Abstract
Organization of transgenes in rice transformed through direct DNA transfer strongly suggests a two-phase integration mechanism. In the “preintegration” phase, transforming plasmid molecules (either intact or partial) are spliced together. This gives rise to rearranged transgenic sequences, which upon integration do not contain any interspersed plant genomic sequences. Subsequently, integration of transgenic DNA into the host genome is initiated. Our experiments suggest that the original site of integration acts as a hot spot, facilitating subsequent integration of successive transgenic molecules at the same locus. The resulting transgenic locus may have plant DNA separating the transgenic sequences. Our data indicate that transformation through direct DNA transfer, specifically particle bombardment, generally results in a single transgenic locus as a result of this two-phase integration mechanism. Transgenic plants generated through such processes may, therefore, be more amenable to breeding programs as the single transgenic locus will be easier to characterize genetically. Results from direct DNA transfer experiments suggest that in the absence of protein factors involved in exogenous DNA transfer through Agrobacterium, the qualitative and/or quantitative efficiency of transformation events is not compromised. Our results cast doubt on the role of Agrobacterium vir genes in the integration process.
Efficient genetic manipulation of major crops, including cereals, has been made possible by transfer of foreign DNA into the host genome through direct DNA transfer procedures such as particle bombardment (1–3). Recently, Agrobacterium-mediated transformation of rice (4), maize (5), and barley (6) has been reported.
Much research has been undertaken to understand the mechanism of Agrobacterium-mediated transfer and integration of foreign DNA into plant genome (7). However, little is known about equivalent mechanisms for foreign DNA integration into plants generated through bombardment or other direct DNA transfer methods. Understanding the mechanism of integration of transgenes through direct DNA transfer is important in elucidating, at the plant cell level, events that characterize the process of DNA integration, without the bacterial gene products involved in Agrobacterium-mediated transformation. This understanding will be crucial to achieve the targeted gene delivery. As most transgenic plants (certainly all cereals) in field trials and on the market have been generated through particle bombardment, it assumes more importance.
Transgenic plants generated either through Agrobacterium-mediated transformation or direct DNA transfer share a common feature, that of transgenic loci frequently consisting of multiple copies of the transgenes. This varies from species to species, and in general, it is accepted that transgenic solanaceous plants generated through Agrobacterium-mediated transformation contain lower transgene copy numbers and simpler integration patterns (8). In species such as rice, maize, and barley (4–6), however, there is not much difference in transgene copy number and/or the degree of rearrangement of the transforming plasmid as a result of the method used to engineer these plants.
T-DNA-mediated transformation results in integration of the introduced genes at one or more independent loci (7). This feature has not been studied extensively in plants generated through direct DNA transfer. The limited information available is based mostly on segregation analysis at the phenotype level; however, this is not reliable as introduced genes can undergo silencing (9).
We analyzed transgenic plants from 16 lines representing six popular rice cultivars. Particle bombardment was used for plant transformation. Our aim was to characterize these lines for the copy numbers of transgenes and the number of independently segregating loci and more importantly to elaborate the organization of transgenes at the transgenic locus to understand the underlying integration mechanism.
Two cointegrate constructs, pWRG 2426 and pWRG 4517 (Fig. 1 A and B) were used for transformation. Our results indicate that multiple integration events occurred as a cluster giving rise to a single transgenic locus in all the lines we analyzed. Multiple copies of transgenes were separated by plant genomic DNA. Our data suggest a possible two-phase mechanism for integration of transgenes. We propose a sequence of events resulting in stable integration of transgenes in transformed plants.
Figure 1.
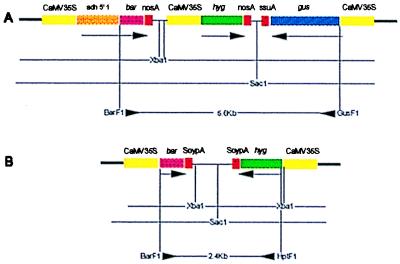
(A) Plasmid pWRG 2426 contains hpt as the selectable gene flanked by two nonselected genes, bar and gusA. All three genes are driven by CaMV35S promoter. The bar and hpt are transcribed in one direction and the gusA in the opposite direction. The bar gene has the maize Adh1 5′ intron between the promoter and coding sequence, and bar and hpt have the nosA terminator but gusA has the ssuA terminator. Restriction enzymes XbaI and SacI separate one gene from the other two as shown by the horizontal bars. This feature of the construct is particularly useful in determining the transgene organization after integration into the genome. By using forward primers for bar (BaF1) and gusA (GusF1) a 5.6-kb fragment is amplified through long PCR. (B) Plasmid pWRG 4517 contains hpt as the selectable gene and bar as the nonselected gene; both genes have CaMV35S promoter and SoypA terminator and transcribe in opposite directions. Restriction enzymes SacI and XbaI both separate the two genes from each other. By using forward primers for bar (BarF1) and hpt (HptF1) a 2.4-kb fragment is amplified through PCR.
Our findings that bombardment-mediated transformation rarely, if ever, gives rise to independently segregating multiple transgenic loci has important implications on map-based selection programs for crop improvement. Finally, data presented here suggest that the importance of Agrobacterium vir genes is system specific, with no particular advantage over direct DNA transfer methods in the process of DNA integration.
MATERIALS AND METHODS
Gene Constructs.
Construction of plasmids WRG 2426 and WRG 4517 and their features are described elsewhere (10).
Transgenic Material.
Rice varieties used were: Bengal, Cypress, Lido, IR72, Koshihikari, and Gulfmont. The transformation process is described elsewhere (11). Variety Gulfmont was transformed with plasmid pWRG 4517, and all other varieties were transformed with pWRG 2426.
DNA Isolation and Southern Hybridization.
Leaf DNA was isolated by using the cetyltrimethylammonium bromide (CTAB) DNA extraction method (12). DNA (5 μg) was digested overnight with the appropriate restriction enzyme(s). It was fractionated on a 0.8% agarose gel and alkali blotted onto Amersham N+ Hybond membrane according to the manufacturer’s instructions. Probe DNA (25 ng) was radiolabeled with α-32P-labeled dCTP (3,000 Ci/mmol) through the random primer method (13). Prehybridization and hybridization were carried out at 65°C in the presence of high salt buffer, pH 6.8 (3 M NaCl/0.1 M Pipes/0.02 M Na2EDTA), Denhardt’s solution, and salmon sperm DNA (100 mg/ml of hybridization mix).
Long PCR.
DNA isolated through the CTAB method was used for PCR, which was performed with the Extend long template PCR kit from Boehringer Mannheim according to the manufacturer’s recommendations.
DNA Sequencing.
PCR or long PCR products were purified from the agarose gel by using a kit from Qiagen (Chatsworth, CA), and the purified products were sequenced with the automated Applied Biosystems sequencing apparatus and reaction kit according to the manufacturer’s instructions.
RESULTS
Multiple Copies of the Transgenes Occur at a Single Locus.
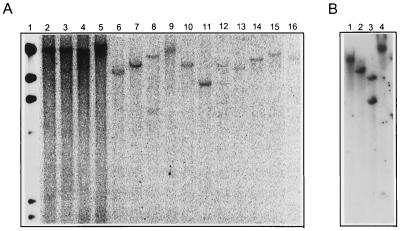
Restriction enzymes that do not cut within the transforming plasmid (noncutters) were used to digest genomic DNA. Southern blot hybridization was performed by using 32P-labeled coding regions of the three genes (bar, hpt, and gusA) as probes. All 16 lines we analyzed resulted in a single band when probed with the coding region of hpt with at least one of the five noncutters used. Four lines (K496-4, K496-3, B550-9, and G517-3) did not give a single band when genomic DNA was digested with the noncutter NheI (Fig. 2A). However, in these four lines a single band was observed when other noncutting enzymes SfiI (K496-4) and ClaI (K496-3, B550-9, and G517-3) were used to digest genomic DNA. Gel electrophoresis at 0.3% agarose concentration was performed to separate fragments within the range of 5–50 kb. This did not result in more than one band for any of the lines in which hybridizations were observed at high molecular weight regions after gel electrophoresis at 0.8% concentration of agarose. The presence of a single band after digestion with at least one noncutting restriction enzyme indicates that transgenic sequences were present at a single locus in all the lines we analyzed. The presence of sites for noncutters such as NheI (K496-4, K496-3, B550-9, and G517-3) and BstXI (K496-2 and K495-1) within the transgenic locus was an indication of the presence of intervening genomic sequences between the transgenic sequences (Fig. 2B).
Figure 2.
(A) Digestion of genomic DNA of 15 transgenic lines with restriction enzyme NheI and subsequent Southern hybridization with the hpt coding region as probe results in a single band for 11 lines. As NheI does not have a restriction site on the construct this indicates integration of the transgenes at the single locus. The four plant lines K496-3 (lane 3), K496-4 (lane 4), B550-9 (lane 8), and G517-3 (lane 12) show more than one band. However, these lines result in a single band with other restriction enzymes that do not cleave within the construct (see Results) as exemplified by (B) where genomic DNA of K496-2 was digested with restriction enzymes ClaI (lane 1), NheI (lane 2), BstXI (lane 3), and SfiI (lane 4) and probed with the coding region of gusA. Only BstXI results in two bands.
Organization of Transgenes.
The two plasmids used for transformation were pWRG 2426 and pWRG 4517 (Fig. 1 A and B). The simplest case of a transgenic locus generated with plasmid pWRG 4517 is an intact integration event of a single copy of the cointegrate unit comprising one copy of each of the two genes (bar and hpt). The two genes would be visualized as separate bands after digestion of genomic DNA with XbaI or SacI followed by Southern hybridization. Digestion of DNA with restriction enzyme XbaI would result in a band of 1900 bp for hpt. The size of the band containing the bar gene would be unpredictable, depending on the nearest XbaI site in the genome. DNA digestion with SacI would result in a unique band for each gene. We did not observe such a pattern for any of the transformants engineered with pWRG 4517. However, we identified three lines (G517-1, G517-2, and G517-5) with single-copy inserts for bar and hpt. Southern analysis of the genomic DNA after restriction with XbaI and SacI allowed us to define the organization of the transgenic locus in these lines (Fig. 3). The 1900-bp band expected for hpt after digestion of the genomic DNA with XbaI was obtained only for G517-5.
Figure 3.
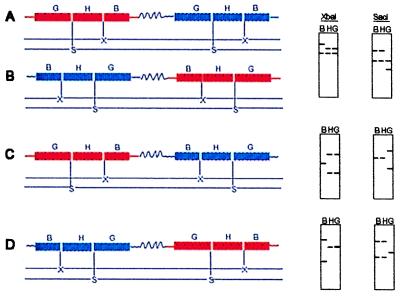
Molecular organization of the transgenic locus in five independent transgenic lines of Gulfmont variety transformed with pWRG 4517. (A) G517-1, (B) G517-2, (C) G517-5, (D) G517-7, and (E) G517-3. H, hpt; B, bar; X, XbaI; S, SacI. (See Results for details.)
A 2.4-kb fragment expected to be amplified by using forward primers for bar and hpt (Fig. 1B) was not observed in any of the three lines. This suggested that the single copy of the bar and hpt gene present in these three lines was not the result of integration of an intact unit of pWRG 4517. In G517-2 and G517-5 a PCR fragment of ca. 3 kb was observed with the forward primers for the two genes, indicating that the orientation of transgenes was maintained as in the transforming construct, but an additional sequence of ca. 500 bp was present between them. Additionally Southern hybridization analysis for plant lines G517-1, G517-2, and G517-5 indicated that the XbaI and SacI sites present between the two (bar and hpt) genes in the original transforming plasmid were now absent from the transgenic locus in all three. Evidence for this is provided by the single common band for both bar and hpt following Southern hybridization between either of the probes (bar/hpt) and genomic DNA digested with restriction enzymes XbaI or SacI. Methylation of the XbaI and/or SacI site, which may cause these sites to remain uncut, is unlikely as it would lead to transgene silencing. Both transgenes, however, were shown to be expressing the protein product in all three lines. For G517-1 we obtained a fragment of 1.6 kb with reverse primer for bar and a forward primer for hpt, indicating that orientation of the genes was switched so that both were now transcribing in the same direction. By using similar Southern and PCR analyses, structures for organization of transgenes in plant lines G517-3 and G517-7 were defined. These lines contained two copies each of the bar and hpt genes. G517-7 had 2 units of the construct integrated as direct repeat, whereas in G517-3 the construct was rearranged and at least three molecules of the transforming plasmid were involved. These structures indicated that rearrangement of the exogenous DNA may be involved even in simple integrations of one to two copies of the transgenes.
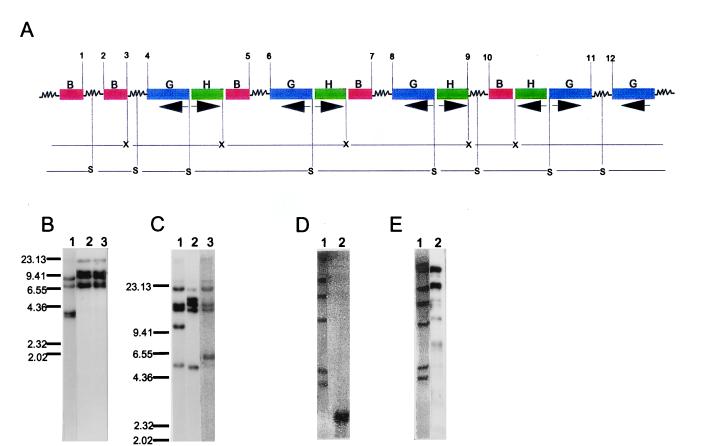
The simplest integration event we observed with pWRG 2426 (Fig. 1A) comprised one intact unit of the three genes, bar, hpt and gusA (referred to as cointegrate unit from hereon), and one additional copy of the bar gene. Multicopy insertion events for pWRG 2426 were analyzed based on different possible combinations in which two molecules of the cointegrate unit could integrate at the same site (Fig. 4). Southern analysis after digestion of the genomic DNA with XbaI and SacI would result in a predictable combination of unique and/or common bands for the three genes in each of the three possibilities (Fig. 4). The sizes of the bands in all the cases would be unpredictable. Similarly, common and/or unique bands for the three genes can be predicted for a combination of more than two molecules of the cointegrate unit. Intervening sequences between the two or more cointegrate units may contain sites for XbaI and SacI. This would suggest that such sequences are most likely plant genomic sequences. By using these criteria and also knowing that all the transgenes were clustered at a single locus, we postulated tentative models for the organization of transgenic loci in different lines. These were subsequently validated by using long PCR with specific sets of primers that would result in a predictable number of amplified fragments. Additional validation of the model was provided by Southern hybridizations following double digestion of genomic DNA with XbaI and SacI. Band patterns were predictable for such digests. We observed a relatively simple organization for some loci but also more complex situations in other lines. XbaI or SacI digestions of genomic DNA of K496-4 and subsequent probing with the three genes allowed us to propose a structure for the transgenic locus in this line (Fig. 5A). The results of the hybridization are shown in Fig. 5 B and C. The proposed structure would give rise to one band for hpt after double digestion with XbaI and SacI, if all the XbaI and SacI sites of the integrated DNA are maintained. The predicted single band of the correct size (1.2 kb) was observed (Fig. 5D) indicating that the sites had not been disrupted. The structure was further validated through long PCR by amplification of all nine predicted bands by using reverse primers for hpt and gusA. Fig. 5E shows the bands after Southern hybridization of the long PCR products with the 32P-labeled gusA probe.
Figure 4.
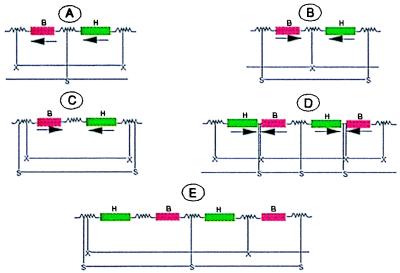
Diagrammatic representation of integration of two copies of plasmid pWRG 2426 as direct repeats (A and B) or indirect repeats, with bar as two adjacent genes (C) or gusA as two adjacent genes (D) and the corresponding Southern hybridization band pattern after restriction digest of the genomic DNA with XbaI (X) or SacI (S) probed with bar (B), hpt (H), or gusA (G). The jagged lines represent intervening vector or plant DNA. It is assumed that the intervening DNA does not possess sites for XbaI and SacI. However, the altered band patterns can be predicted if sites for one or both enzymes are present. Although the sizes of the bands are arbitrarily represented as they would not be predictable, the bands of identical size for two or more genes represent the same fragment as shown by the horizontal lines between X or S for different arrangements. Following similar reasoning band patterns can be predicted for more than two copies.
Figure 5.
(A) Molecular organization of the transgenes at a single locus in line K496-4. This represents one of the most complex cases. The locus has three units of the cointegrate plasmid integrated in tandem and one in an inverted configuration; in addition one gusA and twobar genes are present. Arrows under gusA and hpt indicate direction of transcription and therefore the direction in which the reverse primers for the two genes extend over the template in PCR. Numbers 1–12 delimit the regions of intervening sequences. X, XbaI; S, SacI; B, bar; G, gusA; H, hpt. (B) and (C) Southern hybridization results for K496-4 DNA digested with XbaI and SacI, respectively. Lanes 1–3, genomic DNA probed with coding regions of bar, gusA, and hpt, respectively. (D) Double digestion ofgenomic DNA of K496-4 with XbaI and SacI. All four copies of the hpt comigrate at the expected size of 1.2 kb. Lane 1, DNA size marker λ phage DNA digested with HindIII. Lane 2, genomic DNA probed with hpt coding region. (E) Southern hybridization of long PCR products obtained with reverse primers for hpt and gusA. Lane 1, DNA size marker λ phage DNA digested with HindIII. Lane 2, long PCR products probed with gusA coding region. The two dark bands represent two bands each. Numbers indicated on the side of B and C represent size marker in kb. These numbers apply for the size marker shown in lane 1 of D and E also.
We observed an array of rearrangements of the transforming plasmid. In lines with multicopy inserts, however, at least one intact copy of the insert was present in all but one (L551-4) line transformed with pWRG 2426. This was confirmed by amplification, through long PCR, of the expected 5.6-kb fragment with forward primers for bar and gusA (Fig. 1A). The rearrangement and multimerization of the exogenous DNA resulted in direct and inverted repeats of the cointegrate unit; integration of the partial unit was also observed.
Characterization of Intervening Sequences.
By using long PCR, fragments ranging from 1.5 to 8.8 kb were amplified with reverse (3′) primers in hpt and gusA from eight transformants of pWRG 2426. As these primers run opposite to each other, the amplified fragments are most likely to result from two molecules of the cointegrate unit integrated close to one another. Such fragments would therefore contain the “linker” DNA toward their center. In all eight lines, a number of sets of nested primers were used to move toward the center of the fragments by obtaining smaller fragments of predictable sizes. The uncharacterized portions of the original fragments were then sequenced by using the last set of primers that amplified a fragment of predictable size. In seven of the eight lines the linker was part of the plasmid, but in one line a 136-bp sequence was not a part of the plasmid. A nucleic acid sequence database search (blast, fasta, and tags) with this sequence did not show similarity to any known sequences.
Determination of Copy Number of Transgenes.
Elucidating the structure of the transgenic locus gave the correct number for the copies of different integrated transgenes. Southern analysis performed after digestion of genomic DNA with restriction enzyme(s), which have a unique site in the plasmid, did not always give the correct transgene copy number. This was because of the integration of multiple copies of the transgenes as direct or inverted repeats. Table 1 shows (i) the number of bands for different transgenes, in different lines, by Southern analysis of the genomic DNA after restriction with enzymes XbaI and SacI and the number of bands hybridizing to the specific probe suggesting the number of copies for that gene; (ii) the number of copies as determined by the organization of transgenes at the transgenic locus. In 4 of 11 multicopy lines (C549-1, I533-3, L551-8, and L551-10), there was agreement in the number of transgene copies estimated through XbaI and SacI digest and through organization model. In five lines only one enzyme XbaI or SacI digest gave the correct number. In two lines (K496-3 and B550-9) neither restriction enzyme indicated the correct number. We obtained transgenic plants with low (1–3), intermediate (4–6), and high (7–10 or more) copy numbers of integrated genes. The total number of integrated genes was generally high (Table 1), but this result was a reflection of the cointegrate construct used. For individual genes more than 70% of the lines were in the low copy number category, with 1–3 copies of the transgenes integrated.
Table 1.
Determination of copy number of transgenes for transformants of pWRG 2426
| Plant line | Copy number estimation with XbaI
|
Copy number estimation with SacI
|
Copy number determination using transgene organization
|
Total number of transgenes*
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| bar | hpt | gus | bar | hpt | gus | bar | hpt | gus | bar + hpt + gus | |
| K495-1 | 3 | 3 | 3 | 3 | 3 | 4 | 3 | 3 | 4 | 10 |
| K496-2 | 1 | 2 | 2 | 2 | 3 | 3 | 2 | 3 | 3 | 8 |
| K496-3 | 2 | 5 | 4 | 5 | 5 | 6 | 4 | 5 | 6 | 15 |
| K496-4 | 4 | 4 | 4 | 5 | 4 | 5 | 5 | 4 | 5 | 14 |
| C549-1 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 1 | 4 |
| I533-1 | 3 | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 3 | 9 |
| I533-3 | 3 | 2 | 3 | 3 | 2 | 3 | 3 | 2 | 3 | 8 |
| B550-9 | 5 | 4 | 4 | 5 | 3 | 3 | 6 | 4 | 4 | 14 |
| L551-4 | 2 | 3 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 7 |
| L551-8 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 6 |
| L551-10 | 3 | 4 | 3 | 3 | 4 | 3 | 3 | 4 | 3 | 10 |
*Based on transgene organization.
DISCUSSION
The two most popular methods for foreign DNA insertion into plants are Agrobacterium-mediated transformation (16–19) or direct gene transfer through bombardment (20, 21). Genetic engineering of cereals has been facilitated by using bombardment methodologies. Even though Agrobacterium has been successfully used to introduce foreign genes into rice (4) maize (5) and barley (6) bombardment remains the method of choice for the introgression of foreign genes into important crops (2). The complexities of numerous steps involved in Agrobacterium-mediated transformation (binding of the bacteria to the plant cell, induction of the vir genes, transfer of the T-DNA to the plant cell, the subsequent nuclear targeting of the T-DNA as a single-stranded moiety, and finally integration) have been highlighted by differences in susceptibility of different ecotypes of Arabidopsis to agroinfection (22). This observation argues in favor of simpler, ecotype-independent direct DNA transfer methods.
The mechanism of integration of exogenous DNA into the host genome is poorly understood. Elucidating the organization of transgenic loci has been the main approach to understanding events that determine the mechanism of integration. Such studies have been limited to transgenic plants generated through Agrobacterium (23–25). In one study, however, 112 transformed maize callus clones obtained through particle bombardment were analyzed for rearrangements of the plasmid (26). We characterized transgenic loci in 16 transgenic rice plant lines generated through particle bombardment. Our results shed light on some of the events that may be involved in integration of exogenous DNA into the plant genome. We present evidence to support our model in which a two-phase integration mechanism appears to operate. This appears to be mediated by the establishment of temporary “hot spots” for integration of foreign DNA.
Multicopy integration events at the same locus have been reported for transformants generated through both Agrobacterium-mediated transformation (27–29) and particle bombardment-mediated transformation (30, 31). In Agrobacterium-mediated transformation transgenic multimers are believed to form within the plant cell as no multimerization was seen in vir-induced Agrobacteria (32). In bombardment-mediated transformation, plasmid isolation protocols preclude the inclusion of linear multimers in the DNA used to coat metal particles. Therefore, multimeric transgenes seen in plants generated through bombardment support the notion of the plant cell itself being the site for multimerization.
In Agrobacterium-mediated transformation, two alternative mechanisms have been suggested for the formation of multimeric units; these involve replication or ligation (29). De Neve et al. (25) favor the ligation model, and they demonstrated that T-DNA molecules from two different strains of Agrobacterium carrying two different marker genes (kanamycin and hygromycin resistances) integrate as direct or inverted repeats in the same locus. Our results also support a ligation model for multimerization of the transgenes. Determination of transgene organization in K496-4 (Fig. 5) revealed a complex integration pattern comprising not only direct or inverted repeats of the three-gene cointegrate unit but also parts of this transforming plasmid in the same locus. A replication model would give rise only to direct repeats, although invoking the model may account for the presence of parts of the insert. The change in orientation of the two transgenes was detected for line G517-1 (see Results). This implies a rearrangement and ligation reaction between the transgenes, most probably involving two different molecules of the transforming construct. Cotransformation experiments involving bombardment of two different plasmids also support a ligation model as transgenes on two plasmids are seen to integrate in the same locus (33).
Exogenous DNA transferred through bombardment does not involve any proteins or enzymes attached to it unlike T-DNA, which has the VIRD2 protein attached to the 5′ right border end of the complex (34, 35). VIRD2 has been shown to have ligase-like activity (36). Ligation of T-DNA molecules into multimers may therefore be favored because of the presence of the VIRD2 protein. This is supported by higher frequencies of tandem and inverted repeat configuration in the vicinity of the right border of the T-DNA (37). However, after transfer of foreign DNA through bombardment, similar extrachromosomal ligations leading to direct and inverted repeats of the transforming construct have been observed in the absence of VIRD2 in a number of lines we analyzed. This suggests that ligation is mediated by plant enzymes alone.
Multimerization by ligation may occur either before or during integration. De Neve et al. (25) suggest that T-DNA repeats are formed by extrachromosomal ligation before integration. Our results suggest that ligation may be operating in a two-phase mechanism. In the preintegration phase, ligation occurs between molecules of the transforming plasmid extrachromosomally, resulting in rearranged transgenic sequences. In the second phase, such rearranged sequences integrate at the same locus through a second round of ligations. We envisage the following sequence of events underlying the process of integration of foreign DNA into the host genome through bombardment.
Penetration of cells with DNA-coated metal particles elicits a wound response. Hunold et al. (38) showed that penetration was accompanied by callose formation in the wound area. Induction of wound response is known to activate the enzymatic machinery for host DNA repair/foreign DNA degradation. Wound induction was also shown to be necessary for Agrobacterium-mediated T-DNA transfer (39). Thus, the combined action of DNA repair and degradation enzymes on the introduced DNA gives rise to rearranged transgenic sequences. These may not be limited to complete multimers of tandem direct or inverted repeats but may include incomplete parts of the insert. A detail characterization of the transgenic locus in rice transformed through the calcium phosphate method revealed rearrangement of the plasmid spanning 10 and 29 kb in two transgenic lines (40). Register et al. (26) found that certain sequences were more frequently rearranged than others. The preintegration phase is thus dominated by extrachromosomal rearrangements and ligations. For Agrobacterium-mediated transformation, extrachromosomal rearrangement through homologous recombination was reported (41). Such extrachromosomal recombination occurs within 30 min of entry of the plasmid molecules into the plant cell (42). Fragmentation and reassembly of DNA occurs in vitro, and it was shown that identity between four bases was sufficient to bring about reassembly between DNA fragments through recombination. Even two bases of uninterrupted identity could result in a crossover with sufficient homology in the flanking sequence (43).
In its second phase, our model postulates initiation of integration of the scrambled and/or intact transgenic sequences in competent cells at specific target sites. Competence of the cells and target sites is most likely dependent on the functional and developmental stage of the cell. It was shown for Agrobacterium-mediated transformation of Arabidopsis that transgenes integrated in transcriptionally active regions of the genome (44–46). However, transformation efficiency with particle bombardment was seen to be higher in M and G2 phases of the cell cycle (47). A higher efficiency of transformation during the M phase in comparison with the S phase implies that a relaxed conformation of the chromatin may not be required for foreign DNA integration. On the basis of increased frequency of recombination in plants subjected to double-stranded break-inducing agents such as x-rays, methyl methanesulfonate, and mitomycin C (48–50), it was speculated that induction of recombination may be a general stress response and may serve as one line of DNA repair pathway (51). Double-stranded break points in the genome may serve as integration sites through the mechanism of illegitimate recombination based on microhomology as suggested for Agrobacterium-mediated transformation (23, 24). This idea is again supported by in vitro recombination experiments (43). It appears that once the process of integration is initiated at a specific site, this site is even more receptive to further integration events, becoming a temporary hot spot for integration. Multicopies of transgenes at such a locus are the result of secondary ligations, separated spatially if not temporally from the primary ligations, which give rise to rearranged transgenic sequences before integration. The secondary ligations result in plant genomic sequences intervening in the transgenic sequences, which are characterized by the presence of sites for restriction enzymes that do not cut within the plasmid. The presence of sites in DNA “linking” the transgenes for enzymes that either have a unique or no site in the plasmid indicates a high probability of the linker being plant genomic DNA rather than being created as a result of deletion or filling in activities of the repair enzymes (52), particularly if such sites occur more than once in the same line as is the case for the SacI sites between points 1–2, 3–4, and 11–12 in K496-4 (Fig. 5A). With our approach of characterizing intervening sequences through long PCR and sequencing, we found that at least in one line, a stretch of 136 bp did not belong to the transforming plasmid.
Plants resulting from Agrobacterium-mediated transformation contain transgenes organized either in one or multiple independently segregating transgenic loci. Reports suggesting multiple transgenic loci for plants generated through bombardment frequently rely on segregation analysis at the phenotype level (33). This is most likely misleading as transgenic plants may contain transgenes in a silenced state (53–55). Register et al. (26) found that 48 of the 55 multicopy clones had all transgenes at a single locus. However, they used one “noncutter.” By using five noncutters we found that all copies of the transgenes in any line were physically linked at a single locus, though in some cases plant genomic DNA was interspersed with the transgenes. A detailed molecular analysis is therefore important to assess the usefulness of the transgenics. This notion is supported by our observations that (i) a large proportion of multicopy lines (up to 6 copies) we recovered expressed the transgenes at high levels over a number of generations. (ii) Specific transgenes in a given locus were subject to silencing, although others expressed at high levels, e.g., in plant line K496-4 the gusA is silenced but bar and hpt are not. This implies that copy number and site of integration may not have a profound influence on transgene silencing in isolation of transgene configuration and integrity (56). Additionally, the correct copy number of integrated transgenes could be obtained only after a detailed molecular analysis.
Arabidopsis mutants deficient in stable T-DNA integration lack DNA damage repair enzymes (22, 57). The presence of vir gene products was not sufficient to bring about stable transformation. Reports on integration of non-T-DNA sequences (6, 56, 58) dispel the notion of “clean” T-DNA integration through VIRD2. In contrast, transformation through bombardment offers the advantage of recovering transgenic plants following delivery of minimal gene cassettes (lacking vector sequences) into the cell (L. T. Duc, D. Suahakar, A.K., M.L., P.V., D.A.L., and P.C., unpublished data).
Correlation between delivery of double-stranded DNA-coated metal particles to nucleus and stable transformation events through bombardment (38, 59) obviate nuclear trafficking of DNA as a single-stranded moiety (60, 61) subsequently converted to double-stranded form before integration into the genome (41).
Efficiency of transformation with Agrobacterium reported for rice, maize, and barley (4–6) is comparable with that obtained with bombardment. The frequency of low copy inserts in our study was comparable with Agrobacterium-mediated transformation. We get comparable transformation efficiencies for tobacco through either method.
Ecological risks of biological vectors persisting in transgenic plants are nil for direct DNA transfer but high for Agrobacterium-mediated transfer (62, 63).
We suggest that bombardment-mediated transformation has unique advantages in mechanical delivery of double-stranded minimal gene cassette at high efficiency, generating ecologically safe transgenic material with a single transgenic locus. This renders breeding programs simpler with no need to eliminate populations having segregating transgenes. Genetic analysis, mapping of transgenes, and map-based selection programs are also simpler. Finally, our data suggest that through a two-phase ligation mechanism for integration of exogenous DNA, even in the absence of vir gene products, integration patterns for bombardment-mediated transformation are analogous to those obtained through Agrobacterium-mediated methods.
Acknowledgments
We thank Drs. John Snape, Eva Stoger, Roy Dunford, Simon Griffiths, and Michael Roberts for useful discussions and Dr. Katrien Devos for useful comments on the manuscript. Plasmids pWRG 2426 and pWRG 4517 were supplied by Agracetus Inc. A.K. thanks The Overseas Development Administration, United Kingdom, for financial support. This document is an output from a project funded by the Overseas Development Administration Plant Sciences Program managed by the Centre for Arid Zone Studies, University of Wales, Bangor, for the benefit of developing countries.
ABBREVIATION
- CTAB
cetyltrimethylammonium bromide
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF033193).
References
- 1.Christou P. Euphytica. 1995;85:13–27. [Google Scholar]
- 2.Jahne A, Becker D, Lorz H. Euphytica. 1995;85:35–44. [Google Scholar]
- 3.Potrykus I. In: Rice Genetics II: Proceedings of the Second International Rice Genetics Symposium. Khush G S, editor. Philippines: International Rice Research Institute; 1991. pp. 637–646. [Google Scholar]
- 4.Hiei Y, Ohta S, Komari T, Kumashiro T. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 5.Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T. Nat Biotechnol. 1996;14:745–750. doi: 10.1038/nbt0696-745. [DOI] [PubMed] [Google Scholar]
- 6.Tingay S, McElroy D, Kalla R, Fieg S, Wang M, Thornton S, Bretell R. Plant J. 1997;11:1369–1376. [Google Scholar]
- 7.Tinland B. Trends Plant Sci. 1996;1:178–183. [Google Scholar]
- 8.Chilton M D. Proc Natl Acad Sci USA. 1993;90:3119–3120. doi: 10.1073/pnas.90.8.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matzke M A, Matzke A J M. Plant Physiol. 1995;107:679–685. doi: 10.1104/pp.107.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooley J, Ford T, Christou P. Theor Appl Genet. 1995;90:97–104. doi: 10.1007/BF00221001. [DOI] [PubMed] [Google Scholar]
- 11.Christou P, Ford T, Kofron M. Bio/Technology. 1991;9:957–962. [Google Scholar]
- 12.Rogers S O, Bendlich A L. In: Molecular Biology Manual. Gelvin S B, Schilperoort A R, editors. Vol. 2. Dodrecht, the Netherlands: Kluwer; 1994. pp. 1–8. [Google Scholar]
- 13.Feinberg A P, Vogelstein B. Anal Biochem. 1994;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 14.Peterhans A, Schlupmann H, Basse C, Paszkowski J. EMBO J. 1990;9:3437–3445. doi: 10.1002/j.1460-2075.1990.tb07551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tovar J, Lichtenstein C. Plant Cell. 1992;4:319–332. doi: 10.1105/tpc.4.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barton K A, Binns A N, Matzke A J M, Chilton M D. Cell. 1983;32:1033–1043. doi: 10.1016/0092-8674(83)90288-x. [DOI] [PubMed] [Google Scholar]
- 17.Bevan M W, Flavell R B, Chilton M D. Nature (London) 1983;304:184–187. [Google Scholar]
- 18.Herrera-Estrella L, Depicker A, Van Montagu M, Schell J. Nature (London) 1983;303:209–213. [Google Scholar]
- 19.Hoekema A, Hirsch P R, Hooykaas P J J. Nature (London) 1983;303:179–180. [Google Scholar]
- 20.Klein T M, Wolf E D, Wu R, Sanford J C. Nature (London) 1987;327:70–73. [Google Scholar]
- 21.Sanford J C. Trends Biotechnol. 1988;6:299–302. [Google Scholar]
- 22.Nam J, Matthysse A G, Gelvin S B. Plant Cell. 1997;9:317–333. doi: 10.1105/tpc.9.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gheysen G, Villarroel R, Van Montagu M. Genes Dev. 1991;5:287–297. doi: 10.1101/gad.5.2.287. [DOI] [PubMed] [Google Scholar]
- 24.Mayerhofer R, Koncz-Kalman Z, Nawrath C, Bakkeren G, Crameri A, Angelis K, Redei G P, Schell J, Hohn B. EMBO J. 1991;10:697–704. doi: 10.1002/j.1460-2075.1991.tb07999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Neve M, De Buck P, Jacobs A, Van Montagu M, Depicker A. Plant J. 1997;11:15–29. doi: 10.1046/j.1365-313x.1997.11010015.x. [DOI] [PubMed] [Google Scholar]
- 26.Register J C, III, Peterson D J, Bell P J, Bullock P W, Evans I J, Frame B, Greenland A J, Higgs N S, Jepson I, Jiao S, et al. Plant Mol Biol. 1994;25:951–961. doi: 10.1007/BF00014669. [DOI] [PubMed] [Google Scholar]
- 27.Holsters M, Villarroel R, Gielen J, Seurink J, De Greve H, Van Montagu M, Schell J. Mol Gen Genet. 1983;190:35–41. [Google Scholar]
- 28.Kwok W W, Nester E W, Gordon M P. Nucleic Acids Res. 1985;13:459–471. doi: 10.1093/nar/13.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorgensen R, Snyder C, Jones J D G. Mol Gen Genet. 1987;207:471–477. [Google Scholar]
- 30.Klein T M, Harper E C, Svab Z, Sanford J C, Fromm M E, Maliga P. Proc Natl Acad Sci USA. 1988;85:8502–8505. doi: 10.1073/pnas.85.22.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein T M, Kornstein L, Sanford J C, Fromm M E. Plant Physiol. 1989;91:440–444. doi: 10.1104/pp.91.1.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zambriski P. Annu Rev Genet. 1988;22:1–30. doi: 10.1146/annurev.ge.22.120188.000245. [DOI] [PubMed] [Google Scholar]
- 33.Qu R, De Kochko A, Zhang L, Marmey P, Li L, Tian W, Zhang S, Fauquet C M, Beachy R N. In Vitro Cell Dev Biol-Plant. 1996;32:233–240. [Google Scholar]
- 34.Ward E R, Barnes W M. Science. 1988;242:927–930. [Google Scholar]
- 35.Howard E A, Winsor B A, De-Vos G, Zambryski P. Proc Natl Acad Sci USA. 1989;86:4017–4021. doi: 10.1073/pnas.86.11.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pansegrau W, Schoumacher F, Hohn B, Lanka E. Proc Natl Acad Sci USA. 1993;90:11538–11542. doi: 10.1073/pnas.90.24.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grevelding C, Fantes V, Kemper E, Schell J, Masterson R. Plant Mol Biol. 1993;23:847–860. doi: 10.1007/BF00021539. [DOI] [PubMed] [Google Scholar]
- 38.Hunold R, Bronner R, Hahne G. Plant J. 1994;5:593–604. [Google Scholar]
- 39.Lippincott B B, Lippincott J A. J Bacteriol. 1969;97:620–628. doi: 10.1128/jb.97.2.620-628.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takano M, Egawa H, Ikeda J E, Wakasa K. Plant J. 1997;11:353–361. doi: 10.1046/j.1365-313x.1997.11030353.x. [DOI] [PubMed] [Google Scholar]
- 41.Offringa R, de Groot M J A, Haagsman H J, Does M P, van den Elzen P, Hooykaas P J J. EMBO J. 1990;9:3077–3084. doi: 10.1002/j.1460-2075.1990.tb07504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puchta H, Kocher S, Hohn B. Mol Cell Biol. 1992;12:3372–3379. doi: 10.1128/mcb.12.8.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stemmer W P C. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coates D, Taliercio E W, Gelwin S B. Plant Mol Biol. 1987;8:159–168. doi: 10.1007/BF00025327. [DOI] [PubMed] [Google Scholar]
- 45.Koncz C, Martin N, Mayerhofer R, Konkz-Kalman Z, Korber H, Redei G P, Schell J. Proc Natl Acad Sci USA. 1989;86:8467–8471. doi: 10.1073/pnas.86.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herman L, Jacobs A, Van Montagu M, Depicker A. Mol Gen Genet. 1990;244:248–256. doi: 10.1007/BF00271558. [DOI] [PubMed] [Google Scholar]
- 47.Iida A, Yamashita T, Yamada Y, Morikawa H. Plant Physiol. 1991;97:1585–1587. doi: 10.1104/pp.97.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lebel E G, Masson J, Bogucki A, Paszkowski J. Proc Natl Acad Sci USA. 1993;90:422–426. doi: 10.1073/pnas.90.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schulman M J. EMBO J. 1995;14:4102–4107. doi: 10.1002/j.1460-2075.1995.tb00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puchta H, Swoboda P, Hohn B. Plant J. 1995;7:203–210. [Google Scholar]
- 51.Puchta H, Hohn B. Trends Plant Sci. 1996;1:3340–3348. doi: 10.1016/S1360-1385(03)00004-9. [DOI] [PubMed] [Google Scholar]
- 52.Roth D B, Wilson J H. In: Genetic Recombination. Kucherlapati R, Smith G R, editors. Washington, DC: Am. Soc. Microbiol.; 1988. pp. 621–653. [Google Scholar]
- 53.Flavell R B. Proc Natl Acad Sci USA. 1994;91:3490–3496. doi: 10.1073/pnas.91.9.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finnegan J, McElroy D. Bio/Technology. 1994;12:883–888. [Google Scholar]
- 55.Meyer P. Trends Biotechnol. 1995;13:332–337. [Google Scholar]
- 56.Cluster P D, O’Dell M, Metzlaff M, Flavell R B. Plant Mol Biol. 1996;32:1197–1203. doi: 10.1007/BF00041406. [DOI] [PubMed] [Google Scholar]
- 57.Sonti R V, Chiurazzi M, Wong D, Davies C S, Harlow G R, Mount D W. Proc Natl Acad Sci USA. 1995;92:11786–11790. doi: 10.1073/pnas.92.25.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramanathan V, Veluthambi K. Plant Mol Biol. 1995;28:1149–1154. doi: 10.1007/BF00032676. [DOI] [PubMed] [Google Scholar]
- 59.Yamashita T, Iida A, Morikawa H. Plant Physiol. 1991;97:829–831. doi: 10.1104/pp.97.2.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tinland B, Hohn B, Puchta H. Proc Natl Acad Sci USA. 1994;91:8000–8004. doi: 10.1073/pnas.91.17.8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zupan J R, Citovsky V, Zambryski P. Proc Natl Acad Sci USA. 1996;93:2392–2397. doi: 10.1073/pnas.93.6.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matzk A, Mantell S, Schiemann J. Mol Plant–Microbe Interact. 1996;9:373–381. [Google Scholar]
- 63.Barrett C, Cobb E, McNicol R, Lyon G. Plant Cell Tissue Organ Cult. 1997;47:135–144. [Google Scholar]