Abstract
Millions of people worldwide suffer from nutritional imbalances of essential metals like zinc. These same metals, along with pollutants like cadmium and lead, contaminate soils at many sites around the world. In addition to posing a threat to human health, these metals can poison plants, livestock, and wildlife. Deciphering how metals are absorbed, transported, and incorporated as protein cofactors may help solve both of these problems. For example, edible plants could be engineered to serve as better dietary sources of metal nutrients, and other plant species could be tailored to remove metal ions from contaminated soils. We report here the cloning of the first zinc transporter genes from plants, the ZIP1, ZIP2, and ZIP3 genes of Arabidopsis thaliana. Expression in yeast of these closely related genes confers zinc uptake activities. In the plant, ZIP1 and ZIP3 are expressed in roots in response to zinc deficiency, suggesting that they transport zinc from the soil into the plant. Although expression of ZIP2 has not been detected, a fourth related Arabidopsis gene identified by genome sequencing, ZIP4, is induced in both shoots and roots of zinc-limited plants. Thus, ZIP4 may transport zinc intracellularly or between plant tissues. These ZIP proteins define a family of metal ion transporters that are found in plants, protozoa, fungi, invertebrates, and vertebrates, making it now possible to address questions of metal ion accumulation and homeostasis in diverse organisms.
Zn is an essential catalytic component of over 300 enzymes, including alkaline phosphatase, alcohol dehydrogenase, Cu-Zn superoxide dismutase, and carbonic anhydrase. Zn also plays a critical structural role in many proteins. For example, several motifs found in transcriptional regulatory proteins are stabilized by Zn, including the Zn finger, Zn cluster, and RING finger domains. Proteins containing these domains are very common; the yeast genome sequencing project has determined that almost 2% of all yeast gene products contain these types of Zn binding domains (1, 2). Inside cells, Zn is neither oxidized nor reduced; thus, the essential roles of Zn in cells is based largely on its behavior as a divalent cation that has a strong tendency to form stable tetrahedral complexes (for a review see ref. 3).
Despite the importance of Zn as an essential micronutrient for plant growth, relatively few studies have examined the mechanisms and regulation of Zn absorption by roots. Zn is taken up from the soil solution as a divalent cation (4). Currently, there is little agreement as to whether uptake is via ion channels or via a divalent cation carrier protein and whether there is a link between uptake and metabolic energy transduction. Studies of Zn uptake in plants mainly have been focused on hyperaccumulators, i.e., plants that can grow in soils containing high levels of Zn and accumulate high concentrations of Zn in their shoots. Certain populations of Thlaspi caerulescens can tolerate up to 40,000 μg of Zn g−1 tissue in their shoots whereas the normal Zn concentration for most plants is between 20 and 100 μg of Zng−1 tissue (4). Radiotracer studies with T. caerulescens and a closely related nonhyperaccumulating species, T. arvense, suggested that Zn uptake is controlled by regulating the number of active transporters in the membrane (5). The Vmax for the uptake of Zn was 4.5-fold greater for T. caerulescens than for the nonhyperaccumulator T. arvense whereas their Km values were not significantly different. Once in the root, Zn is believed to be transported into the xylem, taken up by leaf cells, and then stored in the vacuoles of leaf cells thus preventing the buildup of toxic levels in the cytoplasm. These observations indicate that there are also Zn transporters that transport the metal between cells and into subcellular compartments within the plant. We describe here the successful isolation and characterization of Zn transporter genes from Arabidopsis thaliana. To our knowledge, these are the first zinc transporter genes to be identified in plants.
MATERIALS AND METHODS
Yeast Growth Conditions and Library Screening.
Yeast cells were grown in 1% yeast extract, 2% peptone supplemented with 2% glucose (yeast extract/peptone/dextrose), and in synthetic-defined medium (6.7 g/liter yeast nitrogen base without amino acids) supplemented with 2% glucose, 0.1% casamino acids, 0.01% adenine, and 0.01% tryptophan. Media for plate cultures were prepared with 1.5% agar. Strain ZHY3 (MATα zrt1∷LEU2 zrt2∷HIS3 ade6 can1 his3 leu2 trp1 ura3) (6) was transformed (7) with a plasmid library containing A. thaliana cDNAs inserted under the control of the yeast phosphoglycerate kinase promoter in pFL61 (8). The poly(A) RNA used to construct this library was obtained from whole young seedlings (stage two leaves). Transformants were inoculated directly onto 39 synthetic-defined agar plates at a density of ≈7,000 transformants/plate. Because of the mutations in the ZRT1 and ZRT2 genes, synthetic-defined medium is zinc-limiting for ZHY3. Plasmid dependence of the zinc uptake effects was confirmed by selecting for transformants that had lost the plasmids by using 5-fluoroorotic acid (9).
DNA Manipulations and Sequence Analysis.
DNA was prepared from each of the three yeast transformants, and the plasmids were transformed into Escherichia coli TOP10F′ cells (Stratagene). Plasmid DNA was prepared and digested with NotI, and the insert fragments were cloned into NotI-digested Bluescript SK+ (Stratagene). Nucleotide sequences were obtained on an Applied Biosystems model 373A Automated Sequencer (Dartmouth College Molecular Biology Core Facility, Hanover, NH) by using the Ready-reaction Dye-Terminator kit with AmpliTaq FS (Applied Biosystems). Computer database comparisons were performed by using blast (10); potential transmembrane domains were identified by using top-predii (11); and multiple sequence alignment was performed by using pileup and pretty (GCG). A cDNA version of the ZIP4 coding sequence, constructed by using PCR, was cloned into pFL61 and used in yeast complementation and uptake assays.
Zinc Uptake Assays.
Zinc uptake assays were performed as described for iron uptake (12) except that 65ZnCl2 (Amersham) and low zinc medium–EDTA (13) were substituted for 59FeCl3 and low iron medium-EDTA (14), respectively. Because of their different pH optima for uptake activity, the ZIP1 and ZIP3 activities were assayed at pH 4.7 and ZIP2 activity was assayed at pH 6.0. Michaelis–Menten kinetic values were determined by using kinetasyst software (IntelliKinetics, State College, PA). Stock solutions of the chloride salts of cadmium, cobalt, copper, manganese, and nickel were prepared in distilled water at a concentration of 100 mM. A ZnCl2 stock solution was prepared at 100 mM in 0.02 N HCl, and the FeCl3 stock solution was prepared at 50 mM in 0.1 N HCl.
Plant Growth Conditions.
Seeds of A. thaliana (Columbia gl-1) were surface-sterilized and plated on Gamborg’s B5 medium (Sigma) (pH 5.8) containing 2% sucrose, 1 mM Mes, and 0.7% agar. Plates were kept at 4°C in the dark for 4 days before being incubated under constant light at 21°C for 10 days. Seedlings then were transferred to either zinc-sufficient or zinc-deficient hydroponic medium, formulated by using the geochem-pc program (15). This medium (pH 6.0) contains 2 mM Ca(NO3)2, 1 mM KNO3, 500 μM MgSO4, 80 μM KH2PO4, 1 mM Mes, 10 μM H3BO3, and 0.1 μM Na2MoO4; the micronutrients CuSO4, Fe2(SO4)3, MnCl2, and NiCl2 were added in the presence of 100 μM excess EDTA to give final concentrations of 10 μM CuEDTA, 50 μM FeEDTA, 0.6 μM MnEDTA, and 0.1 μM NiEDTA. For the zinc-sufficient medium, ZnEDTA was present at a final concentration of 10 μM. The seedlings then were grown hydroponically for an additional 10 days. The zinc status of the plants was confirmed by using inductively coupled argon plasma spectrometry.
Nucleic Acid Analysis.
Poly(A) RNA was extracted from root and shoot fractions of plants grown either in zinc-sufficient or zinc-deficient media. Samples (1 μg) of RNA were denatured, electrophoresed on a 0.8% agarose/6.2% formaldehyde gel, and then transferred to a nylon membrane (Biotrans, ICN). RNA was bound to the membrane by UV crosslinking (Stratalinker, Stratagene). The membrane was prehybridized, hybridized, and washed as described by Pilgrim and McClung (16) with the exception that washes were performed at 42°C. Membranes were stripped for reprobing with a boiling solution of 0.1% SDS. DNA fragments used as hybridization probes were radiolabeled by random priming (17).
RESULTS AND DISCUSSION
To identify potential zinc transporters from plants, an A. thaliana cDNA expression library was screened for clones that restored zinc-limited growth when expressed in a Saccharomyces cerevisiae zrt1 zrt2 mutant. This mutant lacks both high and low affinity zinc uptake systems and is extremely sensitive to zinc limitation because of its reliance on less efficient uptake pathways (6). From the ≈270,000 transformants obtained, three independent transformants were isolated in which the zrt1 zrt2 phenotype was suppressed. Restriction mapping and Southern blot hybridization analysis of the plasmids obtained from these transformants demonstrated that cDNA clones of three different genes had been isolated. The entire cDNA insert of each plasmid was sequenced. Comparison of these sequences with the current databases revealed that these cDNAs each encoded proteins similar to the products encoded by the ZRT1 and ZRT2 genes of S. cerevisiae and the IRT1 gene of A. thaliana (18) (Fig. 1). For this reason, the genes were named ZIP1, ZIP2, and ZIP3 (for ZRT, IRT-like Protein). ZRT1 and ZRT2 encode the high and low affinity zinc transporters in yeast, respectively (6, 13), and IRT1 encodes a probable iron transporter (18). A fourth ZIP gene, ZIP4, was identified during sequencing of the Arabidopsis genome (GenBank accession no. U95973); given the similarity of its deduced protein product to those encoded by the other ZIP genes, we included ZIP4 in subsequent analyses.
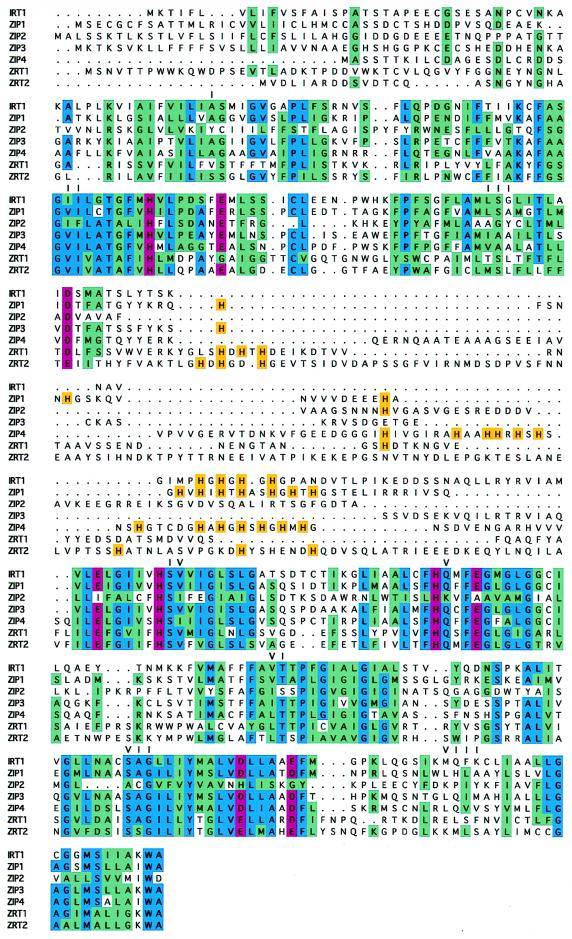
Figure 1.
Alignment of the amino acid sequences of seven full length Arabidopsis and Saccharomyces ZIP proteins (IRT1, ZIP1, ZIP2, ZIP3, ZIP4, ZRT1, and ZRT2). The eight transmembrane domains predicted for each these proteins are numbered I–VIII. Residues highlighted in blue are identical, and residues highlighted in green represent conservative substitutions. The histidines found in the variable region between transmembrane domains III and IV are highlighted in gold. Residues highlighted in pink are conserved histidines or acidic residues found within predicted transmembrane helices.
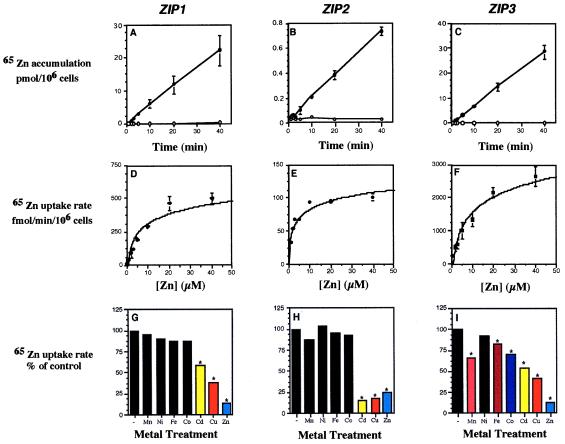
The ability of ZIP1, ZIP2, and ZIP3 to suppress the growth defect of a zrt1 zrt2 yeast mutant suggested that these genes may encode zinc transporters. To test this hypothesis, we examined 65Zn accumulation in a zrt1 zrt2 strain expressing each of these genes. At 0°C, no zinc accumulation was detected in ZIP1- or ZIP3-expressing strains (Fig. 2 A and C) or in an untransformed control strain (data not shown). At 30°C, no zinc accumulation was detected in the untransformed control strain (ref. 6 and data not shown) whereas a high rate of accumulation was observed in the ZIP1- and ZIP3-expressing strains (≈270 and 710 fmol/min/106 cells, respectively). The rate of ZIP1- or ZIP3-dependent zinc accumulation was not altered greatly by variations in pH of the assay solution between 4.0 and 6.0 (data not shown). In contrast, ZIP2-dependent zinc accumulation was highly pH-dependent; no uptake activity was detectable in the ZIP2-expressing strain when assayed at pH values below 5.0 (data not shown), yet at pH 6.0, temperature-dependent zinc accumulation was observed (≈19 fmol/min/106 cells) (Fig. 2B).
Figure 2.
Biochemical properties of zinc uptake in yeast expressing the ZIP1, ZIP2, or ZIP3 genes. zrt1 zrt2 (ZHY3) transformants expressing ZIP1, ZIP2, or ZIP3 were grown to exponential phase (≈5 × 106 cells/ml) in synthetic-defined medium and assayed for zinc uptake with 65Zn. Because of their different pH optima for uptake activity, the ZIP1 and ZIP3 activities were assayed at pH 4.7, and ZIP2 activity was assayed at pH 6.0. (A–C) Time and temperature dependence of zinc accumulation assayed with 10 μM (ZIP1, ZIP3) or 1 μM 65Zn (ZIP2) at 0°C (open symbols) and at 30°C (closed symbols). (D–F) Concentration dependence of zinc uptake was determined by measuring zinc accumulation for 5 min over a range of substrate concentrations. Each point in A–F represents the mean of two experiments each performed in duplicate, and the error bars represent ±1 SD. (G–I) Inhibition of ZIP-dependent uptake in yeast by other metals. Cells were assayed for zinc uptake rate with 10 μM (ZIP1, ZIP3) or 1 μM (ZIP2) 65Zn in the absence (−) or presence of 10-fold excess of the chloride salts of the indicated metal ions. Each value represents the mean of four replicates. The asterisks indicate significant differences from control values (P < 0.05) as determined by one-way ANOVA followed by Scheffe’s test.
A cDNA version of the ZIP4 coding sequence, constructed by using PCR, was cloned into pFL61 and was used in yeast complementation and uptake assays. No uptake could be detected in yeast cells expressing the ZIP4 gene nor could ZIP4 complement the growth defect of the zrt1 zrt2 mutant on zinc-limited medium. This lack of uptake may be due to subcellular rather than plasma membrane localization of ZIP4 in yeast.
When assayed over a range of zinc concentrations, uptake activity dependent on expression of the ZIP1, ZIP2, and ZIP3 genes was concentration-dependent and saturable (Fig. 2 D–F). The Michaelis–Menten kinetic values derived from these data are described in Table 1. Of interest, the apparent Km values of these transporters are similar to the levels of zinc commonly found in plant rhizospheres (19).
Table 1.
Michaelis–Menten kinetic constants of zinc uptake in yeast
| Gene | Apparent Km, μM | Vmax, fmol/min/106 cells |
|---|---|---|
| ZIP1 | 13 ± 2 | 693 ± 43 |
| ZIP2 | 2 ± 0.3 | 107 ± 4 |
| ZIP3 | 14 ± 2 | 3528 ± 207 |
To assess whether metals in addition to zinc are substrates for ZIP1, ZIP2, or ZIP3, we tested other metal ions for their ability to inhibit zinc uptake mediated by these proteins. Zinc uptake by ZIP1 was not inhibited by a 10-fold excess of Mn, Ni, Fe, or Co (Fig. 2G). Zinc was the most potent competitor, demonstrating that ZIP1 prefers zinc as its substrate over these other metal ions. Cd and Cu also inhibited zinc uptake but to a lesser extent. Although the mechanism of these inhibitory effects is not yet known, these results suggest that Cd and Cu also may be substrates for ZIP1. Zinc uptake mediated by ZIP3 was inhibited by Mn, Fe, Co, Cd, and Cu (Fig. 2I). Again, zinc had the greatest effect, indicating that ZIP3 also prefers zinc. Cd and Cu inhibited zinc uptake mediated by ZIP2 to as great a degree as zinc (Fig. 2H) suggesting that ZIP2 also may have a high affinity for Cd and/or Cu. In summary, these three transporters show unique sensitivities to other metal ions that may reflect differences in their substrate specificities. We have found that expression of ZIP1, ZIP2, ZIP3, and ZIP4 in a fet3 fet4 mutant [DEY1453 (20)] failed to alter the iron-limited growth defect of this strain or alter its iron uptake activity. Thus, the ZIP proteins cannot transport iron.
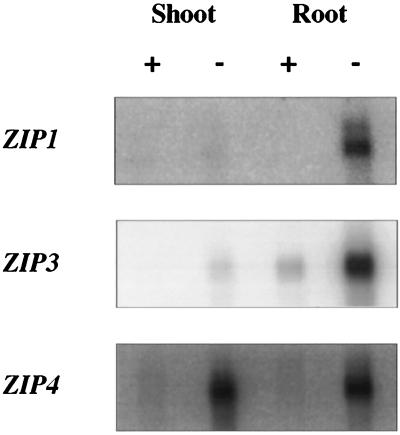
Why does Arabidopsis have multiple zinc transporters? Zinc, like other metal ions, is transported from the soil into the root and then must cross both cellular and organellar membranes as it is distributed throughout the plant. Specific zinc transporters may play different roles in this process. Northern blot analysis demonstrated that ZIP1 and ZIP3 are zinc-responsive. These genes are expressed predominantly in the roots of zinc-deficient plants; little or no mRNA was detected in the roots of zinc-sufficient plants or in the shoots of zinc-sufficient or zinc-deficient plants (Fig. 3). Despite repeated attempts, ZIP2 mRNA could not be detected in plants grown under either condition. ZIP4 also responds to zinc deficiency, but, unlike ZIP1 and ZIP3, this gene is induced in both the shoots and roots of zinc-deficient plants. This result is especially interesting given the predicted targeting of ZIP4 to chloroplasts (see below). The results of our Northern blot analyses are consistent with a role for ZIP1 and ZIP3 in the uptake of zinc from the rhizosphere and a role for ZIP4 in the transport of zinc in plastids. It is not surprising that ZIP gene expression would be regulated; zinc is toxic when present in excess. We also examined ZIP gene expression in iron-sufficient and iron-deficient plants and saw no response of any of the ZIP genes to the same conditions of iron deficiency that induce IRT1 expression (data not shown). Thus, ZIP1, ZIP3, and ZIP4 respond specifically to a lack of zinc.
Figure 3.
Regulation of ZIP1, ZIP3, and ZIP4 mRNA levels by zinc availability. Gene probes were hybridized to a Northern blot containing 1 μg of poly(A) mRNA prepared from the roots or shoots of hydroponically grown plants. The same blot was used sequentially (after stripping) for hybridization to each of the gene probes.
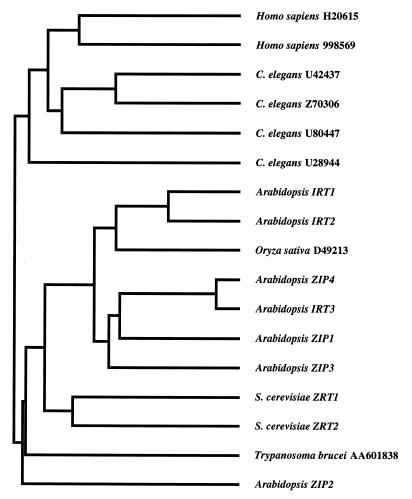
The predicted products of the ZIP1, ZIP2, ZIP3, and ZIP4 genes are 355, 353, 339, and 374 aa in length, respectively. The ZIP proteins contain eight potential transmembrane domains, numbered I–VIII in Fig. 1. ZIP1, ZIP2, and ZIP3 each have a potential signal sequence and are predicted to be plasma membrane proteins; ZIP4 contains a potential chloroplast targeting sequence [PSORT, (21)]. To date, we have identified a total of 18 ZIP family members, including nine plant genes (including eight from Arabidopsis alone), two yeast genes, one gene in the protozoan Trypanosoma brucei variety rhodesiense, four genes in the nematode C. elegans, and two in humans. Family members range in length from 309 to 476 aa; this range is largely due to variation in the number of residues between transmembrane domains III and IV, a domain we have designated the “variable region.” The amino acid sequences of all known ZIP family members were aligned, and a dendrogram describing their sequence similarities was generated (Fig. 4). The family can be divided into two subfamilies, one containing sequences from animals and the other containing the plant, fungal, and protozoan members. Within the plant/fungal/protozoan subfamily, the two zinc transporters from yeast form one group and all but one of the plant sequences form a second group.
Figure 4.
Dendrogram showing amino acid sequence relationships among the ZIP family members. GenBank accession numbers for sequences designated by name are as follows: IRT1, U27590; IRT2, T04324; ZIP4, U95973; IRT3, M35868; ZRT1, P32804; and ZRT2, X91258. The amino acid sequence of human 998569 is only available in the Entrez database. The Oryza sativa D49213, IRT2, IRT3, and Homo sapiens H20615 clones have not been sequenced completely.
A potential metal binding motif, containing multiple histidine residues, is found in the variable regions of almost all of the fully sequenced members of this family including IRT1, IRT2, ZIP1, ZIP4, ZRT1, and ZRT2. In contrast, ZIP2 and ZIP3 contain only a single histidine in this region. Studies of ZRT1 suggest that the histidine-rich motif is located on the intracellular face of the plasma membrane and also indicate that it is essential for transporter function (D.E. and M. Broderius, unpublished work). Three of the transmembrane domains (II, IV, and V) contain a histidine residue that is fully conserved among all family members. These histidines are predicted to lie on the polar face of amphipathic helices, suggesting a possible role for these residues in substrate transport through the membrane. Furthermore, conserved acidic amino acids present in the transmembrane domains of several of the ZIP proteins also may be important for substrate movement.
The ZIP genes of Arabidopsis represent four members of a rapidly growing family of eukaryotic proteins. To date, six of the family members have been implicated in metal ion transport. These results demonstrate that the ZIP transporter family plays roles in metal ion metabolism in a diverse array of eukaryotic organisms. The ZIP family is structurally distinct from other metal ion transporters such as the CDF family (22), which includes the recently identified mammalian zinc effluxers P-type ATPases (23) that are involved in the transport of a variety of cations and the Nramp proteins recently implicated in divalent cation transport (24–26). The ZIP genes then offer a good starting point not only for understanding how metals cross membranes but also for engineering plant-based solutions to nutrient deficiencies and to environmental remediation.
Acknowledgments
We thank Ann Thering, Elizabeth Rogers, and Rob McClung for critical reading of the manuscript, Raad Gitan for analyzing ZIP4 in yeast, and Michael Rutzke for performing the inductively coupled argon plasma analyses. This work was supported by grants from the National Science Foundation to M.L.G. (IBN-9318093) and to D.E. (MCB-9405200).
Footnotes
References
- 1.Böhm S, Frishman D, Mewes H W. Nucleic Acids Res. 1997;25:2464–2469. doi: 10.1093/nar/25.12.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schjerling P, Holmberg S. Nucleic Acids Res. 1996;24:4599–4607. doi: 10.1093/nar/24.23.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg J M, Shi Y. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 4.Marschner H. Mineral Nutrition of Higher Plants. 2nd Ed. Boston: Press Academic; 1995. [Google Scholar]
- 5.Lasat M M, Baker A J M, Kochian L V. Plant Physiol. 1996;112:1715–1722. doi: 10.1104/pp.112.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao H, Eide D. J Biol Chem. 1996;271:23203–23210. doi: 10.1074/jbc.271.38.23203. [DOI] [PubMed] [Google Scholar]
- 7.Schiestl R H, Gietz R D. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 8.Minet M, Dufour M E, Lacroute F. Plant J. 1992;2:417–422. doi: 10.1111/j.1365-313x.1992.00417.x. [DOI] [PubMed] [Google Scholar]
- 9.Boeke J D, Trueheart J, Natsoulis G, Fink G R. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 10.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Claros M G, Von Heijne G. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 12.Eide D, Davis-Kaplan S, Jordan I, Sipe D, Kaplan J. J Biol Chem. 1992;267:20774–20781. [PubMed] [Google Scholar]
- 13.Zhao H, Eide D. Proc Natl Acad Sci USA. 1996;93:2454–2458. doi: 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eide D, Guarente L. J Gen Microbiol. 1992;138:347–354. doi: 10.1099/00221287-138-2-347. [DOI] [PubMed] [Google Scholar]
- 15.Parker D R, Chaney R L, Norvell W A. In: Chemical Equilibrium and Reaction Models. Loeppert R H, Schwab A P, Goldberg S, editors. Madison, WI: Soil Society of America; 1995. pp. 163–199. [Google Scholar]
- 16.Pilgrim M L, McClung C R. Plant Physiol. 1993;103:553–564. doi: 10.1104/pp.103.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feinberg A P, Vogelstein B. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 18.Eide D, Broderius M, Fett J, Guerinot M L. Proc Natl Acad Sci USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welch R M. Crit Rev Plant Sci. 1995;14:49–82. [Google Scholar]
- 20.Dix D R, Bridgham J T, Broderius M A, Byersdorfer C A, Eide D J. J Biol Chem. 1994;269:26092–26099. [PubMed] [Google Scholar]
- 21.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulsen L T, Saier M H. J Membrane Biol. 1997;156:99–103. doi: 10.1007/s002329900192. [DOI] [PubMed] [Google Scholar]
- 23.Axelsen K B, Palmgren M G. J Mol Evol. 1998;46:84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- 24.Belouchi A, Kwan T, Gros P. Plant Mol Biol. 1997;33:1085–1092. doi: 10.1023/a:1005723304911. [DOI] [PubMed] [Google Scholar]
- 25.Fleming M D, Trenor C C, Su M A, Foernzler D, Beier D R, Dietrich W F, Andrews N C. Nat Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 26.Gunshin H, Mackenzie B, Berger U V, Gunshin Y, Romero M F, Boron W F, Nussberger S, Gollan J L, Hediger M A. Nature (London) 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]