure of the TAP RNA-binding domain. Because this domain is predicted to be unstructured, adaptor binding may induce a more structured conformation in TAP, conducive to RNA binding. Whether adaptors are capable of enhancing the TAP RNA-binding activity in vivo, thus providing a second means to regulate TAP activity, is an open question. Overexpression of REF does not lead to increased mRNP association for TAP using an mRNP capture assay (data not shown and ref. 25). However, other proteins may be limiting in this situation, and the stable recruitment of TAP to mRNA may require factors such as U2AF or the Cap-binding complex. Interestingly, 9G8 has recently been shown to enhance expression of genes containing a constitutive transport element (CTE) (31), which may arise through enhanced TAP–CTE interaction. Clearly, further experiments are required to resolve this issue and further establish whether loss of an export adaptor during the late stages of export destabilizes TAP–RNA interactions, priming TAP for displacement from mRNA in the cytoplasm.
Experimental Procedures
Plasmids.
Plasmids used in this work are described in Table S1.
GST Pulldown Assays and Co-IPs.
Pulldown assays were performed in the presence of 5 μg of RNase as described in ref. 26 in PBS + 0.1% Tween or in buffer RB100 [25 mM Hepes (pH 7.5), 100 mM KOAc, 10 mM MgCl2, 1 mM DTT, 0.05% Triton X-100, 10% (vol/vol) glycerol] for Fig. S1. Co-IP assays from transfected 293T extracts treated with RNase were performed as described in ref. 12. GST-TAP and p15 were produced by coexpression of pGEX6P1-TAP and pET9a-p15 in Escherichia coli. The two proteins copurify on glutathione–Sepharose.
RNA Analysis.
EMSA used a γ-32P end-labeled CAGUCGCAUAGUGCA RNA as described in ref. 32. In vitro cross-linking assays used 32-mer 32P continuously labeled RNA synthesized from 1 μg of XbaI-restricted pBluescript-KS, 5 μg of GST-REF or GST-9G8 1–122, or free TAP-p15 in 18 μl of RNA-Cl buffer [15 mM Hepes (pH 7.9), 8 mM NaCl, 100 μM KCl, 0.2 mM EDTA, 5 mM MgCl2, 0.05% Tween 20, 10% (vol/vol) glycerol] were mixed with 2 μl of RNA for 10 min on ice and 10 min at room temperature. One to five molar excess of TAP-p15 was incubated with GST-REF–9G8-RNA complexes for 10 min at room temperature and then UV-irradiated on ice. Reactions were treated with 5 μg of RNase A for 30 min at 37°C for 32-mer and incubated for 15 min with 10 μl of GSH beads. For in vitro cross-linking assays with 32P end-labeled 5′-CAGUCGCAUAGUGCA or 5′-UCAUCGAUC (Dharmacon), 5 μg of REF, SRP20 1–90, or 9G8 12–98 was incubated with 40 ng of RNA for 10 min at room temperature before adding 2.5 μg of immobilized GST-TAP-p15. Beads were washed, and eluted complexes were UV-irradiated on ice. Complexes were analyzed by SDS/PAGE stained with Coomassie blue and PhosphorImaging. RNA affinities of GST-TAP-p15 and GST-TAP-p15–REF complex were measured by using reactions containing 5 μg of immobilized proteins and 0.31–10 μM 15-mer 32P end-labeled RNA in 50 mM NaP (pH 7), 50 mM NaCl, 1 mM MgCl2, 0.1% Tween 20 (Sigma). Beads were washed before Cerenkov counting the bound radioactivity with a Beckman counter.
For in vivo cross-linking assays, PBS-washed transfected 293T cells were UV-irradiated on ice with 0.120 J/cm2. FLAG-Myc-REF was immunoprecipitated with FLAG-agarose (Sigma) in 50 mM Hepes (pH 7.5), 1 mM EDTA, 0.5% Triton, 10% glycerol, 1 M NaCl, and complexes were eluted with 100 μg/ml FLAG peptide for 30 min at 4°C. mRNA bound to REF was treated with 5 μg of RNaseA for 30 min at 37°C and end-labeled with polynucleotide kinase in the presence of 5 mM MgCl2 followed by SDS/PAGE, Western blotting, and PhosphorImaging analysis. UV-cross-linking mRNP capture assays were performed as described in ref. 33. FISH experiments were performed as described in ref. 26. When indicated, 12.5 μM WT or MUT REF peptides were added to HeLa cells and incubated for 72 h before FISH analysis. The MS2-tethered mRNA export assays were carried out as described in ref. 12.
Acknowledgments.
We thank V. Porteous for technical assistance and P. Mitchell for comments on the manuscript. This work was supported by the Biotechnology and Biological Sciences Research Council, United Kingdom. Microscopy equipment was funded by Wellcome Trust Grant GR077544A1A.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709167105/DCSupplemental.
References
- 1.Kistler AL, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libri D, Graziani N, Saguez C, Boulay J. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev. 2001;15:36–41. doi: 10.1101/gad.852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strasser K, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 4.Masuda S, et al. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng H, et al. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 6.Nojima T, Hirose T, Kimura H, Hagiwara M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J Biol Chem. 2007;282:15645–15651. doi: 10.1074/jbc.M700629200. [DOI] [PubMed] [Google Scholar]
- 7.Strasser K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- 8.Fribourg S, Braun IC, Izaurralde E, Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- 9.Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Kiesler E, Miralles F, Visa N. HEL/UAP56 binds cotranscriptionally to the Balbiani ring pre-mRNA in an intron-independent manner and accompanies the BR mRNP to the nuclear pore. Curr Biol. 2002;12:859–862. doi: 10.1016/s0960-9822(02)00840-0. [DOI] [PubMed] [Google Scholar]
- 11.Gatfield D, Izaurralde E. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J Cell Biol. 2002;159:579–588. doi: 10.1083/jcb.200207128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hargous Y, et al. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J. 2006;25:5126–5137. doi: 10.1038/sj.emboj.7601385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tintaru AM, et al. Structural and functional analysis of RNA and TAP binding to SF2/ASF. EMBO Rep. 2007;8:756–762. doi: 10.1038/sj.embor.7401031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 15.Zenklusen D, Vinciguerra P, Strahm Y, Stutz F. The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol Cell Biol. 2001;21:4219–4232. doi: 10.1128/MCB.21.13.4219-4232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segref A, et al. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katahira J, et al. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18:2593–2609. doi: 10.1093/emboj/18.9.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell. 2004;13:201–212. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
- 19.Golovanov AP, Hautbergue GM, Tintaru AM, Lian LY, Wilson SA. The solution structure of REF2-I reveals interdomain interactions and regions involved in binding mRNA export factors and RNA. RNA. 2006;12:1933–1948. doi: 10.1261/rna.212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodrigues JP, et al. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci USA. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: Binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golovanov AP, Hautbergue GM, Wilson SA, Lian LY. Assignment of (1)H, (13)C, and (15)N resonances for the REF2-I mRNA export factor. J Biomol NMR. 2006;36(Suppl 5):41. doi: 10.1007/s10858-006-9012-2. [DOI] [PubMed] [Google Scholar]
- 23.Braun IC, Herold A, Rode M, Conti E, Izaurralde E. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J Biol Chem. 2001;276:20536–20543. doi: 10.1074/jbc.M100400200. [DOI] [PubMed] [Google Scholar]
- 24.Liker E, Fernandez E, Izaurralde E, Conti E. The structure of the mRNA export factor TAP reveals a cis arrangement of a noncanonical RNP domain and an LRR domain. EMBO J. 2000;19:5587–5598. doi: 10.1093/emboj/19.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zolotukhin AS, Tan W, Bear J, Smulevitch S, Felber BK. U2AF participates in the binding of TAP (NXF1) to mRNA. J Biol Chem. 2002;277:3935–3942. doi: 10.1074/jbc.M107598200. [DOI] [PubMed] [Google Scholar]
- 26.Williams BJ, et al. The prototype γ-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA via the cellular mRNA export pathway. Biochem J. 2005;387:295–308. doi: 10.1042/BJ20041223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruter P, et al. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, et al. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. 2006;443:234–237. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- 31.Swartz JE, Bor YC, Misawa Y, Rekosh D, Hammarskjold ML. The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element. J Biol Chem. 2007;282:19844–19853. doi: 10.1074/jbc.M701660200. [DOI] [PubMed] [Google Scholar]
- 32.Wilson SA, Brown EC, Kingsman AJ, Kingsman SM. TRIP: A novel double-stranded RNA-binding protein which interacts with the leucine-rich repeat of flightless I. Nucleic Acids Res. 1998;26:3460–3467. doi: 10.1093/nar/26.15.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanford JR, Ellis JD, Cazalla D, Caceres JF. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc Natl Acad Sci USA. 2005;102:15042–15047. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]