Abstract
The CCR5 co-receptor binds to the HIV-1 gp120 envelope glycoprotein and facilitates HIV-1 entry into cells. Its N terminus is tyrosine-sulfated, as are many antibodies that react with the co-receptor binding site on gp120. We applied nuclear magnetic resonance and crystallographic techniques to analyze the structure of the CCR5 N terminus and that of the tyrosine-sulfated antibody 412d in complex with gp120 and CD4. The conformations of tyrosine-sulfated regions of CCR5 (α-helix) and 412d (extended-loop) are surprisingly different. Nonetheless, a critical sulfotyrosine on CCR5 and on 412d induces similar structural rearrangements in gp120. These results now provide a framework for understanding HIV-1 interactions with the CCR5 N terminus during viral entry and define a conserved site on gp120, whose recognition of sulfotyrosine engenders posttranslational mimicry by the immune system.
Entry of human immunodeficiency virus type 1 (HIV-1) into host cells requires its gp120 envelope glycoprotein to bind to two cell-surface receptors, CD4 and a co-receptor, either CCR5 or CXCR4 [reviewed in (1, 2)]. CCR5 and CXCR4 are members of a family of chemokine receptors that are G protein–coupled receptors (3) characterized by seven transmembrane helices, an extracellular N terminus, which is variable in length, and three extracellular loops (ECLs) (Fig. 1A). The structure of the co-receptor has not been determined, but some insight has come from the crystal structures of other family members (4).
Fig. 1.
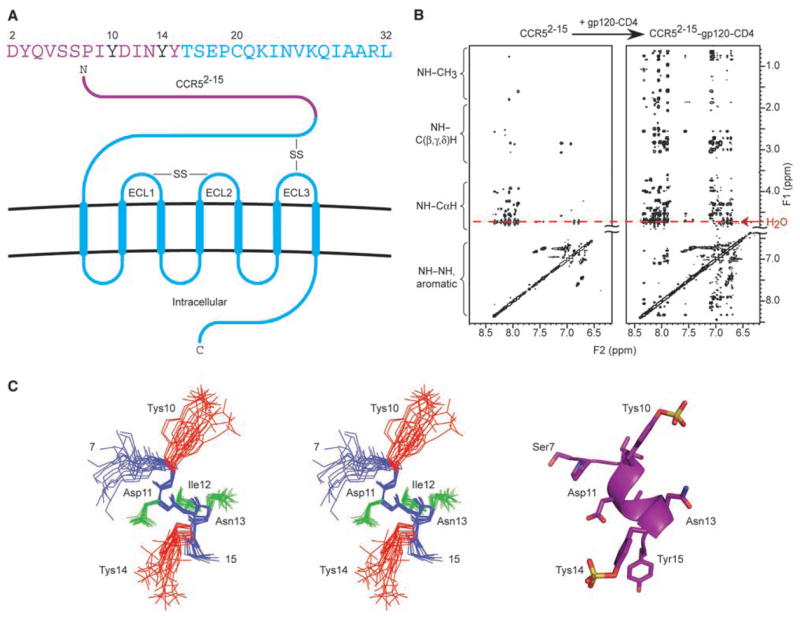
Structure of the tyrosine-sulfated N terminus of CCR5 in the gp120-bound conformation. (A) CCR5 sequence and schematic of its insertion in the cell membrane. Sequence letters in purple correspond to residues in CCR52-15, with sulfotyrosines (Tys) critical for interaction with HIV-1 highlighted in black. ECLs are labeled, and disulfide bridges (-SS-) depicted. (B) 2D NOESY spectra for CCR52-15 free in solution (left) and in the presence of gp120–CD4 (right). NMR samples (20 mM phosphate, 50 mM NaCl, pH 6.85) contained 800 μM CCR52-15 in the presence of 20 μM gp120-CD4 and were recorded at 500 MHz, 300 K, mixing time = 150 msec. Sequential NH(i)–CαH(i-1) NOEs were observed between every residue, thereby confirming sequential assignments, and predicted intraresidue NOEs were observed for all residues. No correlations beyond sequential NOEs were observed between residues 2 and 7, indicating that this region of CCR5 was extended or disordered. In contrast, NOEs from CαH(i) to NH(i + 1,2,3) and from NH(i) to NH(i + 1,2,3) were observed for residues 9 to 15 (fig. S1), indicating an ordered α-helical structure (33). (C) Structure of the ordered region of gp120-bound CCR52-15. Stereoview (left) of 25 lowest energy-simulated annealing structures superimposed by fitting to the backbone of residues 9 to 15. Structural statistics are provided in table S2. Backbone appears in blue, amide hydrogens (9 to 15) in blue, side chains (11 to 13) in green, and Tys 10 and Tys 14 in red. Ribbon diagram (right) of restrained minimized mean structure with side chains in stick representations.
Elements critical to interactions with HIV-1 are located in the co-receptor N terminus and around its second extracellular loop (ECL2) (5–8). The co-receptor N terminus interacts with a highly conserved 4-stranded bridging sheet in gp120, which assembles upon CD4 binding, whereas the ECL2 region of the co-receptor interacts with the tip of the immunodominant V3 loop in gp120. Considerable distance separates these two interactive regions, which suggests that they are independent (9–12).
The N-terminal interaction of co-receptor with HIV-1 requires an unusual posttranslational modification, O-sulfation of tyrosine (13). On CCR5, tyrosines at residues 3, 10, 14, and 15 may be O-sulfated, but sulfations at residues 10 and 14 are sufficient to facilitate interaction with HIV-1 (14). Interestingly, many CD4-induced antibodies that react with the bridging sheet region are also modified by O-sulfation (15). To define structurally the interaction of HIV-1 with the N terminus of CCR5 and to understand the molecular details of the mimicry of this interaction by CD4-induced antibodies, we used a combination of nuclear magnetic resonance (NMR) and x-ray crystallography to determine the structures of the N terminus of CCR5 and of a functionally sulfated antibody, 412d, in complex with HIV-1 gp120. Analysis of these structures, combined with molecular docking and saturation transfer difference NMR, identified a conserved site on gp120, which recognizes sulfotyrosine with high selectivity.
We used NMR techniques that exploit the transfer of information from bound to ligand-free states (16, 17) to analyze the interactions of a 14-residue peptide (CCR52-15), which consisted of residues 2 to 15 of CCR5 with sulfotyrosine (Tys) at positions 10 and 14 (Fig. 1) (18). We collected two-dimensional (2D) nuclear Over-hauser enhancement spectroscopy (NOESY) spectra of solutions containing CCR52-15 either free or in the presence of gp120, CD4, or a gp120-CD4 complex (peptide:protein ratio of 40:1). Whereas spectra containing free CCR52-15 or CCR52-15 with either gp120 or CD4 contained few cross peaks, CCR52-15 in the presence of the gp120-CD4 complex gave rise to high-quality spectra containing numerous NOEs (Fig. 1B and fig. S1). Complete 1H, 13C, and 15N assignments of CCR52-15 (table S1) were made on the basis of standard 2D homonuclear and heteronuclear NMR experiments that measure scalar and dipolar couplings.
The NOESY data of CCR52-15 in the presence of gp120-CD4 (Fig. 1B) were sufficient for calculating a high quality ensemble of NMR structures (Fig. 1C). Structure calculations were carried out on the ordered region comprising residues 7 to 15. A total of 70 distance restraints (corresponding to 35 intraresidue and 35 inter-residue NOEs), and 56 dihedral angle restraints were included in the final round of structure calculations, which gave rise to an ensemble of 40 structures with a backbone root-mean-square deviation (rmsd) of 0.46 Å and an rmsd of 1.39 Å for all atoms in the ordered region (residues 9 to 14) (table S2). Superpositions of the final ensemble defined a helical conformation for residues 9 to 15, which deviated from the ideal by a backbone rmsd of only 0.26 Å (Fig. 1D). Sulfotyrosines 10 and 14 extended from the same face of the helix, with sulfate moieties separated by ~10 Å and an ~90° rotation around the helix axis.
We were unable to obtain crystals of CCR52-15 in complex with HIV-1 gp120-CD4, and the size and glycosylation of the ternary complex hindered direct determination by NMR. We were, however, able to obtain ~3.5 Å diffraction from crystals of the antigen-binding fragment (Fab) of the 412d antibody, in complex with gp120 (core with V3, CCR5-dependent isolate YU2) and CD4. The 412d antibody is functionally tyrosine-sulfated, binds to a CD4-induced epitope that overlaps the site of co-receptor binding on HIV-1 gp120, and recognizes preferentially CCR5-dependent strains of HIV-1 gp120 (15). Moreover, the tyrosine-sulfated region of 412d can be substituted for the tyrosine-sulfated region of CCR5 to create a chimeric 412d/CCR5 receptor that supports HIV-1 entry (19).
We solved the 412d-gp120-CD4 structure by molecular replacement. Despite less than optimal resolution and completeness, initial unbiased maps showed clear definition of important antibody features (fig. S2). Structure refinement resulted in an Rcryst of 20% (Rfree 27%) (Fig. 2, table S3, and fig. S3). The overall mode of binding of 412d resembles that of 17b, which shares a heavy chain of similar genomic origin (fig. S4) (20). A hydrophobic interaction pins the second complementarity-determining region of the heavy chain (CDR H2) to a conserved hydrophobic surface on the bridging sheet of gp120, whereas the acidic CDR H3 binds a basic gp120 surface. Antibody 412d, however, interacts with a much larger overall surface area than either 17b or X5 (fig. S4). The increased 412d interaction surface is due primarily to an increase in buried surface associated with its CDR H3. Comparison of free (20) and bound structures of 412d shows that extensive ordering occurs in CDR H3 when bound to gp120 (fig. S5).
Fig. 2.
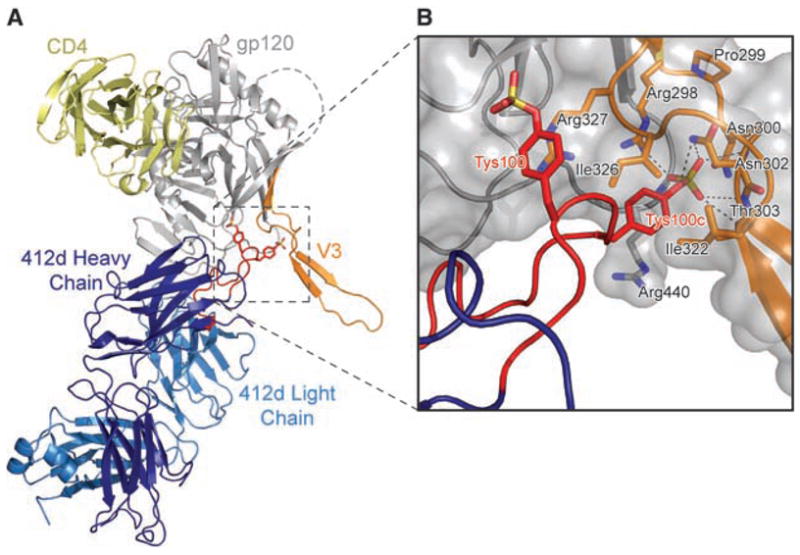
Structure of the tyrosine-sulfated antibody 412d in complex with HIV-1 gp120 and CD4. (A) Ribbon representation. CD4 is yellow, the heavy chain of Fab 412d is dark blue, the light chain is cyan, and gp120 is gray, except for the V3 loop, which is orange. The CDR H3 loop of 412d is red, with sulfotyrosines depicted in stick representation. (B) Close-up, with molecular surface of gp120 in gray and sulfotyrosines of 412d (red labels) and select residues of gp120 (black labels) in stick representation. Dotted lines represent coordinating hydrogen bonds between gp120 and the sulfate group of Tys100c412d. The sulfate of Tys 100c412d makes a full complement of ionic interactions: a salt bridge to Arg 298gp120 and hydrogen bonds to the side-chain nitrogen of Asn 302gp120, the side-chain hydroxyl of Thr 303gp120, and the main-chain amides of 302gp120, 303gp120, and 441gp120 (34).
The two sulfotyrosines in the CDR H3 region of 412d bind to gp120 in quite different ways (Fig. 2). The sulfotyrosine at residue 100 of 412d (Tys 100412d) [Kabat numbering (21)] is mostly exposed, with its aromatic ring making π-cation interactions with the guanidinium of Arg 327gp120 and its sulfate group making only peripheral electrostatic interactions. By contrast, the side-chain of Tys 100c412d is mostly buried, with Ile 322gp120 and Ile 326gp120 embracing one face of the tyrosine ring, while the aliphatic base of Arg 440gp120 supports the other. Together, the two sulfotyrosines account for about 20% of the total buried surface on 412d, with almost 100 Å2 derived from Tys 100c412d.
To facilitate interactions with the sulfotyrosines in 412d, the V3 stem is rearranged. The conserved Arg 298gp120 and Pro 299gp120 at the base of the V3 loop are mostly unchanged, but the subsequent Asn residues at 301gp120 and 302gp120 shift ~7 Å to form one wall of the Tys 100c412d sulfate-binding pocket. Residue 301gp120 is N-glycosylated, but the glycan faces solvent, and its presence should have little impact on the ability of the binding pocket to form. Meanwhile, in the returning strand (22), Ile 322gp120 shifts 10 Å to encase the 100c412d tyrosine ring. Overall, the incoming and outgoing strands of the V3 stem are brought closer together, so that a β-hairpin is formed that replaces the previously flexible V3 stem (23). Thus, whereas most of the gp120-CD4 complex remains unchanged, binding of sulfotyrosine at the bridging sheet-V3 interface results in formation of a more rigid V3.
By employing molecular docking and saturation transfer difference NMR, we sought to use the 412d-gp120-CD4 structure to ascertain how gp120 interacts with the N terminus of CCR5. We first tested whether docking [Autodock 3.0 (24)] of the CDR H3 loop of 412d to gp120 would recapitulate the 412d-gp120 crystal structure. Starting from random initial positions and orientations, multiple runs of the excised CDR H3 loop (residues 97 to 100f) produced an energetically favorable interaction (−16.04 kcal/mol), which closely resembled its location and contacts in the crystal structure (Cα rmsd between crystal and docked CDR H3 was 1.03 Å) (fig. S6). We next docked the NMR structure of the CCR5 N terminus to the crystal structure of gp120-CD4. Multiple runs produced a cluster of energetically favorable solutions (−17.60 kcal/mol for the optimal solution), which placed CCR52-15 at the bridging sheet-V3 interface (Fig. 3, A and B). The top 10% of the solutions (20 best solutions from 200 runs) had rmsds of 1.04 Å (Cα) and 2.24 Å (all atoms).
Fig. 3.
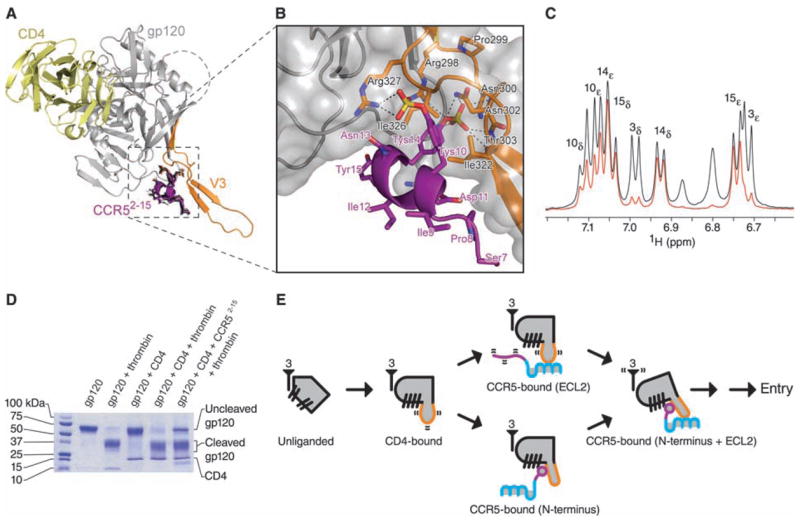
Interaction of the N terminus of CCR5 with HIV-1 gp120-CD4. (A) Molecular docking. The 20 lowest energy structures (black) from 200 docking runs of CCR52-15 are shown in stick representation. Despite initial random orientations, all favorable docking solutions had Tys 14 binding at the bridging sheet-V3 interface; none had Tys 10 at this cleft. Ribbon representations illustrate CD4 in yellow, gp120 in gray (with V3 in orange), and the lowest energy structure of CCR57-15 in purple. (B) Close-up, with molecular surface of gp120 in gray and select residues of gp120 (black labels) and CCR5 (purple labels) in stick representation. (C) Saturation transfer difference NMR spectrum of CCR52-15 in the presence of gp120–CD4 (red) overlaid on a control 1H spectrum (black). Experimental conditions were identical to those used for NOE experiments, except that the carrier was set at –1 and 50 parts per million for on- and off-resonance saturation, respectively. The intensities of the most strongly enhanced peaks (Tys 14 and Tyr 15) have been normalized to the corresponding signals in the control spectrum. Peak assignments made by 2D NMR (table S1) appear above their corresponding doublet signals. Tys 14 and Tyr 15 show strong saturation transfer difference effects, whereas Tys 10 shows a medium effect and Tyr 3 a very weak effect. These effects correlate directly with the buried surface area of each tyrosine ring in the docked structure. See fig. S9 for overlaid spectra employing 1 to 7 s saturation. (D) Effect of CCR52-15 on the proteolytic sensitivity of the V3. Electrophoresis on an 8 to 25% gradient SDS polyacrylamide gel shows the results of thrombin digestion on gp120 (core with V3; YU2 R5 strain of HIV-1) alone, or in the presence of sCD4 or sCD4 and CCR52-15 (35). (E) Structural intermediates of HIV-1 entry. At far left, a single monomer of unliganded gp120 (gray) is shown with separated β-hairpins. The threefold axis, from which gp41 interacts in the functional oligomer, is labeled with the number 3. In the CD4-bound state, the bridging sheet assembles, and the V3 (orange) is exposed and flexible. The next state involves either (upper pathway) the interaction of the CCR5-ECL2 region with the V3 tip or (lower pathway) the interaction of the CCR5 N terminus, which induces rigidification of the V3 stem. Engagement of CCR5 at both N terminus and ECL2 region triggers additional conformational changes leading to HIV-1 entry.
To validate the docked CCR5-gp120 structure, we performed saturation transfer difference NMR (17) on CCR52-15 in the presence of gp120-CD4. Control and difference spectra are shown in Fig. 3C. Contact surfaces of Tys and Tyr residues of CCR5 in the docked orientation correlated well with saturation transfer difference enhancements (Fig. 3C). We also observed good correlation between interacting residues in the docked gp120-CCR5 interface and gp120 and CCR5 substitutions (9, 25–27) that affect gp120-CCR5 binding (fig. S7).
The N terminus of CCR5 approaches from the same face of gp120 as CD4 but binds to an orthogonal surface at the intersection of the bridging sheet and the V3 loop (Fig. 3). The first CCR5 residues (Ser 7 and Pro 8) that are ordered in the NMR structure interact with the V3 stem. In the helix (residues 9 to 15), Tys 10 interacts with the gp120 core and forms a salt bridge with Arg 327gp120, Asp 11 forms an ionic interaction with Arg 440gp120, Tys 14 is completely sequestered in the crevice between V3 and the bridging sheet, and the aromatic ring of Tyr 15 packs against Ile 439gp120 on the bridging sheet.
The structural rearrangements required to form the Tys 14 binding pocket would be expected to rigidify the V3 stem. We tested V3-proteolytic susceptibility (Fig. 3D). CD4 enhances V3-proteolytic susceptibility to thrombin (28, 29), whereas the combination of CD4 and CCR52-15 reduced proteolytic susceptibility (Fig. 3D), consistent with CCR5-rigidification of V3.
Overall, the gp120 recognition surface for CCR52-15 is much more highly conserved for CCR5-dependent isolates compared with those that use CXCR4. Good electrostatic complementarity is found between the acidic CCR52-15 and gp120, where the negatively charged C-terminal helix dipole is oriented toward the basic bridging sheet (fig. S8). The docked structure provides an explanation for the observed lack of order at the N terminus of CCR52-15, where CCR5 appears to extend away from gp120. At the C terminus, Tyr 15 points toward the target cell membrane where, in five residues, a disulfide would normally be made between the N terminus (Cys 20) and the third extracellular loop (Cys 269).
Despite the highly divergent tyrosine-sulfated structures of 412d and CCR5, a single sulfotyrosine (residue 100c in 412d and residue 14 in CCR5) is recognized in a similar manner by gp120 (Fig. 4). We used mutagenesis to probe the degree of similarity in this recognition (fig. S10). The alteration of a single nitrogen in a contact residue (Asn302Asp) in the conserved binding pocket ablates recognition of both 412d and CCR5, whereas a similar substitution (Asn300Asp), just outside the binding pocket, had little effect (30). The observed convergence of recognition likely reflects the high selectivity of this site for sulfotyrosine (a 7 Å deep pocket, with hydrophobic walls and a cationic floor, which is unlikely to interact favorably with other nonmodified amino acids). Such selectivity and favorable energetics bode well for design of therapeutics targeted at this site, because the gp120 residues that line the sulfotyrosine binding pocket are highly conserved for co-receptor binding.
Fig. 4.
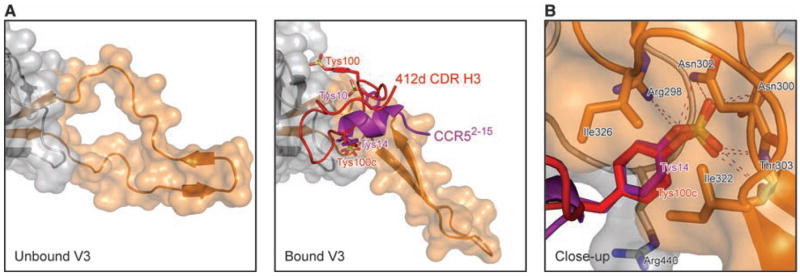
A conserved site for binding sulfotyrosine on HIV-1 gp120. (A) Alterations of the V3 base to accommodate binding of sulfotyrosine. The gp120 (gray) region around the V3 loop (orange) is illustrated in ribbon diagram, with an overlying semitrans-parent surface for unbound (left panel) and bound (right panel) conformations. Binding of the CCR5 N terminus (purple) or the 412d CDR H3 (red), each with two sulfotyrosines (stick representation, with red and purple labels), alters the V3 base, forming a sulfotyrosine binding pocket and a rigid β-hairpin. (B) Close-up of the conserved sulfotyrosine binding pocket. The orientation shown is similar to that in Figs. 2B and 3B [~90° from (A) about a diagonal axis, as defined by the long axis of the V3 from (A)]. Tys 14CCR5 is shown in purple, with Tys 100c412d in red. Select residues of gp120 are shown in stick representation and labeled in black. Hydrogen bonds coordinating the buried sulfate groups in each are depicted with dotted lines.
The structure of the CCR5 N terminus with gp120-CD4 provides a further snapshot of the HIV-1 entry pathway (Fig. 3E). Before binding CD4, the bridging sheet is not formed and the V3 loop is occluded. Binding of CD4 induces bridging sheet assembly and V3 exposure. At this stage, the V3 is flexible and poised close to the target cell membrane. Subsequent interactions with CCR5 are still being elucidated. We show structural details for one: engagement by gp120 of the CCR5 N terminus, which requires formation of a conserved pocket for sulfotyrosine binding and converts the flexible V3 stem into a rigid β-hairpin. It will be interesting to integrate the order and timing of the rearrangements revealed here into the HIV-1 entry mechanism.
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/317/5846/1930/DC1
Materials and Methods
Figs. S1 to S10
Tables S1 to S3
References
References and Notes
- 1.Wyatt R, Sodroski J. Science. 1998;280:1884. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 2.Berger EA, Murphy PM, Farber JM. Annu Rev Immunol. 1999;17:657. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 3.Bockaert J, Pin JP. EMBO J. 1999;18:1723. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palczewski K, et al. Science. 2000;289:739. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 5.Liu R, et al. Cell. 1996;86:367. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 6.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. J Exp Med. 1997;71:4744. doi: 10.1128/jvi.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doranz BJ, et al. J Virol. 1997;71:6305. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dragic T. J Gen Virol. 2001;82:1807. doi: 10.1099/0022-1317-82-8-1807. [DOI] [PubMed] [Google Scholar]
- 9.Rizzuto CD, et al. Science. 1998;280:1949. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 10.Hartley O, Klasse PJ, Sattentau QJ, Moore JP. AIDS Res Hum Retroviruses. 2005;21:171. doi: 10.1089/aid.2005.21.171. [DOI] [PubMed] [Google Scholar]
- 11.Huang C, et al. Science. 2005;310:1025. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cormier EG, Dragic T. J Virol. 2002;76:8953. doi: 10.1128/JVI.76.17.8953-8957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farzan M, et al. Cell. 1999;96:667. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 14.Cormier EG, et al. Proc Natl Acad Sci USA. 2000;97:5762. doi: 10.1073/pnas.97.11.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choe H, et al. Cell. 2003;114:161. doi: 10.1016/s0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- 16.Feeney J, Birdsall B, Roberts GCK, Burgen ASV. Biochemistry. 1983;22:628. doi: 10.1021/bi00272a016. [DOI] [PubMed] [Google Scholar]
- 17.Mayer M, Meyer B. J Am Chem Soc. 2001;123:6108. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 18.Several peptides were tested in initial NMR titration experiments, wherein line broadening of peptide signals was monitored upon addition of CD4. Although a peptide corresponding to CCR5 residues 2 to 18 (25) showed tighter binding than 2 to 15, the shorter peptide exhibited superior quality NOESY spectra. See also (31).
- 19.Xiang SH, et al. J Virol. 2005;79:6068. doi: 10.1128/JVI.79.10.6068-6077.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang C, et al. Proc Natl Acad Sci USA. 2004;101:2706. [Google Scholar]
- 21.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of Proteins of Immunological Interest. 5. U.S. Department of Health and Human Services, National Institutes of Health; Bethesda, MD: 1991. [Google Scholar]
- 22.Stem movements are somewhat imprecise because the returning V3 stem exhibits considerable disorder before interaction with the N terminus of CCR5 (11).
- 23.The nascent β-hairpin in the V3 stem extends ~15 Å; further extension is interrupted by a lattice contact, which occurs with the outgoing portion of the V3 stem. In the absence of this lattice contact, the stem β-hairpin may extend farther, perhaps joining with the V3 tip to zip up much of the V3 loop.
- 24.Morris GM, et al. J Comput Chem. 1998;19:1639. [Google Scholar]
- 25.Cormier EG, Tran DNH, Yukhayeva L, Olson WC, Dragic T. J Virol. 2001;75:5541. doi: 10.1128/JVI.75.12.5541-5549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabut GEE, Konner JA, Kajumo F, Moore JP, Dragic T. J Virol. 1998;72:3464. doi: 10.1128/jvi.72.4.3464-3468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizzuto C, Sodroski J. AIDS Res Hum Retroviruses. 2000;16:741. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- 28.Clements GJ, et al. AIDS. 1991;7:3. doi: 10.1089/aid.1991.7.3. [DOI] [PubMed] [Google Scholar]
- 29.Sattentau QJ, Moore JP. J Exp Med. 1991;174:407. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The functionally sulfated CD4-induced antibody E51, however, was not affected by either Asn300Asp or Asn302Asp substitutions (fig. S10), indicating variation in CD4-induced antibody recognition, perhaps reflective of the ability of E51 to recognize both CCR5- and CXCR4-dependent isolates of HIV-1, unlike the more specific recognition of 412d.
- 31.Materials and methods are available as supporting material on Science Online.
- 32.Quiocho FA. Kidney Int. 1996;49:943. doi: 10.1038/ki.1996.132. [DOI] [PubMed] [Google Scholar]
- 33.Wuthrich K. NMR of Proteins and Nucleic Acids. Wiley, NJ: 1986. [Google Scholar]
- 34.Data incompleteness and resolution (~3.5 Å) made delineation of hydrogen bonds problematic. The current designation is consistent with the substitutional mutagenesis experiments (fig. S10); alternatively, discrimination between sulfotyrosine and phosphotyrosine suggests complete sulfate coordination by hydrogen bond acceptors (32), with the side-chain nitrogen of Asn 302 donating a hydrogen bond instead of the hydroxyl of Thr 303.
- 35.The 24-hour time point that is shown clearly depicts the protective effect of CCR52-15 with sCD4. CCR52-15 without sCD4 does not show this effect, and shorter incubations show that sCD4 enhances V3 cleavage.
- 36.We thank L. Chen for assistance with proteolysis of V3; S. Buchanan, D. Dimitrov, J. Hoxie, and L. Shapiro for discussions and comments on the manuscript; D. Hurt and J. Skinner for assistance with statistics; J. Stuckey for assistance with figures; and the NIH AIDS Research and Reference Reagent Program for CD4. Support for this work was provided by the Intramural Research Program (National Institute of Allergy and Infectious Diseases and National Institute of Diabetes and Digestive and Kidney Diseases) and Intramural AIDS Targeted Antiviral Program (C.A.B., P.D.K., and R.W.), by a grant from the Bill and Melinda Gates Foundation Grand Challenges in Global Heath Initiative (J.R., P.D.K., and R.W.), by the International AIDS Vaccine Initiative (J.S., I.A.W.), and by grants from NIH (J.R., J.S., and I.A.W.). This study used the high-performance computational capabilities of the Biowulf Linux cluster at NIH (http://biowulf.nih.gov). Use of insertion device 22 (Southeast Regional Collaborative Access Team) at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-109-Eng-38. Coordinates of the CCR52-15 NMR structure (2RLL), as well as coordinates and structure factors for the 412d-gp120-CD4 crystal structure (2QAD), have been deposited with the Protein DataBank. Coordinates of the docked CCR5 N terminus with gp120 and CD4 are available from the authors.


