Abstract
The endangered brown bear populations (Ursus arctos) in Iberia have been suggested to be the last fragments of the brown bear population that served as recolonization stock for large parts of Europe during the Pleistocene. Conservation efforts are intense, and results are closely monitored. However, the efforts are based on the assumption that the Iberian bears are a unique unit that has evolved locally for an extended period. We have sequenced mitochondrial DNA (mtDNA) from ancient Iberian bear remains and analyzed them as a serial dataset, monitoring changes in diversity and occurrence of European haplogroups over time. Using these data, we show that the Iberian bear population has experienced a dynamic, recent evolutionary history. Not only has the population undergone mitochondrial gene flow from other European brown bears, but the effective population size also has fluctuated substantially. We conclude that the Iberian bear population has been a fluid evolutionary unit, developed by gene flow from other populations and population bottlenecks, far from being in genetic equilibrium or isolated from other brown bear populations. Thus, the current situation is highly unusual and the population may in fact be isolated for the first time in its history.
Keywords: bottleneck, gene flow, mitochondrial DNA, ancient DNA, serial coalescent simulations
The brown bear (Ursus arctos) is listed by the International Union for the Conservation of Nature and Natural Resources (IUCN) as a threatened species (least concern). The species currently is distributed across three different continents: Asia, Europe, and North America. Home ranges have been drastically reduced in recent history (1). For instance, in the United States, the population was reduced by ≈75% in only one century, mostly as a result of home range reduction (1, 2). Although presently abundant in Alaska and Canada, and once occupying much of present day United States and northern Mexico, few bears remain in some areas in northern United States, and the species is extinct in Mexico (3). As in the Americas, the brown bear is abundant in the northern parts of Europe but has been reduced to small and fragmented populations in southern Europe. Most of the southern European populations are in danger of extinction (4, 5); Italy hosts two small isolated and highly endangered populations (6, 7), and a similar situation is seen in Greece, the Pyrenees, and Spain. The Pyrenean population was reduced to five to six individuals according to 1980s estimates (8–11). Two reintroductions have been made since 1993, translocating bears from the Kocevje reserve in Slovenia. The Cantabrian Mountains in northern Spain, where the two remaining populations live, represent the southwestern limit of the current European brown bear distribution. These two small populations are geographically isolated, the eastern one with only 25–30 individuals and the western one with ≈120 bears (J.L.G-G., unpublished data). Some of these fragmented European populations have been defined as single conservation units (5). The Cantabrian populations are highly threatened and at risk of extinction (12, 13), and the situation for the Pyrenean populations is even worse.
Although presently threatened in southern Europe and extinct from large parts of central Europe, the brown bear has been an integral part of the European fauna for at least the last half million years (14). Mitochondrial DNA (mtDNA) phylogeographic studies on the remaining populations divide them into eastern and western lineages (4), with the western branch further subdivided into two clusters, one comprising bears from the Pyrenees, southern Sweden, and Spain, and the other comprising bears from Italy and the Balkans (15). The Cantabrian populations are the only ones whose genetic diversity has not been altered by human-assisted introgression from other populations. The two western subgroups are believed to have originated from two ancestral glacial refugia (4). The eastern lineage is proposed to have expanded from glacial refugia placed in the Carpathian Mountains (14, 16).
The Iberian brown bears are believed to have been isolated for extensive periods of time and supposedly provided a stock for recurrent recolonizations of Europe after multiple glaciations, including the last glacial maximum (LGM), 23,000–18,000 years ago (4, 17). Gene flow into Iberia is considered to be of minor importance because the inferred ancestral population size in the peninsula, in combination with the recurrent extinction of bear populations in the regions neighboring Iberia, would rule out exotic contributions. Thus, the Iberian brown bear is likely to have continually occupied the peninsula and possibly adapted to local conditions since at least the Pleistocene. If this is the case, the conservation of Iberian bears is of importance not only for Iberia but also for the whole of Europe, for which it has served as postglacial recolonization stock. However, several recent studies suggest that the extent of past gene flow, as inferred from contemporary material, may have been underestimated (18, 19), and there is evidence that this also is true for Iberian bears (20). In fact, Iberian bears may have been a part of a continuous European population, albeit isolated by distance, even during the LGM. To investigate this issue further, we analyzed ancient and modern brown bear mtDNA sequences. We used these serial data to identify potential population size changes, estimate gene flow, and investigate the genetic diversity of the Iberian population from the Pleistocene to its present, albeit reduced, size.
Results
Genetic Affinities of the Spanish Population.
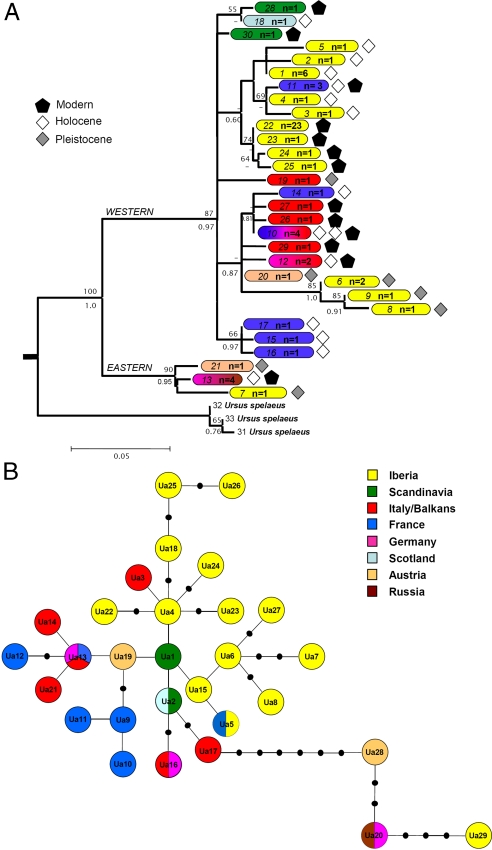
We successfully amplified and sequenced mtDNA from a total of 14 bears using 20 new fossil and subfossil specimens [see supporting information (SI) Dataset 1 and SI Fig. 3]. The dataset contains ancient Iberian brown bear sequences, where some are from sites that have previously yielded DNA (20–22). One sample (Vb9184, see SI Dataset 1) was identified as a cave bear (Ursus spelaeus). Of the remaining 13 individuals, 4 yielded a sequence of 115 bp whereas 9 yielded a sequence of 177 bp (at positions 16589–16650, 16728–16792, and 16800–16849 of the brown bear mtDNA genome; accession no. AF303110). In addition, 24 modern brown bear sequences (177-bp long; accession numbers EU400184–EU400206) were obtained from shed hair samples from the current bear populations in the Cantabrian Mountains in northern Spain. Contamination was not detected in any of the negative controls used during the extraction and amplification processes. To include all of the data generated in the present study, we created two datasets, spanning 177 bp and 115 bp of the mitochondrial control region (referred below as the long and short datasets, respectively). In our analyses, we also included European brown bear haplotypes available from GenBank (4, 18–20, 23). Together, the long and short datasets comprise 67 and 71 brown bear sequences, respectively (see SI Datasets 2 and 3). All analyses were performed by using the two datasets independently and yielded similar results. We inferred a phylogenetic tree from the long dataset (Fig. 1A), to maximize the resolution, and constructed a minimum spanning network using the short dataset (Fig. 1B), to visualize the data.
Fig. 1.
Past and current genetic diversity. (A) Bayesian phylogeny generated by using ancient and modern brown bear European sequences (long dataset) and three cave bears as outgroup. Bootstrap values are shown above the node, and posterior probabilities are shown under the nodes. The color of each square represents the geographic origin, the number in italics indicates the haplotype (see SI Dataset 2), and the “n” is the haplotype frequency. (B) Minimum spanning network showing the haplotype distribution of ancient and modern brown bear sequences (short dataset). Numbers inside the circles indicate the assigned haplotype from a total of 29 haplotypes obtained.
The phylogeny (Fig. 1A) was estimated with the long dataset by using a closely related outgroup [the cave bear, U. spelaeus (24); see SI Dataset 4 for accession numbers]. Although partially unresolved, it has a similar topology to those published previously using mtDNA datasets from both extant and extinct European brown bears (4, 20). In our tree, we identify three statistically supported major clades. One clade comprises modern sequences from eastern (Russia) brown bears together with Pleistocene Iberian and Austrian brown bears and a Holocene German bear. A second clade comprises an extinct group of haplotypes, previously described (20) from a Holocene site in southern France. The third clade previously has been thought to represent two monophyletic groups (Iberian and Italian/Balkan) (4). Although our tree suggests these two clades as monophyletic groups, we fail to find support for them. This last clade comprises both modern and ancient sequences. We note that the Iberian populations do not all cluster together in this last clade, suggesting genetic differentiation between Pleistocene and post-LGM (i.e., recent and Holocene) haplotypes.
The minimum spanning network was constructed by using a total of 29 haplotypes (Fig. 1B). The samples from the sites of Arlanpe and Valdegoba show highly divergent haplotypes, together with previously published data from the sites of Atapuerca (Spain), Grotta Beatrice Cenci (Italy), and Mühlberg (Germany) (20).
Analyses of molecular variance (AMOVAs) were conducted on the short and long datasets to test for evidence of isolation between the Spanish and the large European populations. Interestingly, we found significant levels of geographic substructure in the current and the Holocene populations (long dataset: FSTcurrent = 0.5169, P < 10−5; FSTHolocene = 0.3391, P < 10−4; short dataset: FSTcurrent = 0.4540, P < 10−5; FSTHolocene = 0.3345, P < 10−5), but not in the Pleistocene (large dataset: FSTPleistocene = 0.17691, P = 0.1723; short dataset: FSTPleistocene = 0.1505, P = 0.1885). The results tentatively suggest that gene flow between Europe and Spain was greater during the Pleistocene period than for more recent periods (both at present times and at the Holocene) (23). These observations should be taken as preliminary, however, because (i) the number of haplotypes available for the ancient periods is limited and (ii) the Iberian population clearly depart from the constant size assumption, which is a prerequisite for accurate Fst calculations (see below).
Genetic Diversity of the Spanish Population.
Using the DNA sequences from 24 modern and 13 ancient Iberian brown bears, in combination with published sequences from seven ancient and one modern Iberian bears (see SI Fig. 3), we have been able to estimate the extent of genetic diversity change in the peninsula through time, from the Pleistocene until today. To test whether the changes in diversity show parallel developments in the rest of Europe, we conducted similar analyses for the non-Iberian European population samples, taking advantage of previously reported haplotypes (4, 18–20, 23). It is noteworthy that the genetic diversity of the Spanish brown bear population continuously decreases from the Pleistocene to modern times (Table 1). This pattern is observed for the different statistics we used as estimators of genetic diversity (S, H, and nucleotide diversity). Such a pattern is not observed to the same extent in the European population, which suggests that the reduction in genetic diversity between the Pleistocene and today is specific to the Spanish brown bear population. Such a situation could have resulted from a recent demographic bottleneck followed by population expansion, as suggested by significantly negative Tajima's D and Fu and Li statistics in the extant population (Table 1).
Table 1.
Summary statistics and neutrality test values for the different brown bear populations
| L, nt | Population | N | N haplo. | N poly. | H | Nucleotide diversity | Tajima D | D (P value) | FS | FS(P value) |
|---|---|---|---|---|---|---|---|---|---|---|
| Long | ||||||||||
| 176 | Spain, present | 27 | 5 | 6 | 0.2792 ± 0.1118 | 0.276% ±0.273% | −2.099 | 0.001* | −2.154 | 0.029* |
| 177 | Spain, Holocene | 10 | 5 | 9 | 0.6667 ± 0.1633 | 1.339% ± 0.899% | −1.473 | 0.070 | −0.479 | 0.319 |
| 177 | Spain, Pleistocene | 5 | 4 | 20 | 0.9000 ± 0.1610 | 8.921% ± 5.631% | −0.616 | 0.365 | 1.395 | 0.708 |
| 177 | Spain, ancient | 15 | 9 | 30 | 0.8476 ± 0.0878 | 6.746% ± 3.627% | −0.681 | 0.274 | 0.168 | 0.556 |
| 177 | Europe, present | 11 | 8 | 22 | 0.9273 ± 0.0665 | 8.273% ± 4.532% | 0.870 | 0.849 | −0.005 | 0.464 |
| 177 | Europe, Holocene | 11 | 8 | 26 | 0.9273 ± 0.0665 | 6.190% ± 3.442% | −0.718 | 0.244 | −0.431 | 0.382 |
| 177 | Europe, Pleistocene | 4 | 4 | 17 | 1.0000 ± 0.1768 | 7.664% ± 5.237% | −0.117 | 0.609 | 0.232 | 0.321 |
| 177 | Europe, ancient | 15 | 12 | 27 | 0.9619 ± 0.0399 | 6.599% ± 3.552% | −0.211 | 0.477 | −2.828 | 0.098 |
| Short | ||||||||||
| 114 | Spain, present | 27 | 5 | 5 | 0.2792 ± 0.1118 | 0.355% ± 0.377% | −2.010 | 0.003* | −2.530 | 0.013* |
| 115 | Spain, Holocene | 14 | 6 | 8 | 0.7473 ± 0.1114 | 1.625% ± 1.109% | −1.284 | 0.108 | −1.474 | 0.137 |
| 115 | Spain, Pleistocene | 5 | 4 | 13 | 0.9000 ± 0.1610 | 8.358% ± 5.408% | −0.279 | 0.484 | 0.780 | 0.560 |
| 115 | Spain, ancient | 19 | 10 | 19 | 0.8596 ± 0.0696 | 4.917% ± 2.757% | −0.962 | 0.145 | −1.774 | 0.186 |
| 115 | Europe, present | 11 | 8 | 15 | 0.9273 ± 0.0665 | 8.002% ± 4.499% | 0.698 | 0.803 | −1.014 | 0.263 |
| 115 | Europe, Holocene | 11 | 8 | 18 | 0.9273 ± 0.0665 | 6.177% ± 3.543% | −0.825 | 0.233 | −1.452 | 0.189 |
| 115 | Europe, Pleistocene | 4 | 4 | 11 | 1.0000 ± 0.1768 | 7.563% ± 5.293% | 0.000 | 0.661 | 0.255 | 0.553 |
| 115 | Europe, ancient | 15 | 12 | 19 | 0.9619 ± 0.0399 | 6.598% ± 3.658% | −0.396 | 0.360 | −4.531 | 0.009 |
L, length of the dataset (in nucleotides); N, number of sequences; N haplo., number of haplotypes; N poly., number of polymorphic sites; H, gene diversity (i.e., haplotype diversity); FS, Fu and Li statistics; *, statistically significant.
We further estimated the population mutation parameter, θ, and used this in conjunction with mutation rate estimates to infer the effective population sizes (see Materials and Methods) of the Spanish brown bear population at present and during the Holocene and Pleistocene (SI Table 2; it should be remembered that such values are probably overestimated because not all samples of the Holocene or Pleistocene are of the same age). First, we calculated two θ estimators (θS and θπ). It is noteworthy that although θS and θπ values remain constant for the European population over the three different time periods, they decrease by 60–85% post-LGM (Pleistocene/Holocene transition) and by 48–80% further reduction in recent times (Holocene/current times) in Iberia. These results suggest two successive bottlenecks that affected the Spanish brown bear population at the Pleistocene/Holocene transition (≈10,000 years ago) and during the last 350 years (as the most recent haplotype considered as a member of the Holocene population is ≈350 years old in our datasets; see Materials and Methods). A similar pattern is observed if the θ parameter is estimated by using a Markov chain Monte Carlo coalescent genealogy sampler (LAMARC 2.0), in a maximum likelihood framework and in a Bayesian framework, except that we note a possible reduction of the European brown bear population at the end of the Holocene (SI Table 3). Accordingly, it is most likely that shifts in θ are indicative of a 54–69% reduction in the effective size of the Spanish brown bear population post-LGM and 20–31% reduction in recent times.
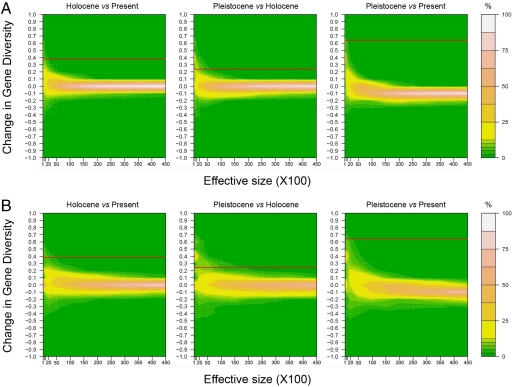
Serial coalescence simulations were conducted to further test whether the shifts in gene diversities at both transitions could be observed under a model of constant population size (Fig. 2, SI Fig. 4, and SI Tables 4–9). Interestingly, it was not possible to detect a shift equal to or greater than the one we observe at the Holocene/present time transition in >5% of our serial coalescent simulations, as long as the effective size was higher than ≈500–1,000 individuals (SI Tables 4 and 5). Moreover, a shift similar to what is observed post-LGM (transition Pleistocene/Holocene) appears in >5% of our serial coalescent simulations only for effective size values below ≈5,000–10,000 or ≈10,000–15,000 individuals, depending on the mutation rate we consider (fast and slow, respectively; SI Tables 4 and 5).
Fig. 2.
Changes in gene diversity simulated between different time periods assuming a constant population size. Mutation rate: 29.8% substitution per site per million years (A) and 10.0% substitution per site per million years (B). The colors refer to the proportion of simulations leading to the corresponding shift in gene diversity. Thirteen possible effective sizes were simulated (100, 500, 1,000, 2,000, 5,000, 10,000, 15,000, 20,000, 25,000, 30,000, 35,000, 40,000, and 45,000 individuals). Red line indicates observed changes in gene diversity (large dataset).
Discussion
We present a serial dataset of brown bear mtDNA sequences based on 177-bp and 115-bp mtDNA, respectively, ranging in age from the Pleistocene until today. Although they are short fragments, they provided enough data for statistical significance and support in a range of analyses. The results indicate a possible shift in population size since the Pleistocene and the presence of eastern lineage haplotypes in Iberia during prehistory.
Under traditional glacial refugia hypotheses (4, 17), the extant brown bear phylogeographic structure derives from ancestral glacial refugia: the western lineage originating from Iberia, Italy, and the Balkans, and the eastern lineage possibly derived from a Carpathian refugium (14, 16). In contrast to such a strict refugial model, but in concordance with a continuous European prehistoric population, we have identified a sequence from a Pleistocene Iberian brown bear from Arlanpe site (the Basque country) that belongs to the eastern clade. In our analyses, such a phylogenetic assignment is supported by maximal posterior probabilities (Fig. 1A). This pattern is further supported by three Pleistocene brown bear sequences from Valdegoba (northern Spain), which cluster with a previously published sequence from Atapuerca (northern Spain) and with several sequences from modern Italian and Balkan bears. Furthermore, AMOVAs suggest little geographic substructure among Spanish and European Pleistocene populations. These new data confirm the lack of phylogeographic discontinuity in European brown bears before the LGM (23). Although Spanish and European Holocene populations appear geographically differentiated in our AMOVAs, a recent study has suggested that gene flow could have continued from the Pleistocene to the Holocene (20). An Iberian brown bear, dated to the time of the LGM from the site of Atapuerca in Burgos in the north of Spain, was more closely related to Italian/Balkan bears than to the Iberian ones. Moreover, during the Holocene in Mont Ventoux (southern France) three mitochondrial groups are found between 1,570 to 6,525 years B.P.: one belonging to the Iberian group, another one to the Italian/Balkan one, and yet a third one not associated with any of the three main glacial refugia (20). Note, however, that support for the Spanish and the Italian/Balkan clades are low in our tree. In this study, we have found three different individuals from Valdegoba, a Late Pleistocene site also in Burgos, that group together with the sample from Atapuerca.
Our data also indicate that population size has varied significantly over time. The decrease in population size over the last centuries (XV–XX) is well documented and also is visible in our analyses of genetic diversity. However, there also is a possible shift in population size at the Pleistocene/Holocene transition, as indicated by changes in values over time for the different θ estimators. This shift is statistically significant, assuming plausible effective population size ranges (>2,000–10,000 or >10,000–15,000 considering the fast or slow mutation rates, respectively).
We consider such a population size to be likely for the following reasons. Using the fast mutation rate, we estimated that the Spanish brown bear population may have reached on average 7,300 and 12,000 effective individuals (depending on the dataset and the estimation method; Table 1 and SI Table 2). Assuming such effective sizes (≈10,000 individuals), only a negligible percentage (0.8% and 0.0% for the long and short datasets, respectively) of the serial coalescent simulations could replicate the shift observed in gene diversity at the Pleistocene/Holocene transition without any change in effective population sizes (SI Tables 4 and 5). The same holds true for the slow mutation rate, as <0.6–0.9% of the serial coalescent simulations lead to the shift observed in gene diversity at the Pleistocene/Holocene transition, assuming constant effective sizes ≈25,000–35,000 individuals (21,818–35,686; SI Tables 4 and 5). Furthermore, it should be noted that the Spanish Pleistocene effective population size does not refer solely to the Spanish population because we have demonstrated significant gene flow between Spain and both the eastern and western parts of Europe during the Pleistocene. In addition, the different θ estimates for the Spanish population are in the range of what is observed for the European population taken as a whole (Table 1 and SI Table 2). Therefore, the effective size is expected to be large and probably well >26,000 individuals [the minimum of the 95% confidence range, estimated in Saarma et al. (16)].
Historical and paleontological sources provide additional information on changes in the brown bear habitat range and thus can be used as an additional tool for inferring population sizes. As recently as the XIVth century, brown bears inhabited most of the mountains and forests of the Iberian Peninsula (25). Their habitat ranged from the Galaico-Portuguese massif (26, 27) to the forests in Andalucia in southern Spain, where they are no longer extant, as well as the Cantabrian and Pyrenees mountain ranges, which they still inhabit today. The use of firearms for hunting from the 16th century, among other human activities, may have been the cause for the reduction of habitat range, restricting brown bears to the northern fringe of Spain by the 18th century (26). Between the 16th and 20th, centuries it is estimated that ≈3,000 bears were killed by hunting in the French Pyrenees alone (8), and thus before this critical period of hunting it is likely that the population of bears for the entire Iberian peninsula would have been in the range of tens of thousands of individuals.
Furthermore, the distribution of sites yielding brown bear fossils in Iberia during the Late Pleistocene is in accordance with such an assumption. Their presence in many localities of northern Iberia (28), together with their occurrence in the rest of the Iberian territory [i.e., Valdegoba (Burgos), Pinilla (Madrid), Bolomor (Valencia), Zafarraya (Granada), and Gorham's cave (Gibraltar) among others], could represent a substantial population size before the LGM. Thus, we estimate that the two demographic bottlenecks post-LGM and in the last centuries would have reduced the effective size of the Spanish population by factors of 1.3–5.0 and 2.2–6.7 respectively, but we do acknowledge the preliminary nature of our estimate and the need to confront it with more data, including data from other loci.
The most straightforward interpretation of our results is that of an Iberian brown bear population in constant change until recently. Genetic diversity measures indicate that several bottlenecks have occurred, including the pre-Holocene one, that have participated in shaping the present day patterns of variation. Further, even though specific Iberian haplotypes have arisen, there also is a clear pattern of exotic influence throughout history. The bear populations present in contemporary Iberia may have evolved here but under a constant flux of external influence and population size change. Thus, the brown bear populations in the Iberian Peninsula would have been isolated only during a recent period along their history.
Materials and Methods
Twenty ancient brown bear bones and teeth from different localities in Spain (see SI Dataset 1) were sampled. Additionally, 24 contemporary brown bears previously identified as 24 different individuals using microsatellites at the Museo Nacional de Ciencias Naturales in Madrid (J.L.G.-G. and I.D., unpublished data) from the western population in the Cantabrian Mountains in northern Spain were sampled by using shed hairs. Four samples (Aketegi II, Hortiguela, Maza Cotina, and La Paloma) were radiocarbon-dated by accelerator mass spectrometry (AMS) at the Ångström Laboratory, Uppsala University in Sweden. The three samples from Valdegoba have a paleontological context dated to 75,000 to 90,000 years old (29). The sample belonging to Arlanpe, in the Basque Country, dates back to the Pleistocene period. Because a radiocarbon date is still not available at the moment, two extreme dates, chosen in a range compatible with the information about the excavation context, have been assigned for this sample (40 KY or 80 KY). Therefore, all of the serial coalescent analyses (see below) were performed twice. Importantly, this did not affect the change in gene diversity observed between the Pleistocene and Holocene periods, as shown in SI Tables 2 and 3 (80 KY) and SI Tables 6 and 7 (40 KY).
DNA Extraction from Ancient Brown Bears.
Samples (Ua1–Ua4).
Bone and teeth were ground to powder under liquid nitrogen with a Spex 6700 Freezer Mill according to the manufacturer's instructions and mortar and pestle.
DNA was extracted from 150 mg of powder by using a silica-binding approach (30). These four samples (Ua1–Ua4) were extracted, amplified twice, and directly sequenced at Centro Mixto Universidad Complutense de Madrid–Instituto de Salud Carlos III de Evolución y Comportamiento Humanos, Madrid, and replicated by using a solvent approach (18) at University College London. Both times the amplifications were performed according to Leonard et al. (18) and directly sequenced. Primers used were L16164 and H16299 from ref. 31. In this case, when conflict was observed among the data, a majority rule consensus was applied, considering two of three the correct sequence for analysis. Contamination was monitored by using four extraction water blanks and six PCR negative controls.
Samples (Ua5–Ua20).
From every specimen, ≈150 mg of bone powder was obtained by grinding the sample with mortar and pestle. DNA extraction was performed following previously published protocols (32, 33). Five microliters of extract were used for a single PCR. PCR conditions consisted of an 11-min activation step at 95°C followed by 55 cycles at 94°C for 30 s, corresponding annealing temperature for 30 s and 74°C for 30 s. All samples, with the exception of Aketegi II, Maza Cotina, La Paloma, and Villavieja, were amplified by using primers URSUSF1_136–156/URSUSR1_273–290, OP54, and OP55. DNA of these four samples was amplified by using primers L16164/H16299 (31). This procedure was carried out as part of a previous study, and extracts were not available for secondary amplifications using the other primer sets used in this study.
Primer used (forward/reverse, Tm°, number of bp) were: Valdiosera 2007, 5′-CAGCACCCAAAGCTAATGTTC-3′/5′-GCACGAKMTACATAGGGG-3′, 52°C, 111 bp; Hänni 1994, 5′-GCCCCATGCATATAAGCATG-3′/5′-GGAGCGAGAAGAGGTACACGT-3′, 55°C, 139 bp; OP54, 5′-GTATAGTCTGTAAGCATGTAT-3′/5′-AGG TAC ACG TAC TCG CAA-3′, 54°C, 50 bp; and OP55, 5′-CCCCATGCATATAAGCATGTACAT-3′/5′-CTCCCGGACTAAGTGAAATAC ATG-3′, 55°C, 64 bp.
Every fragment was amplified twice on each individual, followed by cloning (16 clones in total, 8 per each of the two independent PCR products) using the TOPO TA Cloning Kit (Invitrogen) and sequenced with M13 Universal primers. Purification of colony PCR products was done using Millipore filtration plates (Montage PCRu96 Plate). When sequences from the same sample differed among PCR products, samples were amplified twice and cloned again (six clones per PCR product) to choose the correct sequence. Because of the exceptional nature of the result and its implications, four additional amplifications were performed for sample Asieko (Ua10) (see SI Tables 4, 5, 8 and 9) followed by cloning (eight clones per independent amplification). From a total of 48 cloned sequences, 8 clones (all from one single amplification) differed from the others. Thus, we applied the majority rule consensus considering 40 of 48 sequences the correct ones. Contamination was monitored by using two extraction water blanks for every four samples and four to five PCR blanks in every amplification.
DNA Extraction from Modern Brown Bears.
Naturally shed wild brown bear hair roots were used as a DNA source for the analysis of modern brown bears currently inhabiting the Cantabrian Mountains in northern Spain. All samples were collected by using masks and disposable gloves and were placed into paper envelopes for the sample conservation.
DNA was isolated from hair roots using a phenol-chloroform method (18). A minimum of six hair roots per sample (n = 24) was used to ensure sufficient amounts of DNA. The extraction was conducted in a laboratory dedicated to ancient DNA extractions at the Museo Nacional de Ciencias Naturales in Madrid. Water was used as negative controls for extraction and PCR. PCR conditions were performed at 95°C for 10 min followed by 55 cycles at 95°C for 30 s, 45 s at annealing temperature and 72°C for 45 s each cycle, and 10 min at 72°C for the final elongation. The primers used to amplify a fragment of 271 bp of the mitochondrial control region were TabUa_F as forward (4) and H16299 as reverse (31) primers.
Data Analyses.
Two datasets were generated as different sequence lengths were recovered among ancient specimens: (i) a short dataset spanning over 115 nt and including all of the new specimens extracted in this study (i.e., 26 contemporary and 19 ancient bears) and (ii) a long dataset consisting of the longest sequence information (177 nt) available among our new specimens (i.e., 26 contemporary and 15 ancient bears). For population genetic analyses, all of the ancient and modern haplotypes available in GenBank for western European brown bears were added (see SI Datasets 1–4). Some eastern haplotypes from Russia and Slovakia were aligned, and they are closely related to one of our Pleistocene Iberian sequence (Fig. 1). Three cave bear haplotypes (accession nos. AY149268, AY149271, and AY149273) were used as outgroups in phylogenetic analyses. Maximum likelihood and Bayesian phylogenetic trees were generated under a HKY+G+I model of molecular evolution with PhyML online (500 bootstrap replicates) (34) and MrBayes software (35, 36), respectively. The Bayesian analysis was based on 7,500 trees sampled over 10,000,000 generations (sample frequency 1/1,000 and burn in-value 2,500). In the following, the parameters of the different models were estimated with PhyML online and PAUP* from both phylogenetic datasets but removing outgroups to best fit the mutational process within brown bears. Standard diversity indices (S, H, and nucleotide diversity), neutrality tests (Tajima's D, Fu and Li Fs), minimum spanning networks, and AMOVAs were performed with Arlequin (Ver. 3.1) (37). For AMOVA and molecular diversity calculations, genetic distances were calculated assuming uneven transition/transversion ratios (K2 correction) and heterogeneous mutation rates among sites (alpha-parameter 0.271 and 0.236 for the short and long datasets, respectively). Fst significance was tested over 20,000 permutations. Additionally, the genetic polymorphism was used to estimate the mitochondrial effective population size (Ne) for the Spanish brown bear population at present and different past periods of time (350–7,500 ± 55 years ago and 15,000 to 80,000 years ago designed as the Holocene and Pleistocene periods, respectively). Because θH is biased for single locus estimates and θK is measured independently from sequence information, only θS and θp were estimated from Arlequin. A Markov chain Monte Carlo coalescent genealogy sampler [LAMARC 2.0 (38)] was used to provide θ estimates under a maximum likelihood (θMLE) and Bayesian framework (θMPE). The search among genealogies (and population parameters for the Bayesian analyses) was conducted by using default parameters but heating supplements (four simultaneous Markov chains). Likelihood ratio tests were conducted to choose among the two models of molecular evolution provided in LAMARC 2.0 (F84 or GTR). Finally, a F84 model of molecular evolution with four classes of mutation rates (alpha-shape parameter = 0.275 and 0.289, transition/transversion ratio = 47.806 and 70.209 for the short and long datasets, respectively) was used. The effective size then was calculated assuming a neutral model of molecular evolution (θ = 2Neμ, where μ is the mutation rate for the complete sequence per generation for θS and θπ, but the mutation rate per site per generation for θMLE and θMPE). We assumed two possible mutation rates (10% and 29.8% substitution per site per million years) according to the literature (4, 16).
Finally, we took advantage of the Serial SimCoal software developed by Ramakrishnan and colleagues (39, 40) to assess the significance of the observed changes in gene diversity between different periods of time using serial coalescent simulations. When available, the radiocarbon ages of each specimen was converted in number of generations assuming a value of 12.5 years as mean generation time, in agreement with what is reported for current populations (41). Otherwise, the age of a specimen was inferred from the excavation context; when such information was missing, the age was assumed as equal to the average of absolute ages of the specimens belonging to the same time period. Because the specimen from Arlanpe (the Basque country) presented the most divergent haplotype in Iberia, we performed parallel simulations with two extreme dates to ensure that the result was not an artifact of an erroneous datation (see SI Tables 4, 5, 8 and 9). Simulations were repeated 1,000 times to generate genetic data and changes in gene diversity for the null hypothesis of constant size over the whole history of the population. The simulations were run assuming different possible effective sizes (from 100 individuals to 45,000). The maximum limit of 45,000 individuals was chosen with reference to the most probable effective size of the western lineage European population estimated in ref. 16 under a Bayesian framework. Interestingly, this value is in the range of the maximum effective size estimated from θS, θπ, θMLE, and θMPE (Table 1 and SI Table 2). DNA sequences were generated from the coalescence trees assuming an uneven transition/transversion ratio (47.470 and 100.000 for the short and long datasets, respectively) and heterogeneous mutation rates among sites as a model of molecular evolution (0.271 and 0.236, respectively). Other parameters (same transition/transversion ratio and gamma-shape as for the LAMARC calculations) and additional mutation rates (e.g., 6.5% and 19.5% substitutions/MY) were used but did not change the final results (e.g., SI Tables 6–9). The distributions of changes in gene diversity between the different time periods were calculated, plotted, and compared with the observed values using the R package (ref 42; www.R-project.org).
Supplementary Material
Acknowledgments.
We acknowledge Asier Gómez, Eneko Iriarte, and Joseba Ríos for kindly providing information and material of interest from their excavation site and Marie Sémon for help with R subroutines. We also thank Alfonso Hartasanchez for providing modern samples from the Cantabrian bears, Universidad de Oviedo (Diego Alvarez Lao), Sociedad de Ciencias Aranzadi (Jesús Altuna), Museo Nacional de Ciencias Naturales (Begoña Sánchez), and Universidad de Burgos (Ana I. Ortega) for access to museum collections and permissions for sampling. Tom Gilbert and Uma Ramakrishnan provided valuable comments that helped to improve the text. Financial support was provided by the Consejería de Medio Ambiente, Ordenación del Territorio e Infraestructuras del Principado de Asturias (DGVI 1253/03), Ministerio de Ciencia y Tecnología of the government of Spain (Project no. CGL2006-13532-C03-02), and Consejo Superior de Investigaciones Científicas (UAC2003-0034) and Fundación Atapuerca.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU400176–EU400211).
This article contains supporting information online at www.pnas.org/cgi/content/full/0712223105/DC1.
References
- 1.Servheen C. Status and conservation of the bears of the world. Proceedings of the 8th International Conference on Bear Research and Management; British Columbia, Canada. 1990. Monograph Series No. 2. [Google Scholar]
- 2.Mattson DJ, Merrill T. Extirpations of grizzly bears in the contiguous United States, 1850–2000. Cons Biol. 2002;16:1123–1136. [Google Scholar]
- 3.Waits LP, Talbot SL, Ward RH, Shields GF. Phylogeography of the North American brown bear and implications for conservation. Cons Biol. 1998;12:408–417. [Google Scholar]
- 4.Taberlet P, Bouvet J. Mitochondrial DNA polymorphism, phylogeography, and conservation genetics of the brown bear (Ursus arctos) in Europe. Proc Biol Sci. 1994;255:195–200. doi: 10.1098/rspb.1994.0028. [DOI] [PubMed] [Google Scholar]
- 5.Randi E. Conservation genetics of carnivores in Italy. Comptes Rendus Biol. 2003;326(Suppl 1):S54–S60. doi: 10.1016/s1631-0691(03)00038-6. [DOI] [PubMed] [Google Scholar]
- 6.Posillico M, Meriggi A, Pagnin E, Lovari S, Russo L. A habitat model for brown bear conservation and land use planning in the central Apennines. Biol Conserv. 2004;118:141–150. [Google Scholar]
- 7.Posillico M, Petrella A, Sammarone L. Preliminary plan for conservation of the brown bear (translated from Italian) In: Commission E, editor. Project LIFENAT99/IT/006244. Rome: Ministero delle Politiche Agricole e Forestali; 2002. [Google Scholar]
- 8.Alonso M, Toldrá LX. The brown bear in Catalunya. In: Naves J, Palomero G, editors. The Brown Bear (Ursus arctos) in Spain. Madrid: Instituto Nacional Para la Conservación de la Naturaleza; 1993. pp. 339–350. (translated from Spanish) [Google Scholar]
- 9.Camarra JJ, Parde JM. The brown bear in France—Status and management in 1985. Aquilo Serie Zoologica. 1990;27:93–96. [Google Scholar]
- 10.Caussimont G, Herrero J. Brown bears from the Spanish Pyrenean region: Current status and conservation proposals (translated from French) Int Conf Bear Res Manage. 1992;9:26–35. [Google Scholar]
- 11.Taberlet P, Bouvet J. Bear conservation genetics. Nature. 1992;358:197. doi: 10.1038/358197a0. [DOI] [PubMed] [Google Scholar]
- 12.Naves J, Fernández-Gil A. Ursus arctos Linnaeus, 1758. In: Palomo LJ, Gisbert J, editors. Atlas of Terrestrial Mammals of Spain. Madrid: Ministerio de Medio Ambiente, SECEM, y SECEMU; 2002. pp. 282–285. (translated from Spanish) [Google Scholar]
- 13.Naves J, Wiegand T, Fernández A, Stephan T. Risk of extinction of the Cantabrian brown bear: The western population. Oviedo, Spain: Fundación Oso de Asturias; 1999. (translated from Spanish) [Google Scholar]
- 14.Sommer R, Benecke N. The Recolonisation of Europe by brown bears Ursus arctos Linnaeus 1758 after the Last Glacial Maximum. Mammal Rev. 2005;35:156–164. [Google Scholar]
- 15.Frankel O, Soule M. Conservation and Evolution. Cambridge: Cambridge Univ Press; 1981. [Google Scholar]
- 16.Saarma U, et al. Mitogenetic structure of brown bears (Ursus arctos L.) in northeastern Europe and a new time frame for the formation of European brown bear lineages. Mol Ecol. 2007;16:401–413. doi: 10.1111/j.1365-294X.2006.03130.x. [DOI] [PubMed] [Google Scholar]
- 17.Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc. 1996;58:247–276. [Google Scholar]
- 18.Leonard JA, Wayne RK, Cooper A. Population genetics of ice age brown bears. Proc Natl Acad Sci USA. 2000;97:1651–1654. doi: 10.1073/pnas.040453097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnes I, Matheus P, Shapiro B, Jensen D, Cooper A. Dynamics of Pleistocene population extinctions in Beringian brown bears. Science. 2002;295:2267–2270. doi: 10.1126/science.1067814. [DOI] [PubMed] [Google Scholar]
- 20.Valdiosera CE, et al. Staying out in the cold: Glacial refugia and mitochondrial DNA phylogeography in ancient European brown bears. Mol Ecol. 2007;16:5141–5149. doi: 10.1111/j.1365-294X.2007.03590.x. [DOI] [PubMed] [Google Scholar]
- 21.Anderung C, et al. Prehistoric contacts over the Straits of Gibraltar indicated by genetic analyses of Iberian Bronze Age cattle. Proc Natl Acad Sci USA. 2005;102:8431–8435. doi: 10.1073/pnas.0503396102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valdiosera C, et al. Typing single polymorphic nucleotides as a way to access Middle Pleistocene DNA. Biol Lett. 2006;2:601–603. doi: 10.1098/rsbl.2006.0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofreiter M, et al. Lack of Phylogeography in European mammals before the last glaciation. Proc Natl Acad Sci USA. 2004;101:12963–12968. doi: 10.1073/pnas.0403618101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orlando L, et al. Ancient DNA and the population genetics of cave bears (Ursus spelaeus) through space and time. Mol Biol Evol. 2002;19:1920–1933. doi: 10.1093/oxfordjournals.molbev.a004016. [DOI] [PubMed] [Google Scholar]
- 25.Nores C, Naves J. Historical distribution of the brown bear in the Iberian Peninsula. In: Naves J, Palomero G, editors. The Brown Bear (Ursus arctos) in Spain. Madrid: Instituto Nacional Para la Conservación de la Naturaleza; 1993. pp. 13–33. (translated from Spanish) [Google Scholar]
- 26.Almazán duque de. History of Hunting in Spain. Barcelona: Instituto Gráfico Oliva de Vilanova; 1934. (translated from Spanish) [Google Scholar]
- 27.López Ontiveros A, Valle B, García Verdugo FR. Hunting and Geographic Landscape of the Bethic Lands According to the Book of the Hunters. Córdoba: Junta de Andalucía; 1991. (translated from Spanish) [Google Scholar]
- 28.Torres T. Pleistocene-Holocene ursids of the Iberian Peninsula. Madrid: Universidad Politécnica de Madrid; 1984. (translated from Spanish) [Google Scholar]
- 29.Quam RM, et al. Human remains from Valdegoba cave (Huérmeces, Burgos, Spain) J Hum Evol. 2001;41:385–435. doi: 10.1006/jhev.2001.0486. [DOI] [PubMed] [Google Scholar]
- 30.Yang DY, Eng B, Waye JS, Dudar JC, Saunders SR. Improved DNA extraction from ancient bones using silica-based spin columns. Am J Phys Anthropol. 1998;105:539–543. doi: 10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 31.Hänni C, Laudet V, Stehelin D, Taberlet P. Tracking the origins of the cave bear (Ursus spelaeus) by mitochondrial DNA sequencing. Proc Natl Acad Sci USA. 1994;91:12336–12340. doi: 10.1073/pnas.91.25.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boom R, et al. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohland N, Hofreiter M. Comparison and optimization of ancient DNA extraction. Biotechniques. 2007;42:343–352. doi: 10.2144/000112383. [DOI] [PubMed] [Google Scholar]
- 34.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 35.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- 36.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference on the mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 37.Schneider S, Roessli D, Excoffier L. Geneva: University of Geneva; 2000. Arlequin: A Software for Population Genetics Data Analysis. Version 2000. [Google Scholar]
- 38.Kuhner MK. LAMARC 2.0: Maximum Likelihood and Bayesian estimation of population parameters. Bioinformatics. 2006;22:768–770. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- 39.Ramakrishnan U, Hadly EA, Mountain JL. Detecting past population bottlenecks using temporal genetic data. Mol Ecol. 2005;14:2915–2922. doi: 10.1111/j.1365-294X.2005.02586.x. [DOI] [PubMed] [Google Scholar]
- 40.Anderson CN, Ramakrishnan U, Chan YL, Hadly EA. Serial SimCoal: A population genetics model for data from multiple populations and points in time. Bioinformatics. 2005;21:1733–1734. doi: 10.1093/bioinformatics/bti154. [DOI] [PubMed] [Google Scholar]
- 41.Bellemain E, Zedrosser A, Manel S, Waits LP, Taberlet P, Swenson JE. The dilemma of female mate selection in the brown bear, a species with a sexually selected infanticide. Proc Roy Soc B. 2006;273:283–291. doi: 10.1098/rspb.2005.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Development Core Team. Vienna, Austria: R Foundation for Statistical Computing; 2007. R: A Language and Environment for Statistical Computing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.