Abstract
According to the amyloid hypothesis, the pathogenesis of Alzheimer's disease is triggered by the oligomerization and aggregation of the amyloid-β (Aβ) peptide into protein plaques. Formation of the potentially toxic oligomeric and fibrillar Aβ assemblies is accompanied by a conformational change toward a high content of β-structure. Here, we report the solution structure of Aβ(1–40) in complex with the phage-display selected affibody protein ZAβ3, a binding protein of nanomolar affinity. Bound Aβ(1–40) features a β-hairpin comprising residues 17–36, providing the first high-resolution structure of Aβ in β conformation. The positions of the secondary structure elements strongly resemble those observed for fibrillar Aβ. ZAβ3 stabilizes the β-sheet by extending it intermolecularly and by burying both of the mostly nonpolar faces of the Aβ hairpin within a large hydrophobic tunnel-like cavity. Consequently, ZAβ3 acts as a stoichiometric inhibitor of Aβ fibrillation. The selected Aβ conformation allows us to suggest a structural mechanism for amyloid formation based on soluble oligomeric hairpin intermediates.
Keywords: Aβ-peptide, engineered binding protein, molecular recognition, protein structure, nuclear magnetic resonance
The amyloid-β (Aβ) peptide is a 39–43 residue cleavage product of the amyloid precursor protein. It is the main component of senile plaques, which are neuropathological hallmarks of Alzheimer's disease (AD), and its critical role for AD etiology has been supported by genetic studies (1, 2). Substantial evidence attributes the role of the principal neurotoxic species to soluble oligomeric forms of Aβ (3–7). The precise cellular and molecular mechanisms underlying their toxicity are currently a matter of intense research (3, 7).
Antiamyloid approaches, which target the production, aggregation, and clearance of Aβ, constitute the focus of current efforts to develop AD therapeutics (1, 2, 8). A promising example is anti-amyloid immunotherapy, which has demonstrated effectiveness in animal models and entailed reduced plaque burden in clinical trials (8–10). Three different modes of action of amyloid-dissolving antibodies have been suggested: (i) dissolution as a consequence of direct binding to oligomeric/fibrillar Aβ in the brain, (ii) phagocytosis by microglial cells, and (iii) increased efflux of Aβ from the brain as a result of binding of monomeric Aβ in the plasma (peripheral sink mechanism) (8, 11). Whereas antibodies to the Aβ N terminus, which is located at the fibril surface, are effective in direct binding to aggregated Aβ (12, 13), the binding and stabilization of monomeric Aβ might be a specific feature of antibodies to its hydrophobic central part (14). Recognition of the hydrophobic central part is also a strategy pursued in the search for peptide or peptidomimetic inhibitors of Aβ fibrillation (15–17). Recently, the structures of the antigen binding fragments of two antibodies to the N-terminal Aβ(1–8) peptide were reported (18). However, the structural basis of interactions with the central and C-terminal parts of Aβ remains elusive.
The peripheral sink mechanism implies that not only anti bodies, but any peripherally administered Aβ binding molecule of sufficiently high affinity could potentially dissolve plaques by shifting the dynamic equilibrium between central nervous system Aβ and plasma Aβ toward the latter (14, 19). On this account, affibody ligands to monomeric Aβ(1–40) have recently been selected (20). Affibody ligands represent one class of engineered affinity proteins with applications in biotechnology, biochemical assays, disease diagnosis, and therapy (21–23). They are based on the Z domain derived from staphylococcal protein A and selected by phage display from a combinatorial protein library in which 13 of the 58 amino acid residues are randomized. The randomized positions, distributed over helix 1 and 2 of the three-helix bundle scaffold, have been chosen because of their location to the binding interface in the complex of the Z domain with its target, the Fc fragment from IgG (22).
Here, we investigate the interaction of Aβ(1–40) with the affibody protein ZAβ3 and report the solution structure of the complex. ZAβ3 binds to the central/C-terminal part of Aβ(1–40), which adopts a β-hairpin conformation reminiscent of the Aβ fibril structure.
Results and Discussion
Disulfide-Linked ZAβ3 Dimer Binds Aβ(1–40) with Nanomolar Affinity.
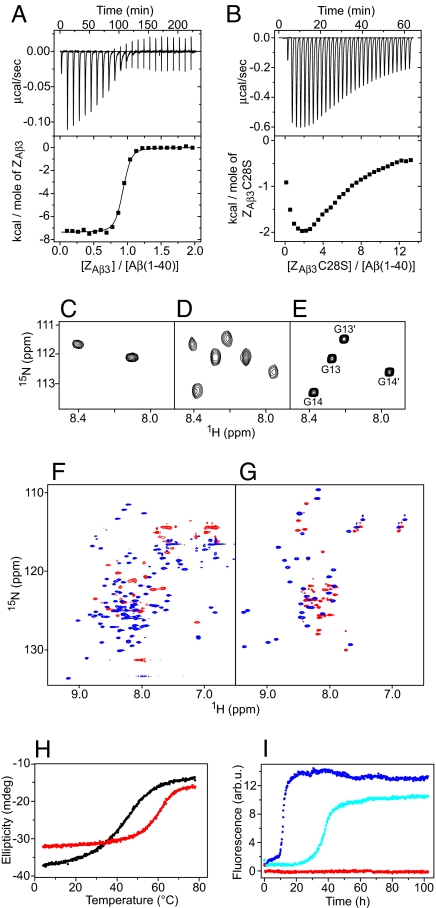
The 16 Affibody variants, for which binding to both Aβ(1–40) and Aβ(1–42) was tested and confirmed (20) (Fig. 1A), all have a cysteine residue at position 28, suggesting that disulfide-linked dimers were selected. Dimeric ZAβ3 binds monomeric Aβ(1–40) with 1:1 stoichiometry and an affinity of Kd = 17 nM as determined by isothermal titration calorimetry (ITC) (Fig. 2A). In contrast, a monomeric mutant, obtained by replacing Cys-28 with serine (ZAβ3C28S), binds Aβ(1–40) with much lower affinity and in agreement with a cooperative association of two ZAβ3C28S molecules with one Aβ(1–40) monomer (Fig. 2B). Cooperative binding of two affibody units to distinct sites on Aβ(1–40) is supported by 15N heteronuclear single quantum correlation (HSQC) NMR spectroscopy, as shown for the glycine region: The two glycines Gly-13 an Gly-14 give rise to one peak each for free ZAβ3C28S (Fig. 2C). Upon titration with unlabeled Aβ(1–40), four new peaks appear with identical intensities, corresponding to Gly-13 and Gly-14 of ZAβ3C28S bound to two distinct Aβ(1–40) sites (Fig. 2D). The chemical shifts of these peaks are practically identical to those observed for saturated ZAβ3 (Fig. 2E), suggesting a common binding mode for the monomeric and dimeric constructs. Binding is coupled to folding of both Aβ(1–40) and the affibody ligand, as indicated by the greatly improved resonance dispersion in HSQC spectra upon complex formation (Fig. 2 F and G). Particularly, several amide proton resonances are shifted downfield to values typical for β-sheet conformation upon binding [supporting information (SI) Fig. S1]. Concomitantly, the thermostability of ZAβ3 increases from a melting temperature of 47°C for free ZAβ3 to 64°C for the complex (Fig. 2H).
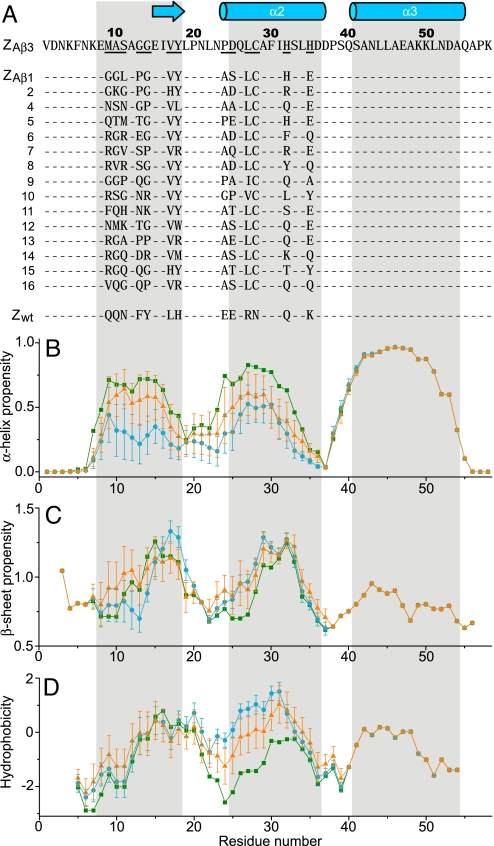
Fig. 1.
Sequences, secondary structure propensities, and hydrophobicity of Aβ-binding affibody proteins. (A) Amino acid sequences of Aβ-binding affibody proteins aligned to ZAβ3 (20). Helical or β-sheet secondary structure in the ZAβ3:Aβ(1–40) complex is indicated by cylinders or arrows, respectively. Areas highlighted in gray correspond to helical structure in the Z scaffold (bottommost sequence). Randomized amino acids are underlined for ZAβ3. (B–D) Averaged α-helix propensity (B), β-sheet propensity (C), and hydrophobicity (D) of the 16 ZAβ affibody protein sequences (blue circles), and 30 previously published affibody protein sequences selected to bind 10 different targets (orange triangles) (see SI Text). The corresponding values of the Z domain are given as green squares. Error bars indicate estimated standard deviations.
Fig. 2.
Biophysical characterization of the ZAβ3:Aβ(1–40) interaction. (A and B) Titration of ZAβ3 dimer (A) or ZAβ3C28S (B) into Aβ(1–40) monitored by ITC at 20 °C. For the experiment in A, N = 0.89 ± 0.01, Kd = 17 ± 2 nM, ΔH = −7.4 ± 0.1 kcal mol−1. (C–E) Glycine region of the 1H-15N HSQC spectrum of 15N-labeled ZAβ3C28S (C and D) or ZAβ3 dimer (E) in the absence (C) or presence (D and E) of an excess of unlabeled Aβ(1–40). Peaks in E are assigned to the subunits Z and Z′ as shown in Fig. 3. (F and G) Sections of the 1H-15N HSQC spectrum of 15N-labeled ZAβ3 dimer (F) or Aβ(1–40) (G) in the absence (red) or presence (blue) of a 15% excess of the respective unlabeled binding partner. (H) Thermal melting of free ZAβ3 dimer (black), and an equimolar mixture of the ZAβ3 dimer and Aβ(1–40) (red) monitored by CD at 220 nm. (I) Aggregation time course of 115 μM Aβ(1–40) in the absence (blue) and presence of 0.5 (cyan) or 1.1 (red) molar equivalents of ZAβ3 dimer monitored by thioflavin T fluorescence.
ZAβ3 Inhibits Aβ(1–40) Fibrillation at Stoichiometric Concentrations.
Thioflavin T fluorescence was used to monitor Aβ(1–40) fibrillation in the absence and presence of ZAβ3 (Fig. 2I). ZAβ3 acts as a potent fibrillation inhibitor. Stoichiometric concentrations of ZAβ3 dimer are required for complete inhibition, revealing that the binding of monomeric Aβ(1–40) is responsible for the inhibitory function.
Structure of the ZAβ3:Aβ(1–40) Complex.
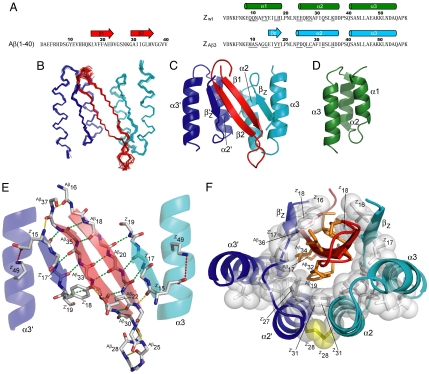
NMR was used to determine the structure of the complex between Aβ(1–40) and the disulfide-linked dimer of ZAβ3 (Fig. 3). The complex consists of a four-stranded antiparallel β-sheet and four α-helices. The selected conformation of Aβ(1–40) is a β-hairpin in which the Aβ17-Aβ23 and Aβ30-Aβ36 fragments make intramolecular backbone hydrogen bonds to form the two central strands of the β-sheet (Fig. 3E). Both faces of the Aβ hairpin are predominantly nonpolar, and both are buried within a large hydrophobic tunnel-like cavity in the ZAβ3 dimer (Fig. 3F). Hence, 1,400 Å2 of surface area, of which 71% is nonpolar, is inaccessible to water in the complex. In the cavity, the Aβ hairpin is flanked on each side by β-strands formed by residues 15–18 of the two ZAβ3 subunits, respectively. This fragment is part of helix 1 in the originating Z domain (24) and affibody complexes reported in refs. 25 and 26. However, it is unfolded in ZAβ3 and presumably in all ZAβ binders, because they contain helix-destabilizing glycine and proline replacements at positions 9–11 and 13–14 (Fig. 1 A and B). Selected residues at positions 17 and 18 in the (former) helix do, however, show β-sheet propensity in agreement with the observed structure (Fig. 1C). A further consequence of helix 1 unfolding is that it opens a large hydrophobic cleft in which the core of the ZAβ3 dimer becomes exposed. The “interior” face of the Aβ hairpin containing the AβLeu-17, AβPhe-19, AβIle-32, AβLeu-34, and AβVal-36 side chains docks into the cleft to form a large intermolecular hydrophobic core.
Fig. 3.
Structure of the ZAβ3:Aβ(1–40) complex. (A) Amino acid sequences of Aβ(1–40), ZAβ3, and the wild-type Z domain (Zwt). Residues randomized in the phagemid library are underlined. Helical or β-sheet secondary structures in the ZAβ3:Aβ(1–40) complex and in Zwt are indicated by cylinders or arrows, respectively. (B) Superimposed simulated annealing structures of the ZAβ3:Aβ(1–40) complex [Aβ(1–40) in red, ZAβ3 subunits in blue or cyan]. (C and D) Ribbon drawings illustrating the topologies of the ZAβ3:Aβ(1–40) complex (C) and the original Z domain scaffold (D). (E) Polar contacts in the ZAβ3:Aβ(1–40) complex. Experimentally validated hydrogen bonds (green), hydrogen bonds observed in >50% of the simulated annealing structures (yellow), and salt bridges (red) are displayed. Residues of the two ZAβ3 subunits are labeled Z or Z′, respectively. (F) The hydrophobic core of the complex. Nonpolar side chains with water exposure <33% are displayed as orange sticks [for Aβ(1–40)] or white sticks and spheres (for ZAβ3). The disulfide bond is shown in yellow.
The ZAβ3 side of the core is held tightly by the selected Cys-28–Cys-28 disulfide linking helices 2 of the two subunits, and it includes the conservatively selected Leu-27, entailing a comparatively strong hydrophobicity in this sequence region (Fig. 1D), and the two Ile-31 side chains. Interestingly, although Ile-31 is not varied in phage display selection, it is still found at the hydrophobic interface of all Z domain and affibody complexes studied so far (25). The “exterior” face of the Aβ hairpin is embraced from both sides by the Ile-16 and (selected) Tyr-18 side chains. In this position, Tyr-18 of one ZAβ3 subunit (Z′18 in Fig. 3E) also forms a hydrogen bond to the AβGlu-22 carboxyl. The N-terminal ZAβ3 β-strands are further anchored against helix 3 in both subunits by nonpolar interactions involving (selected) Val-17 and a salt bridge between Glu-15 and Lys-49. This salt bridge is in fact also present in other Z domain and affibody structures when Glu-15 is in a helical conformation. The selection of alanine or proline at position 24 can be rationalized by the tight packing of the disulfide-linked helices, which requires a small nonpolar residue at this position. Residues 25, 32, and 35 are solvent-exposed and not involved in binding, resulting in a less-conserved selection of polar side chains. The terminal fragments Aβ1-Aβ15 of Aβ(1–40) and 1–13 and 57–58 of both ZAβ3 subunits are not well defined by NMR data; NMR chemical shifts close to random coil values and lack of observable NOEs indicate that they are disordered. The C-terminal AβVal-39 and AβVal-40 of Aβ(1–40) are ordered as judged from several medium- and long-range NOEs, but their conformation is nevertheless not uniquely defined following structure determination by simulated annealing. NOEs supporting the formation of a salt bridge between the side chains of AβAsp-23 and AβLys-28, which is populated in Aβ fibrils formed under certain conditions (27, 28), could not be detected.
Short peptides, which are homologous to Aβ but contain proline residues as β-sheet blockers, have been developed as fibrillogenesis inhibitors (16). The aim of this and related peptide- and protein-based approaches that target Aβ aggregation is to bind the hydrophobic part of Aβ by exploiting the same intermolecular interactions formed in Aβ self-assembly, e.g., β-sheet backbone hydrogen bonds, and to consequently disrupt the potential for further β-sheet extension (15–17, 29). Although no structure of a β-sheet breaker peptide in complex with Aβ has been reported, certain aspects of the concept appear to be reflected in the ZAβ3:Aβ(1–40) interaction: The ZAβ3 β-strands cap the edges of the Aβ(1–40) β-sheet; the strands are short and terminated on their C-terminal side by a proline and on the N-terminal side by a sequence region selected to have little propensity for either α- or β-structure, resulting in an inability to serve as a template for further β-sheet extension.
Relation of the Aβ β-Hairpin to the Conformation Within Amyloid Fibrils.
The structure of Aβ(1–40) bound to ZAβ3 shares important characteristics with fibrillar Aβ. Within amyloid fibrils, Aβ peptides form β-strand-turn-β-strand motifs, with the precise extent of β-strands and turn varying somewhat depending on Aβ variant and preparation conditions (28, 30–32). Overall, the positions of the secondary structure elements are in good agreement with those determined in this study for bound Aβ(1–40). The hydrophobic cluster identified in Aβ(1–40) fibrils (28), including AβLeu-17, AβPhe-19, AβIle-32, AβLeu-34, and AβVal-36, is also present in the Aβ(1–40):ZAβ3 complex (Figs. 3F and 4A). Aβ amyloid fibrils are stabilized by backbone hydrogen bonding within β-sheets and packing of hydrophobic side-chains. Similarly, ZAβ3 forms an intermolecular β-sheet with Aβ(1–40) and provides a hydrophobic interface for both faces of the Aβ(1–40) β-sheet. Thus, ZAβ3 captures Aβ(1–40) in an amyloid-like, but monomeric, conformation and consequently inhibits fibrillation.
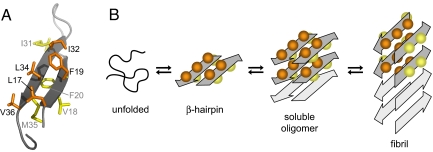
Fig. 4.
Hypothetical aggregation mechanism involving the Aβ β-hairpin. (A) β-hairpin with the interior hydrophobic core side chains (orange) and the exterior side chains (yellow). (B) Schematic of a hypothetical aggregation mechanism that involves the β-hairpin as a transiently populated conformation sampled by the disordered monomer and/or as a constituent of oligomeric Aβ. Soluble oligomers form by hydrophobic stacking of β-hairpins. A concerted conformational transition establishes a fibril seed with in-register, parallel β-sheets. The side chains incorporated in the hydrophobic core within one molecular layer of Aβ(1–40) fibrils correspond to the interior side chains in the ZAβ3:Aβ(1–40) complex. The mechanism is suggested based on the accessibility of the Aβ β-hairpin reported here and ATR-FTIR data reported in reference 35. However, it remains to be proven experimentally.
Although the two Aβ(1–40) β-strands form in-register, intermolecular, parallel β-sheets in amyloid fibrils, they hydrogen bond to each other in the Aβ(1–40):ZAβ3 complex. The two conformations are related by a 90° rotation of both β-strands around their axes (Fig. 4B). Applying a 90° rotation with either direction of rotation to both β-strands in the ZAβ3 bound conformation yields four possible molecular conformations for fibrillar Aβ(1–40) (28, 32). Although the conformation allowing for formation of the hydrophobic cluster AβLeu-17, AβPhe-19, AβIle-32, AβLeu-34, and AβVal-36 within one molecular layer appears to be preferred, other conformations might also be populated depending on the conditions of fibril formation, reflecting the structural plasticity of amyloid fibrils (28, 32). In this context, it can be speculated that the β-hairpin presented here constitutes an intermediate conformation on the pathway to amyloid fibrils, e.g., in the form of a transiently populated conformation sampled by the disordered monomer (33) or as a constituent of oligomeric Aβ (Fig. 4B). In small oligomers, which presumably account for most of the toxicity of amyloidogenic proteins (3, 4, 34), intramolecular β-sheets might be preferred to intermolecular ones because the fibril core structure with its characteristic long-range order is not fully established. Thus, oligomers might form by hydrophobic stacking of β-hairpins and remain soluble as a consequence of the hydrogen bonding capacity of exposed peptide backbones. Fibril seeds could subsequently be generated by a concerted conformational transition toward intermolecular in-register β-sheets (Fig. 4B). The presence of the β-hairpin conformation in oligomers would be in agreement with the observation of a clearly resolvable peak at 1,693 cm−1, indicating an antiparallel β-sheet structure, in the attenuated total reflectance Fourier-transform infrared (ATR-FTIR) spectrum of Aβ oligomers but not fibrils (35).
Amyloid pores, oligomeric assemblies that have been suggested to insert into membranes and confer neurotoxicity by disrupting cell homeostasis (36), could also be composed of Aβ(1–40) hairpins (data not shown). It is unclear, however, whether their dimensions would suffice to span neuronal membranes and whether their mainly nonpolar inner surface would be compatible with a membrane-permeabilizing activity.
Conclusion
This study establishes the β-hairpin as an accessible conformational state of Aβ peptides. Its relation to the conformation of Aβ within amyloid fibrils suggests a role of the β-hairpin in oligomerization and fibrillation. The successful selection of an affibody binding protein that adapts to the conformational preferences of its target by tolerating limited but essential modifications of the scaffold structure highlights the potential of the combinatorial engineering approach. The availability of a binder to monomeric Aβ and its detailed structural and biophysical description will potentiate further investigation of Aβ oligomerization, aggregation, and disaggregation and will help to elucidate to what extent binding and stabilization of monomeric Aβ can interfere with early pathogenic events in AD.
Methods
Proteins.
ZAβ3 and ZAβ3C28S were expressed in Escherichia coli BL21 DE3 cells from plasmid pAY442 containing a T7 promoter (20). In addition to the ZAβ3 affibody sequence displayed in Fig. 1A, the constructs contained an N-terminal (His)6 tag (sequence MGSSHHHHHHLQ) and two C-terminal residues (VD). Cultures were grown at 37°C in LB medium or minimal medium enriched with 1 g/liter 15NH4SO4 and/or 2 g/liter 13C-glucose supplemented with 30 μg/ml kanamycin. Protein expression was induced at OD 0.6–0.8 with IPTG (final concentration 1 mM), followed by further incubation for 4 h. Cells were harvested by centrifugation at 4,000 × g and frozen at −20°C. For purification, cells were resuspended in 50 mM Na-phosphate (pH 7.0), 0.2 mM NaCl, and 1 mM PMSF and lysed by sonication. Insoluble material was removed by centrifugation at 16,000 × g. The (His)6-tagged protein was isolated by TALON metal affinity chromatography (BD Biosciences) according to the manufacturer's instructions. Further purification was achieved by size exclusion chromatography employing an ÄKTA Explorer system equipped with a HiLoad 16/60 Superdex 75 prep grade column (GE Healthcare). The purified affibody proteins were dialysed against 20 mM Na-phosphate (pH 7.2).
Aβ(1–40) was obtained either unlabeled (NaOH-purified) or uniformly 15N or 13C/15N-labeled [trifluoroacetic acid (TFA)-purified] from a commercial source (rPeptide). Aβ(1–40) was dissolved in 30 mM NH4OH (NaOH-purified samples) or 100 mM NH4OH (TFA-purified samples) at a concentration of ≈1.5 mM, centrifuged at 20,800 × g to remove any larger aggregates, and diluted into the final experiment buffer. Protein concentrations were determined by UV absorbance at 280 nm, using extinction coefficients validated by amino acid analysis (Amino Acid Analysis Center, Uppsala University, Uppsala, Sweden). The purities of all proteins were >98% as estimated by SDS/PAGE.
Isothermal Titration Calorimetry (ITC).
ITC was carried out on a VP-ITC calorimeter (MicroCal) at 20°C in 20 mM Na-phosphate (pH 7.2). Aβ(1–40) at concentrations of 7–16 μM was used as titrant in the cell and ZAβ3 at a 9-fold higher concentration (calculated for the dimer) or ZAβ3C28S at a 60-fold higher concentration as titrant in the syringe. All solutions were degassed before experiments. Baseline correction and integration of the calorimeter response was carried out with the Origin software (MicroCal) provided with the calorimeter. To correct for heats of dilution and viscous mixing, the average heat of postsaturation injections was subtracted from each injection. The obtained binding isotherms were fitted to a model in which the variable parameters are the stoichiometry of identical sites (N), an apparent dissociation constant (Kd), and an apparent association enthalpy (ΔH).
Circular Dichroism (CD) Spectroscopy.
CD was performed on a JASCO J-810 spectropolarimeter. Melting curves were recorded at 220 nm, using a 0.1 cm path-length cell containing proteins at concentrations of 17.5 μM [Aβ(1–40), ZAβ3 dimer] or 35 μM (ZAβ3C28S) in 20 mM Na-phosphate, pH 7.2.
Aggregation Assay.
Thioflavin T fluorescence was recorded in 96-well plates (Nunc), using a FLUOstar Optima reader (BMG Labtech) equipped with 440-nm excitation and 480-nm emission filters. The samples contained 100 μl of 115 μM Aβ(1–40) in 50 mM Na-phosphate (pH 7.1), 0.1 M NaCl, 10 μM thioflavin T, and 0.1% Na-azide supplemented with the indicated amount of ZAβ3. Plates were sealed with polyolefin tape (Nunc) and incubated at 37°C. Data points were recorded every 15 min with 5 min of orbital shaking (width 5 mm) before the measurement.
NMR and Structure Determination.
NMR data were collected at 25°C, using Varian Inova 800 and 900-MHz spectrometers, the latter of which was equipped with a cryogenic probe. NMR samples for structure determination contained ≈400 μM 15N-labeled or 13C,15N-labeled Aβ(1–40) or disulfide-linked ZAβ3 dimer, and 15% molar excess of unlabeled ZAβ3 dimer or Aβ(1–40), respectively, in 20 mM Na-phosphate buffer at pH 7.2. Resonance assignments were obtained from standard triple-resonance experiments. Interproton distance constraints were derived from 3D 15N-NOESY and 3D 13C-NOESY spectra recorded with mixing times of 120 ms and calibrated by using known distances in regular secondary structure elements. Intermolecular nuclear Overhauser effects (NOEs) were also identified in 3D F1(13C,15N)-filtered F2(13C or 15N)-edited NOESY experiments (37). Backbone dihedral angle constraints were derived from chemical shifts, using TALOS (38). Backbone hydrogen bond donors were identified in amide hydrogen exchange experiments (see SI Text and Fig. S2) and acceptor carbonyl oxygens were identified based on initial structure calculations (see SI Text). The final constraint dataset (Table S1) contained 3,438 NOE distances, of which 387 are unambiguous intermolecular distances, 160 are backbone dihedral angle constraints, and 39 are hydrogen bonds. Structures were calculated with Xplor-NIH 2.15.0 (39), using ab initio simulated annealing with r−6-averaging. A pseudopotential for the radius of gyration (40) was used on residues 16–40 of Aβ(1–40) and residues 14–56 of the two ZAβ3 subunits to improve packing, and a conformational database potential (41) was used to improve dihedral angle distributions. The full Lennard–Jones potential, an electrostatic potential scaled to 25% of the default value, and parameters for proper covalent disulfide bond geometry were applied during the final slow-cooling refinement. An ensemble of 24 (of 100) structures was selected. Backbone and all-heavy atom rms deviations between structures in the ensemble are 0.32 Å and 0.71 Å, respectively, and 93% of the residues are found in the most favored regions of the Ramachandran diagram (see Table S2 for additional statistics). Hydrogen bonds shown in Fig. 3E have donor hydrogen-acceptor distances <2.4 Å and angles between the donor bond vector and the vector connecting the two heavy atoms of <35° in at least 50% of the structures in the ensemble. Molecular graphics figures were created by using PyMOL (DeLano Scientific).
Acknowledgments.
This work was supported by the Swedish Research Council, the German Academic Exchange Service, and the Mucosal Immunobiology and Vaccine Swedish Foundation for Strategic Research Center.
Footnotes
Conflict of interest statement: A.J. is an employee of Affibody AB.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates and experimental constraints have been deposited in the Protein Data Bank, www.pdb.org [accession no. 2OTK (ZAβ3:Aβ(1–40) complex)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0711731105/DCSupplemental.
References
- 1.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443:774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- 3.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid β-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 4.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 5.Lambert MP, et al. Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. Proc Natl Acad Sci USA. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh DM, et al. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 7.Walsh DM, Selkoe DJ. Aβ Oligomers—a decade of discovery. J Neurochem. 2007;101:1172–1184. doi: 10.1111/j.1471-4159.2006.04426.x. [DOI] [PubMed] [Google Scholar]
- 8.Citron M. Strategies for disease modification in Alzheimer's disease. Nat Rev Neurosci. 2004;5:677–685. doi: 10.1038/nrn1495. [DOI] [PubMed] [Google Scholar]
- 9.Masliah E, et al. Aβ vaccination effects on plaque pathology in the absence of encephalitis in Alzheimer disease. Neurology. 2005;64:129–131. doi: 10.1212/01.WNL.0000148590.39911.DF. [DOI] [PubMed] [Google Scholar]
- 10.Schenk D, et al. Immunization with amyloid-β attenuates Alzheimer-disease-like pathology in the PDAPP mouse. Nature. 1999;400:173–177. doi: 10.1038/22124. [DOI] [PubMed] [Google Scholar]
- 11.Weiner HL, Frenkel D. Immunology and immunotherapy of Alzheimer's disease. Nat Rev Immunol. 2006;6:404–416. doi: 10.1038/nri1843. [DOI] [PubMed] [Google Scholar]
- 12.Frenkel D, Balass M, Katchalski-Katzir E, Solomon B. High affinity binding of monoclonal antibodies to the sequential epitope EFRH of β-amyloid peptide is essential for modulation of fibrillar aggregation. J Neuroimmunol. 1999;95:136–142. doi: 10.1016/s0165-5728(99)00003-x. [DOI] [PubMed] [Google Scholar]
- 13.Solomon B, Koppel R, Frankel D, Hanan-Aharon E. Disaggregation of Alzheimer β-amyloid by site-directed mAb. Proc Natl Acad Sci USA. 1997;94:4109–4112. doi: 10.1073/pnas.94.8.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeMattos RB, et al. Peripheral anti-Aβ antibody alters CNS and plasma Aβ clearance and decreases brain Aβ burden in a mouse model of Alzheimer's disease. Proc Natl Acad Sci USA. 2001;98:8850–8855. doi: 10.1073/pnas.151261398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sciarretta KL, Gordon DJ, Meredith SC. Peptide-based inhibitors of amyloid assembly. Methods Enzymol. 2006;413:273–312. doi: 10.1016/S0076-6879(06)13015-3. [DOI] [PubMed] [Google Scholar]
- 16.Soto C, et al. β-sheet breaker peptides inhibit fibrillogenesis in a rat brain model of amyloidosis: implications for Alzheimer's therapy. Nat Med. 1998;4:822–826. doi: 10.1038/nm0798-822. [DOI] [PubMed] [Google Scholar]
- 17.Tjernberg LO, et al. Arrest of β-amyloid fibril formation by a pentapeptide ligand. J Biol Chem. 1996;271:8545–8548. doi: 10.1074/jbc.271.15.8545. [DOI] [PubMed] [Google Scholar]
- 18.Gardberg AS, et al. Molecular basis for passive immunotherapy of Alzheimer's disease. Proc Natl Acad Sci USA. 2007;104:15659–15664. doi: 10.1073/pnas.0705888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuoka Y, et al. Novel therapeutic approach for the treatment of Alzheimer's disease by peripheral administration of agents with an affinity to β-amyloid. J Neurosci. 2003;23:29–33. doi: 10.1523/JNEUROSCI.23-01-00029.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grönwall C, et al. Selection and characterization of affibody ligands binding to Alzheimer amyloid β peptides. J Biotechnol. 2007;128:162–183. doi: 10.1016/j.jbiotec.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 21.Binz HK, Amstutz P, Plückthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 22.Nord K, et al. Binding proteins selected from combinatorial libraries of an α-helical bacterial receptor domain. Nat Biotechnol. 1997;15:772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- 23.Tolmachev V, et al. Radionuclide therapy of HER2-positive microxenografts using a 177Lu-labeled HER2-specific Affibody molecule. Cancer Res. 2007;67:2773–2782. doi: 10.1158/0008-5472.CAN-06-1630. [DOI] [PubMed] [Google Scholar]
- 24.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-Å resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 25.Lendel C, Dogan J, Härd T. Structural basis for molecular recognition in an affibody:affibody complex. J Mol Biol. 2006;359:1293–1304. doi: 10.1016/j.jmb.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 26.Wahlberg E, et al. An affibody in complex with a target protein: structure and coupled folding. Proc Natl Acad Sci USA. 2003;100:3185–3190. doi: 10.1073/pnas.0436086100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 28.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer's β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith TJ, Stains CI, Meyer SC, Ghosh I. Inhibition of β-amyloid fibrillization by directed evolution of a β-sheet presenting miniature protein. J Am Chem Soc. 2006;128:14456–14457. doi: 10.1021/ja065557e. [DOI] [PubMed] [Google Scholar]
- 30.Lührs T, et al. 3D structure of Alzheimer's amyloid-β(1–42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olofsson A, Sauer-Eriksson AE, Öhman A. The solvent protection of Alzheimer amyloid-β-(1–42) fibrils as determined by solution NMR spectroscopy. J Biol Chem. 2006;281:477–483. doi: 10.1074/jbc.M508962200. [DOI] [PubMed] [Google Scholar]
- 32.Wetzel R, Shivaprasad S, Williams AD. Plasticity of amyloid fibrils. Biochemistry. 2007;46:1–10. doi: 10.1021/bi0620959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou L, et al. Solution NMR studies of the Aβ(1–40) and Aβ(1–42) peptides establish that the Met35 oxidation state affects the mechanism of amyloid formation. J Am Chem Soc. 2004;126:1992–2005. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- 34.Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 35.Habicht G, et al. Directed selection of a conformational antibody domain that prevents mature amyloid fibril formation by stabilizing Aβ protofibrils. Proc Natl Acad Sci USA. 2007;104:19232–19237. doi: 10.1073/pnas.0703793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lashuel HA, Lansbury PT., Jr Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q Rev Biophys. 2006;39:167–201. doi: 10.1017/S0033583506004422. [DOI] [PubMed] [Google Scholar]
- 37.Stuart AC, Borzilleri KA, Withka JM, Palmer AG. Compensating for variations in 1H–13C scalar coupling constants in isotope-filtered NMR experiments. J Am Chem Soc. 1999;121:5346–5347. [Google Scholar]
- 38.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 39.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 40.Kuszewski J, Gronenborn AM, Clore GM. Improving the packing and accuracy of NMR structures with a pseudopotential for the radius of gyration. J Am Chem Soc. 1999;121:2337–2338. [Google Scholar]
- 41.Kuszewski J, Gronenborn AM, Clore GM. Improvements and extensions in the conformational database potential for the refinement of NMR and X-ray structures of proteins and nucleic acids. J Magn Reson. 1997;125:171–177. doi: 10.1006/jmre.1997.1116. [DOI] [PubMed] [Google Scholar]