Abstract
Shugoshin 1 (Sgo1) functions as a protector of centromeric cohesion of sister chromatids in higher eukaryotes. Here, we provide evidence for a new role for Sgo1 in centriole cohesion. Sgo1 depletion via RNA interference induces the formation of multiple centrosome-like structures in mitotic cells that result from the separation of paired centrioles. Sgo1+/− mitotic murine embryonic fibroblasts display split centrosomes. Localization study of two major endogenous splice variants of Sgo1 indicates that the smaller variant, sSgo1, is found at the centrosome in interphase and at spindle poles in mitosis. sSgo1 interacts with Plk1 and its spindle pole localization is Plk1-dependent. Centriole splitting induced by Sgo1 depletion or expressing a dominant negative mutant is suppressed by ectopic expression of sSgo1 or by Plk1 knockdown. Our studies strongly suggest that sSgo1 plays an essential role in protecting centriole cohesion, which is partly regulated by Plk1.
Keywords: Sgo1, Shugoshin, Plk1, cohesin, centriole cohesion centrosome, spindle poles
INTRODUCTION
Accurate chromosome segregation is critical for maintaining genomic stability during cell division. Spindle poles and kinetochores, two major microtubule organization centers (Luders and Stearns, 2007), play an essential role in coordinating chromosome segregation during mitosis. The centrosome duplicates once per cell cycle, which ensures the establishment of bipolar spindle poles during mitosis (Nigg, 2007). The two centrioles separate upon entry into G1 (centriole disengagement) and this step is a prerequisite for centrosome duplication during S phase. As the cell cycle progresses through S and G2, the duplicated centrosomes mature and become disconnected. Separated centrosomes are the structural basis of spindle poles from which bipolar mitotic spindles are formed that coordinate chromosome segregation during mitosis.
Centrosome (as well as centriole) cohesion and separation are tightly regulated during the cell cycle. Centrosome duplication and genome replication are initiated at the onset of S phase. Both events are tightly coordinated, and are triggered by the activity of Cdk2–cyclin E (Hinchcliffe and Sluder, 2002; Sauer and Lehner, 1995). Over the past few years, effort has been made to identify and characterize the molecular entities that both restrict centrosome duplication to once per cell cycle (Wong and Stearns, 2003; Doxsey et al., 2005) and control sister chromatid cohesion before anaphase entry (Uhlmann, 2004; Watanabe, 2005). Recent studies have shed light on the mechanism by which cells coordinate centrosome/centriole cohesion with sister chromatid cohesion during cell division. Tsou and Stearns have shown that separase, a caspase-related protease known to trigger sister chromatid separation by cleavage of cohesin, functions as a licensing factor that controls centriole disengagement at anaphase (Tsou and Stearns, 2006). Supporting this discovery, Thein et al. have recently demonstrated that depletion of astrin, a protein associated with both spindle poles and kinetochores during mitosis, causes premature sister chromatid separation as well as centriole disengagement, leading to the formation of multipolar spindles; moreover, separase is activated in cells that have been depleted of astrin whereas separase-depletion suppresses centriole disengagement and premature sister chromatid separation (Thein et al., 2007). These newly recognized functions of separase and astrin thus underscore the importance of a coordinated control of centrosome dynamics and the chromosome cycle in order to maintain genomic stability during cell division.
Separation of sister chromatids is achieved by sequential removal of the cohesin complex from chromosome arms by the prophase pathway and from centromeres by separase at the anaphase onset (Waizenegger et al., 2000). Work by several independent groups has revealed a new pathway by which centromeric cohesion of sister chromatids is protected in prophase by an evolutionarily conserved protein called Shugoshin (McGuinness et al., 2005; Kitajima et al., 2004; Salic et al., 2004; Tang et al., 2004) Mammalian cells contain two structurally related Shugoshin proteins termed Sgo1 and Sgo2 (Kitajima et al., 2004; Wang and Dai, 2005). Alternatively spliced forms of Sgo1 are present in human and mouse cells (McGuinness et al., 2005; Wang et al., 2006). In human, two major species of Sgo1 mRNA encode a 527 amino acid isoform (Sgo1) and a 292 amino acid isoform (sSgo1), respectively. Sgo1 at centromeres is required for timely chromosomal separation at anaphase entry and is essential for mediating the function of Bub1 in fission yeast and humans (Kitajima et al., 2004; Kitajima et al., 2005; Tang et al., 2004). The shorter isoform, sSgo1, lacks 268 amino acids encoded by exon 6 but contains 33 amino acids encoded by exon 9 (Wang et al., 2006). The unique structural features of sSgo1 suggest that it may have a distinct function in vivo.
The identification of a role for separase in centriole disengagement has prompted us to study whether other molecules critical for regulating centromeric cohesion of sister chromatids also play a role in the centrosome cycle during mitosis. We demonstrate that Sgo1 is not only essential for mediating sister chromatid cohesion but also involved in controlling centriole cohesion and that these bifurcate functions appear to be mediated by two splice variants of Sgo1. Moreover, we show that the polo-like kinase 1 (Plk1), known to be important for the centrosome cycle (Mayor et al., 1999), regulates subcellular localization and function of the centrosomal Sgo1 splice variant.
RESULTS
Sgo1 depletion results in the formation of extra centrosomal foci
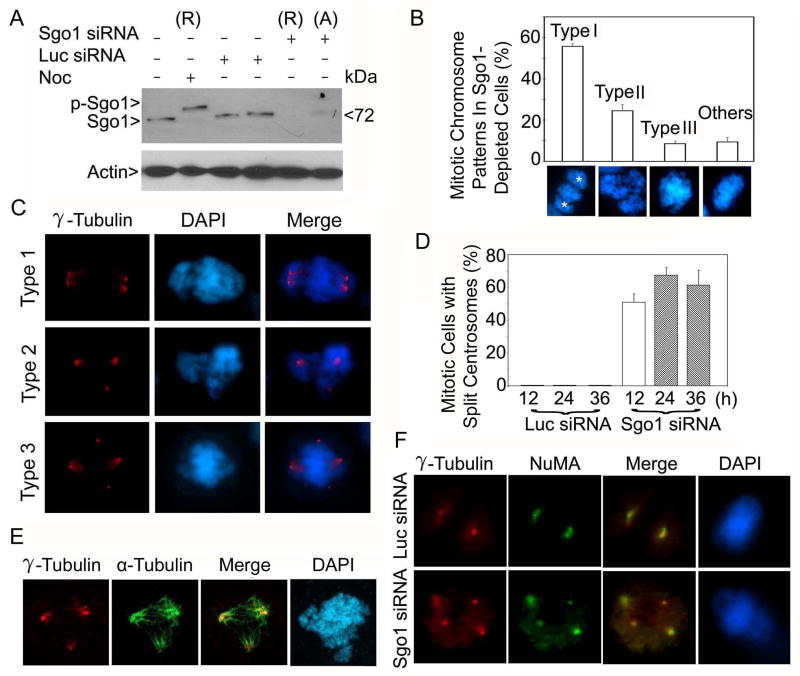
Sgo1 is an evolutionarily conserved protein that functions as a protector of centromeric cohesion during mitosis. Inactivation of Sgo1 function or depletion of Sgo1 in mammalian cells causes missegregation of chromosomes (McGuinness et al., 2005; Tang et al., 2004; Kitajima et al., 2004; Yang et al., 2006). As both spindle poles and kinetochores are important in coordinating chromosome alignment and segregation during mitosis, we designed experiments in which spindle pole integrity as well as chromosome segregation could be monitored simultaneously after Sgo1 depletion via RNAi. Sgo1 was effectively depleted in rounded-up mitotic cells where neither the mitotic (p-Sgo1) nor the interphase form of Sgo1 was detected (Fig. 1A). Evidence that the slower migrating band seen by Western blotting in rounded-up cells after nocodazole treatment is the phosphorylated form is provided by the fact that it was converted to the interphase form after λ-phosphatase treatment (Supplement 1A).
Figure 1.
siRNA depletion of Sgo1 causes formation of extra γ-tubulin foci. (A) HeLa cells were transfected with Sgo1 or luciferase (Luc) siRNA for 24 h. Rounded-up (R) and adherent (A) cells in Sgo1 siRNA transfected dishes were collected separately. An equal amount of each cell lysate was blotted for Sgo1 and β-actin. Nocodazole (Noc)-treated cell lysates were loaded as control. Arrow p-Sgo1 indicates phospho-Sgo1. (B) Rounded-up cells induced as a result of Sgo1 depletion were examined for chromosome patterns after DAPI staining. The data were summarized from more than 300 mitotic cells depleted of Sgo1. The stars (*) denote spindle pole positions. (C) HeLa cells transfected with Sgo1 siRNA were stained with an antibody to γ-tubulin (red). DNA was stained with DAPI (blue). Representative images of cells with centrosome splitting are shown. (D) The percentage of siRNA-transfected cells with extra centrosomal foci was summarized from 200 mitotic cells at each time point. (E) A representative cell transfected with Sgo1 siRNA for 24 h and stained with antibodies to γ-tubulin (red) or α-tubulin (green) is shown. (F) HeLa cells transfected with Sgo1 or Luc siRNA were stained with antibodies to γ-tubulin (red) and NuMA (green). Representative images are shown.
Close examination of DAPI-stained HeLa cells by fluorescence microscopy revealed altered segregation of chromosomes in Sgo1-depleted cells (Fig. 1B). The missegregated chromosomes exhibited several distinct patterns which we have defined as Type I, Type II and Type III in order to distinguish them from one another. Chromosome patterns included those with missegregated chromosomes which formed two clusters at the spindle pole regions in addition to those congregated at the midzone (Type I), triangular shaped chromosome clusters (Type II), and those in which chromosomes were loosely congregated at the metaphase plate (the midzone) with apparent lagging chromosomes (Type III) (Fig. 1B). The “Others” category includes chromosome patterns of relatively normal metaphase appearance or with polyploid contents.
It has been suggested that Sgo1 is involved in generating tension at the kinetochores and affecting microtubule dynamics (Salic et al., 2004). Therefore, Sgo1 depletion might be expected to have an effect on spindle pole integrity. In order to examine this possibility, we stained Sgo1-depleted HeLa cells with an antibody to γ-tubulin, a centrosome marker. Sgo1 depletion caused an increase in γ-tubulin-positive foci in mitotic, but not in interphase cells (data not shown), and these foci exhibited a few distinct patterns (Fig. 1C). Whereas about half the cells contained two apparently separated γ-tubulin foci formed around each spindle pole (Type 1), a significant number of cells contained three or four γ-tubulin foci, some of which were well separated from either spindle pole (Types 2 and 3). The Type 1 spindle pole pattern was tightly associated with Type I missegregated chromosomes. Type 2 spindle pole patterns were often associated with the tripolar arrangement of missegregated chromosomes (Type II). However, there was no apparent correlation between Type 3 spindle pole arrangements and chromosome segregation patterns. Time course studies indicated that the formation of extra γ-tubulin foci occurred within 12 h post-transfection of Sgo1 siRNA, peaking around 24 h (Fig. 1D). These additional foci were likely to be functional for microtubule organization because they usually had associated microtubules (Fig. 1E).
To exclude the possibility that Sgo1 depletion might affect the dynamics of γ-tubulin but not spindle pole integrity, we stained Sgo1-depleted cells with an antibody to NuMA, also a spindle pole protein (Dionne et al., 1999). Whereas NuMA was present as a distinct spot at each spindle pole in normal mitotic cells transfected with luciferase siRNA, in Sgo1-depleted cells NuMA formed additional foci. The extra NuMA foci overlapped with extra γ-tubulin foci (Fig. 1F). Similar phenomena were also observed when other spindle pole markers including Plk1 and Ninein were used (data not shown).
We further confirmed that the formation of extra centrosomal foci also occurred in normal fibroblasts (WI-38) after Sgo1 depletion (supplement 1B), indicating that the observed centrosomal abnormality in HeLa cells was not due to their transformed status. Moreover, individual Sgo1 siRNAs also efficiently induced the formation of extra γ-tubulin foci (Supplement 1C). The formation of extra γ-tubulin foci in Sgo1-depleted cells did not seem to result from tension slack on the mitotic spindles because reducing tension by depolymerizing microtubules by nocodazole essentially eliminated, rather than increased, the number of extra γ-tubulin foci in these cells (Supplements 2A and 2B).
Sgo1 depletion-induced extra centrosomal foci primarily results from centriole splitting
The formation of additional γ-tubulin- or NuMA-positive foci could result from one of two possible mechanisms, centrosome amplification or centriole splitting. Due to the short time interval between Sgo1 depletion and the appearance of these foci (Fig. 1D), we reasoned that they might be derived not from centrosome amplification but from centrosome splitting, perhaps through the separation of paired mother and daughter centrioles (referred as “centriole splitting” hereafter). Consistent with this, the presence of more than four discrete γ-tubulin foci in one mitotic cell was infrequently observed (Supplement 1C, “Others”).
Centrin is a core centriole component, whose signal is confined to individual centrioles. To establish a system with which centrosome amplification or centriole splitting could be examined, we transiently transfected HeLa cells with a plasmid expressing GFP-centrin. Indeed, expressed GFP-centrin was not only localized at the centrosome, but also could discriminate two centrioles within a centrosome (Fig. 2A, boxed areas enlarged). To exclude the possibility that the extra centrosomal foci in Sgo1-depleted mitotic cells were the result of centrosome amplification, we examined the centriole number (GFP-centrin foci) in interphase HeLa cells stably expressing GFP-centrin. We found that the number of interphase cells containing more than 4 centrioles after transfection with Sgo1 siRNAs for 24 h was not increased compared with that of the control cells transfected with luciferase siRNA (Fig. 2B), suggesting that centrosome amplification was not responsible for the increased centrosomal foci in mitotic cells. The mild increase in interphase cells with 4 centrioles (Fig. 2B) may reflect reduction of G1 cell population after Sgo1 depletion.
Figure 2.
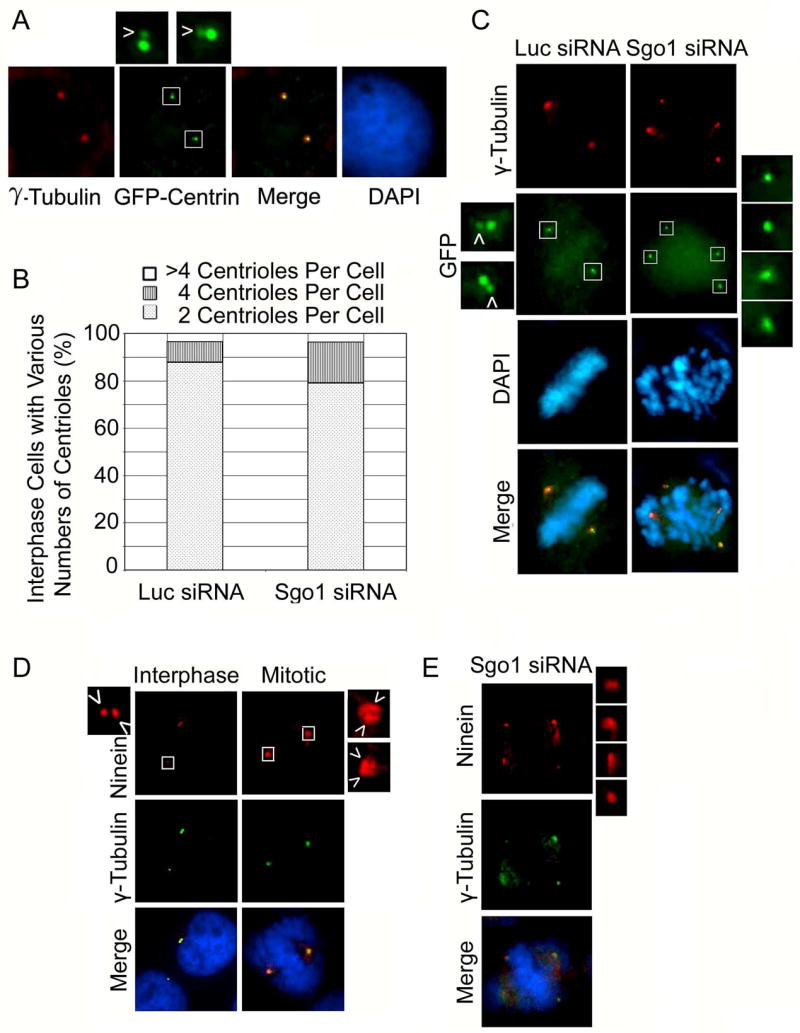
Sgo1 depletion-induced formation of extra centrosomal foci results from centriole splitting. (A) HeLa cells transfected with a GFP-centrin expression plasmid for 24 h were stained with an antibody to GFP (green) or γ-tubulin (red). The daughter centrioles are indicated by arrows. (B) HeLa cells stably expressing GFP-centrin transfected with Sgo1 or Luc siRNA for 24 h were fixed and stained with the antibody to GFP. The number of centrioles in individual interphase cells (100 cells/experiment) was scored under the microscope. The data was summarized from three independent experiments. (C) GFP-centrin-expressing cells were transfected with Sgo1 or Luc siRNA for 24 h. Cells were fixed and stained with antibody to GFP or γ-tubulin. Representative cells are shown. The boxed areas were enlarged. Daughter centrioles are indicated by arrows. (D). HeLa cells were fixed and stained with the antibody to ninein (red) or γ-tubulin (green). Representative cells are shown. Paired centrioles are indicated by arrows. (E) HeLa cells transfected with Sgo1 siRNA for 24 h were stained with the antibody to ninein or γ-tubulin.. The ninein foci were enlarged.
To further confirm the role of Sgo1 in mediating centriole cohesion, we looked at mitotic effects of Sgo1 depletion in HeLa cells stably expressing GFP-centrin. Sgo1 depletion resulted in the formation of extra GFP-centrin foci, which superimposed with γ-tubulin foci (Fig. 2C). A close examination revealed that each spindle pole consisted of a pair of GFP-centrin signals in luciferase siRNA-transfected cells, indicative of the presence of intact mother-daughter centriole pairs. In contrast, GFP-centrin foci detected in HeLa cells transfected with Sgo1 siRNA were discrete and showed no close association of companion centrioles (Fig. 2C, boxed areas enlarged; Supplement Table 1).
Our further study of ninein, another centriole marker, confirms this effect of Sgo1 depletion on centriole splitting. Depletion of Sgo1 in HeLa cells via RNAi caused the formation of extra-ninein foci as well as chromosomal missegregation. Unlike two spindle pole ninein foci in normal mitotic cells (Fig. 2D, boxed areas enlarged), extra-ninein foci in Sgo1-silenced cells were distinct and showed no companion centriole signals (Fig. 2E, insets). Taken together, these experiments provide strong support for the hypothesis that extra spindle pole-like foci induced by Sgo1 silencing results primarily from the separation of paired centrioles.
Sgo1 has a centrosomal localization
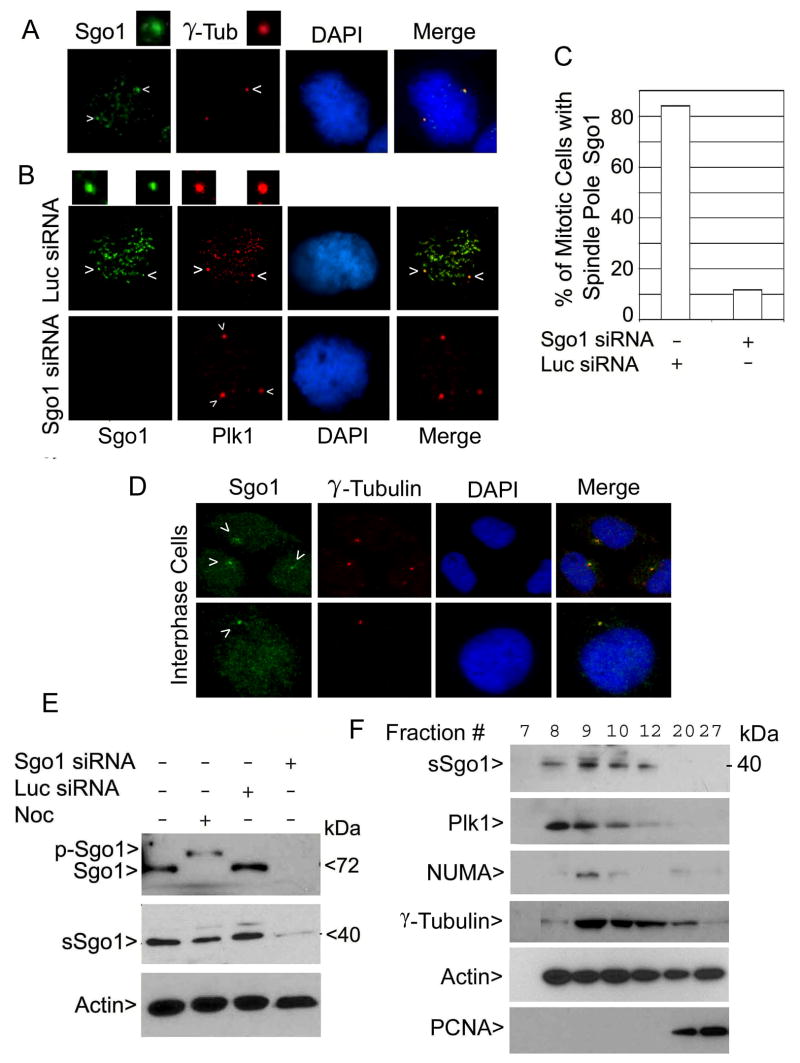
The observation that Sgo1 depletion was linked to spindle pole integrity suggested its physical presence at the centrosome. To detect centrosome/spindle pole localization of Sgo1, we stained HeLa cells with antibodies to Sgo1 and γ-tubulin. Through rigorous extraction of the cells before fixation (Gregson et al., 2001), we were able to detect endogenous Sgo1 co-localized with γ-tubulin at the spindle poles in mitotic HeLa cells (Fig. 3A, the area pointed by the upper right arrow enlarged).
Figure 3.
Sgo1 is localized to the centrosomes/spindle poles. (A) HeLa cells were stained with antibody to Sgo1 (green) or γ-tubulin (red). Arrows indicate spindle pole signal. The area indicated by the upper right arrow was enlarged. (B) U2OS cells transfected with Luc or Sgo1 siRNA for 24 h were stained with the antibody to Sgo1 (green) or Plk1 (red). Arrows indicate spindle pole signal. The areas indicated by arrows were enlarged. (C) Data were summarized from over 100 mitotic cells shown in B after transfection of Sgo1 or Luc siRNA. (D) HeLa cells were stained with antibodies to Sgo1 γ-tubulin. Arrows indicate the centrosome positions. (E) An equal amount of lysates from HeLa cells transfected with Sgo1 or Luc siRNA for 24 h was blotted for Sgo1 and β-actin. Noc-treated cell lysates were loaded as controls. (F) Centrosomes were isolated from HeLa cells.. An equal volume of sample from selected centrosomal isolation fractions was blotted for sSgo1, Plk1, NuMA, γ-tubulin, β-actin, and PCNA.
Plk1 is localized to both spindle poles and kinetochores in mitotic cells (Arnaud et al., 1998). Similar to Plk1, endogenous Sgo1 signals were present both at the spindle poles and the kinetochores in mitotic U2OS cells (Fig. 3B, upper panels, the area pointed by arrow enlarged). The spindle pole Sgo1 signal was specific because transfection with Sgo1 siRNA abolished Sgo1 signals at both mitotic apparatus locations (Fig. 3B and 3C). Endogenous Sgo1 also co-localized with γ-tubulin in interphase cells (Fig. 3D), suggesting that it plays a role in centrosome dynamics throughout the cell cycle. Moreover, the localization of Sgo1 at the centrosome was independent of microtubules because brief treatment with nocodazole did not diminish centrosomal Sgo1 signals (Supplement 2C).
GenBank databases contain numerous Sgo1 cDNA clones which correspond primarily to two major splice variants. One Sgo1 variant codes for the full length protein detected as a 72 kDa protein by Western blotting while the other variant transcript appears to code for a much smaller protein that lacks the sequence encoded by exon 6 (Wang et al., 2006). In order to gain more understanding of the regulation of expression of these two variants and to explore the possibility that the two forms play different roles during cell division, we examined whether HeLa cells expressed both isoforms. Immunoblot analysis revealed that, in addition to the Sgo1 band migrating around 72 kDa as previously reported, HeLa cells also contained a protein of about 40 kDa that was immunoreactive to the Sgo1 antibody; this protein, as well as the 72 kDa one, was specifically depleted upon transfection with Sgo1 siRNA (Fig. 3E). Expression of the short Sgo1 (termed sSgo1 hereafter) was also detected in other cell lines tested including U2OS, A549 and MCF-7 cells (data not shown). To determine which form of Sgo1 was localized or enriched at the centrosome, we isolated the organelle from HeLa cells using density gradient centrifugation. Representative centrosomal fractions were Western blotted with antibodies to Sgo1 as well as to centrosomal markers. The fast migrating form, sSgo1, co-fractionated with NuMA, Plk1, and γ-tubulin; in contrast, sSgo1 did not co-fractionate with PCNA, a nuclear antigen (Fig. 3F), suggesting its absence from kinetochores. The cytoplasm also contains a significant amount of γ-tubulin (Moudjou et al., 1996), which may account for its presence in many fractions of the samples (Fig. 3F). The presence of intact centrosomes in fraction #9 was also confirmed by fluorescence microscopy. The centrosomes isolated from HeLa cells were positive for Sgo1 as well as for γ-tubulin (Supplement 3A).
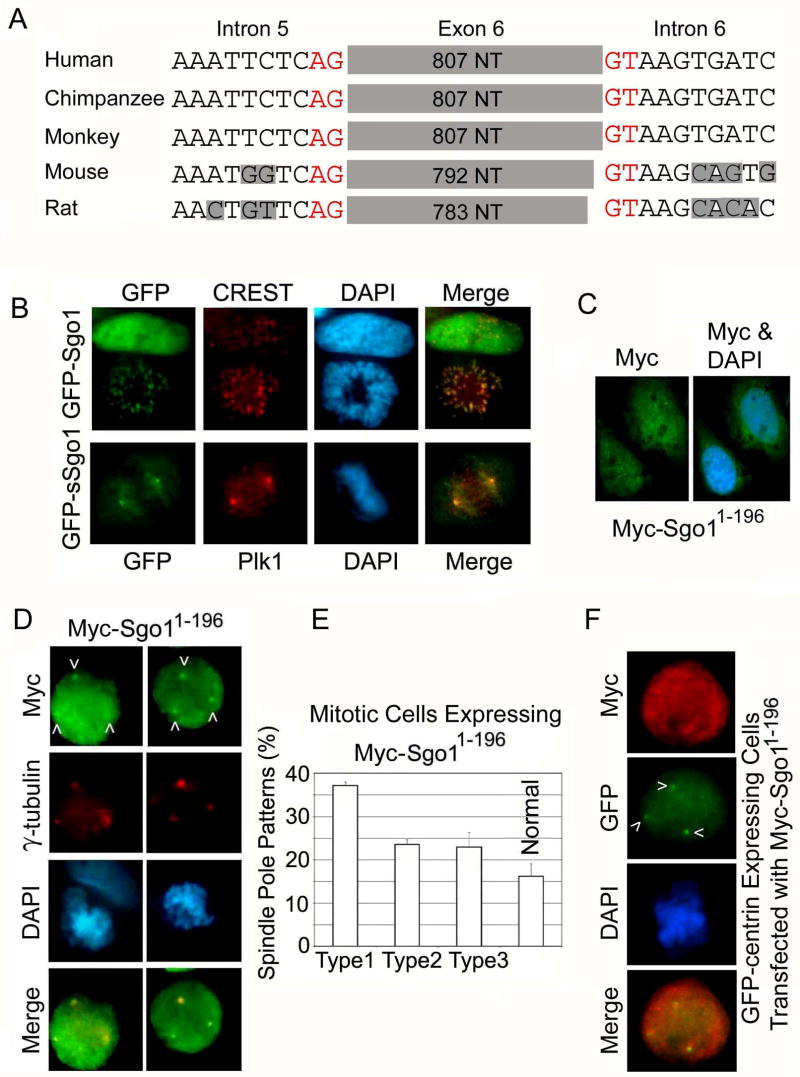
The full length Sgo1 protein contains an exon 6-encoded region that is absent from sSgo1 (Wang et al., 2006). An analysis of intron and exon boundaries of Sgo1 cDNAs reveals that exon 6 (or its structural counterpart) exists as an independent unit among various mammals (Fig. 4A), suggesting a conserved function. To test the possibility that exon 6 might dictate the subcellular localization of Sgo1, we expressed cDNAs coding for the two major splice variants as GFP-fusion proteins. Fluorescence microscopy revealed that whereas GFP-Sgo1 co-localized with CREST at the kinetochores during mitosis, GFP-sSgo1 localized to the spindle poles and the mitotic spindles but not at the kinetochores (Fig. 4B). This supports the hypothesis that these two variants are differentially regulated in the cell.
Figure 4.
sSgo1 functions in regulating spindle pole integrity. (A) The intronic sequences surrounding human exon 6 or its equivalent in other mammals were aligned. Red color marks the invariant splice donor and splice acceptor sequences. The shaded areas indicate non-conserved sequences in introns. (B) HeLa cells transfected with GFP-Sgo1 or GFP-sSgo1 expression constructs for 24 h were fixed and stained with antibodies to GFP (green), CREST (red, upper panel), or Plk1 (red, lower panel). (C) HeLa cells transfected with Myc-Sgo11-196 expression plasmid for 24 h were stained with antibody to Myc. (D&E) HeLa cells transfected with Myc-Sgo11-196 plasmid for 24 h were stained with antibodies to Myc (green) and γ-tubulin (red) (D). Various abnormal spindle pole patterns were summarized from three independent experiments (E) (150 mitotic cells/experiment). (F) HeLa cells expressing GFP-centrin transfected with Myc-Sgo11-196 plasmid for 24 h were stained with antibodies to Myc (red) and GFP (green). Arrows indicate the position of split centrioles.
As a first attempt to study the function of sSgo1 in regulating centrosome integrity, we made and expressed a deletion construct (Myc-Sgo11-196) containing only the N-terminal 196 amino acids of Sgo1. As it does not contain a region coded by a functional exon 6, Sgo11-196 is similar in general sequence structure to sSgo1 (supplement 3B). When ectopically expressed, Myc-Sgo11-196 exhibited both cytoplasmic and nuclear localizations during interphase (Fig. 4C). A significant amount of ectopically expressed Myc-Sgo11-196 co-localized with γ-tubulin during mitosis (Fig. 4D). These cells frequently contained extra γ-tubulin foci (Fig. 4D and 4E, arrows), reminiscent of those induced by transfection of Sgo1 siRNA. These studies indicate that Myc-Sgo11-196 may exhibit a dominant negative function, perhaps by interfering with endogenous sSgo1 activity. Supporting this, when GFP-centrin-expressing cells were transfected with the Myc-Sgo11-196 construct, this mutant Sgo1 protein also caused centriole splitting (Fig. 4F, arrows). No kinetochore Myc-Sgo11-196 was detected in mitotic cells when it was ectopically expressed (data not shown), suggesting that the formation of extra γ-tubulin foci is primarily due to the disruption of sSgo1 function but not due to chromosome missegregation caused by Sgo1 depletion. In fact, Myc-Sgo11-196 did not affect the localization of GFP-Sgo1 at kinetochores (Supplement 3C).
As an additional approach to study sSgo1 function, we designed a Myc-sSgo1 expression construct and an siRNA duplex corresponding to the sequence of the 3′-untranslated region (3′UTR) of Sgo1 mRNA that was absent from Myc-sSgo1. HeLa cells were co-transfected with the Myc-sSgo1 expression construct and the Sgo1 3′UTR siRNA. Immunoblotting revealed that whereas endogenous sSgo1 was efficiently silenced by the 3′UTR siRNA, ectopically expressed Myc-sSgo1 remained in transfected cells (Fig. 5A), indicating that Myc-sSgo1 mRNA was largely resistant to the silencing by the 3′UTR siRNA. Moreover, through examination of mitotic cells expressing transfected Myc-sSgo1, we observed that centriole splitting induced by Sgo1 silencing was significantly rescued (Fig. 5B, Supplement Table 2), indicating a protective function of sSgo1 at the spindle poles. On the other hand, expression of Myc-sSgo1 did not suppress the rate of chromosomal missegregation caused by depletion of endogenous Sgo1 (Fig. 5C, Supplement Table 2), supporting the notion that sSgo1 may not have a direct function at kinetochores. Combined, these observations support the hypothesis that sSgo1 plays a primary role in protecting the integrity of the spindle poles through maintaining centriole cohesion.
Figure 5.
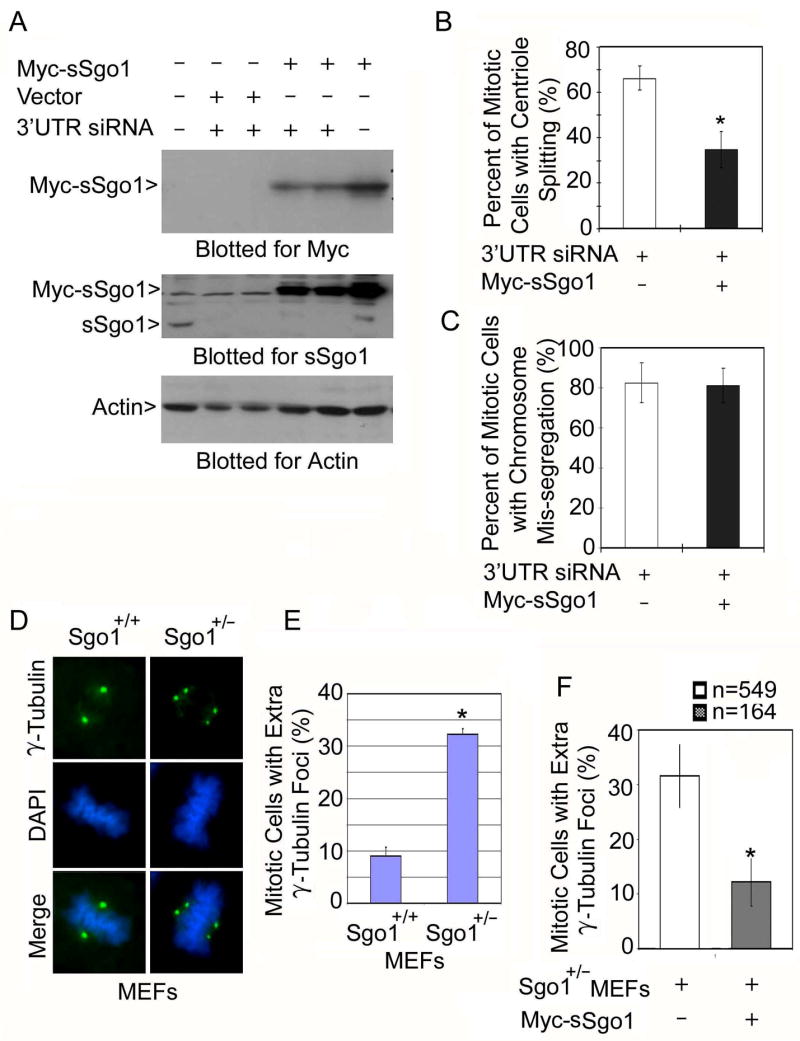
Ectopic expression of sSgo1 suppresses centriole splitting induced by Sgo1 depletion. (A) HeLa cells co-transfected with the Myc-sSgo1 expression construct and Sgo1 3′UTR siRNA for 48 h were collected and an equal amount of cell lysate was blotted for Myc, endogenous sSgo1, and β-actin. (B & C) HeLa cells transfected with the Myc-sSgo1 expression construct and Sgo1 3′UTR siRNA for about 32 h were fixed and stained with antibodies to the Myc tag and γ-tubulin. The data were summarized from over 200 transfected mitotic cells. (D) Wild-type (Sgo1+/+) and Sgo1+/− MEF cells were stained with antibody to γ-tubulin (green). (E) The number of mitotic cells with extra γ-tubulin foci in Sgo1+/+ and Sgo1+/− MEF cells was scored. The data were summarized from three independent experiments (100 mitotic cells/experiment). (F) Sgo1+/− MEFs transfected with a Myc-sSgo1 expression construct for 24 h were stained with antibodies to γ-tubulin and the Myc tag. The number (n) of mitotic cells with extra γ-tubulin foci was scored (2 independent experiments). * denotes significant statistical difference (p<0.01).
We further examined the role of Sgo1 in protecting centriole cohesion using a genetic approach. Through gene-trapping and transgenic mouse genetics, as described in our previous study (Wang et al., 2004), we obtained Sgo1 haploinsufficient mice (unpublished data), from which we derived Sgo1+/− murine embryonic fibroblast (MEF) cells (Supplement 4). Fluorescence microscopy revealed that a significantly higher fraction of Sgo1+/− mitotic MEFs contained extra γ-tubulin foci than did wild-type mitotic MEFs (Sgo1+/+) (Fig. 5D and 5E). Furthermore, expression of transfected Myc-sSgo1 significantly reduced the number of mitotic cells with extra γ-tubulin foci (Fig. 5F). These studies thus provide a line of unequivocal genetic evidence indicating the involvement of Sgo1 in regulating centrosome function.
Plk1 regulates sSgo1 subcellular localization and function
In Drosophila, the Plk1 homolog POLO is involved in regulating Mei-S332, the homolog of mammalian Shugoshin (Clarke et al., 2005). In addition, Plk1 regulates centrosome dynamics during the cell cycle. Hence, we reasoned that Plk1 may regulate the function of sSgo1 during mitosis. To test this possibility, we examined the subcellular localization of Myc-sSgo1 in cells transfected with Plk1 siRNA. Compared with control cells, Plk1 depletion abolished the localization of Myc-sSgo1 at the spindle poles and mitotic spindles (Fig. 6A and 6B). Plk1 down-regulation via RNAi also delocalized GFP-sSgo1 at the spindle poles (Supplement 5A). Plk1 down-regulation was efficient as confirmed by immunoblotting (Fig. 6C).
Figure 6.
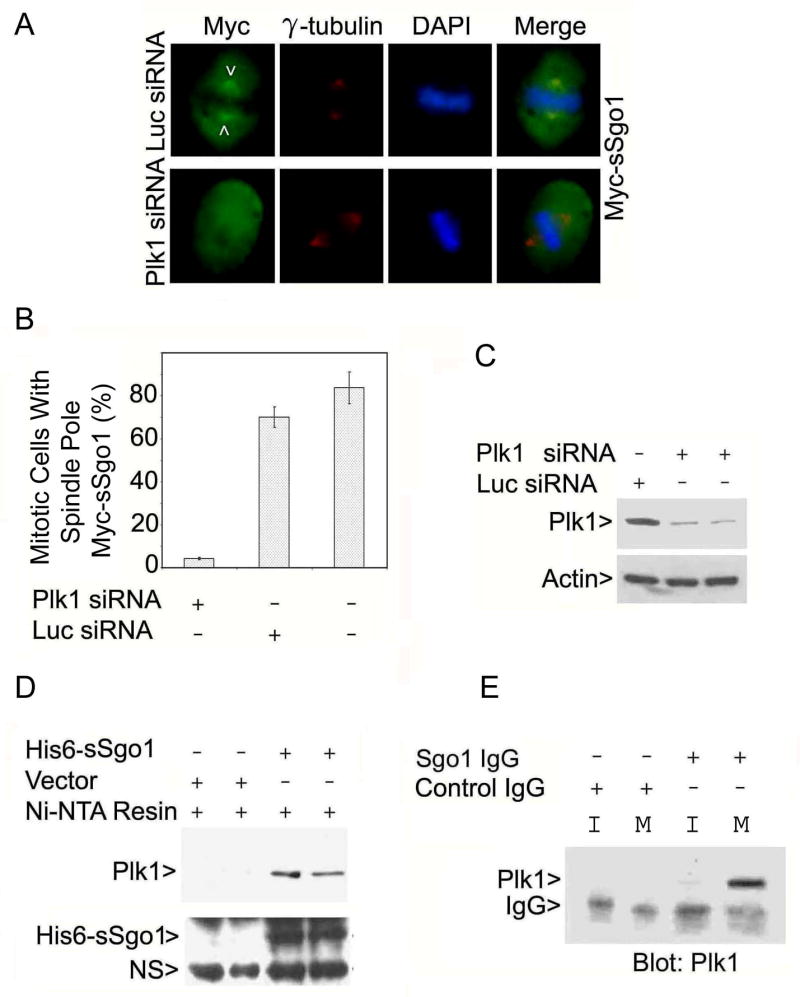
Plk1 is physically associated with sSgo1. (A) HeLa cells co-transfected with Myc-sSgo1 plasmid and Plk1 or Luc siRNA for 24 h were stained with antibodies to the Myc tag (green) and γ-tubulin (red). (B) HeLa cells were cotransfected with Myc-sSgo1 expression plasmid and Plk1 or Luc siRNA. The data were summarized from three independent experiments (100 Plk1-depleted mitotic cells/experiment). (C) HeLa cells transfected with Plk1 or Luc siRNA for 24 h were blotted for Plk1 and β-actin. (D) Aqual amounts of HeLa cell lysates after transfection with a His6-sSgo1plasmid or the vector alone for 48 h were incubated with Ni-NTA resin. Proteins specifically bound to the resin were blotted for Plk1 or Sgo1. Arrow NS denotes a non-specific signal. (E) An equal amount of interphase (I) or mitotic (M) HeLa cell lysate was immunoprecipitated with the anti-Sgo1 antibody or with a control IgG. The immunoprecipitates were blotted for Plk1.
To examine the possibility of a physical interaction between these two proteins, we performed co-precipitation assays using histidine-tagged sSgo1 (His6-sSgo1). We observed that Ni-NTA resin precipitated not only His6-sSgo1 but also Plk1 from HeLa cells transfected with His6-sSgo1 plasmid, but not with the control plasmid (Fig. 6D). To confirm the physical interaction between Sgo1 and Plk1, we performed co-immunoprecipitation experiments using anti-Sgo1 antibodies. The Sgo1 antibodies, but not the pre-immune IgGs, were capable of precipitating a significant amount of Plk1 from mitotic HeLa cells (Fig. 6E).
Plk1 has been shown to directly phosphorylate centrosomal proteins that are important for the regulation of the centrosome cycle or the maintenance of spindle pole integrity during mitosis (Oshimori et al., 2006). The physical interaction between Plk1 and sSgo1 suggests that Plk1 may regulate sSgo1 through phosphorylation. A survey of the sSgo1 amino acid sequence revealed four putative sites that conform to the Plk1 phosphorylation motif. One putative site (serine 260) falls within the region encoded by exon 9, which is spliced out in certain sSgo1 isoforms, suggesting that it may not have a conserved function. Therefore, we first focused on the other 3 motifs in mediating sSgo1 function. We made a series of mutants in which serines or threonines were replaced with alanines; mutant sSgo1 proteins, as well as the wild-type, were expressed as Myc-tagged fusion products. When ectopically expressed, Myc-sSgo1S129A and the wild type Myc-sSgo1 were largely confined to the spindle poles and the mitotic spindles; however, Myc-sSgo1S73A and Myc-sSgo1T146A mutant proteins did not properly localize to the spindle pole regions in a significant fraction of mitotic cells (Fig. 7A and 7B), supporting the idea that phosphorylation by Plk1 is important for the localization and perhaps the function of sSgo1. Immunoblotting revealed that various sSgo1 mutant proteins were expressed at a similar level in HeLa cells (Supplement 5B). We then analyzed if ectopic expression of these putative phosphorylation mutants affected spindle pole integrity. Mitotic cells expressing Myc-sSgo1S73A and Myc-sSgo1T146A, but not Myc-sSgo1S129A, exhibited a significant increase in split spindle poles compared with those expressing wild-type Myc-sSgo1 (Fig. 7C), suggesting that correct localization of sSgo1 is necessary for its spindle pole function during mitosis. The relatively low percentage of split spindle poles in mitotic cells expressing either Myc-sSgo1S73A or Myc-sSgo1T146A suggests that phosphorylation of sSgo1 on multiple sites may be necessary for its full activity.
Figure 7.
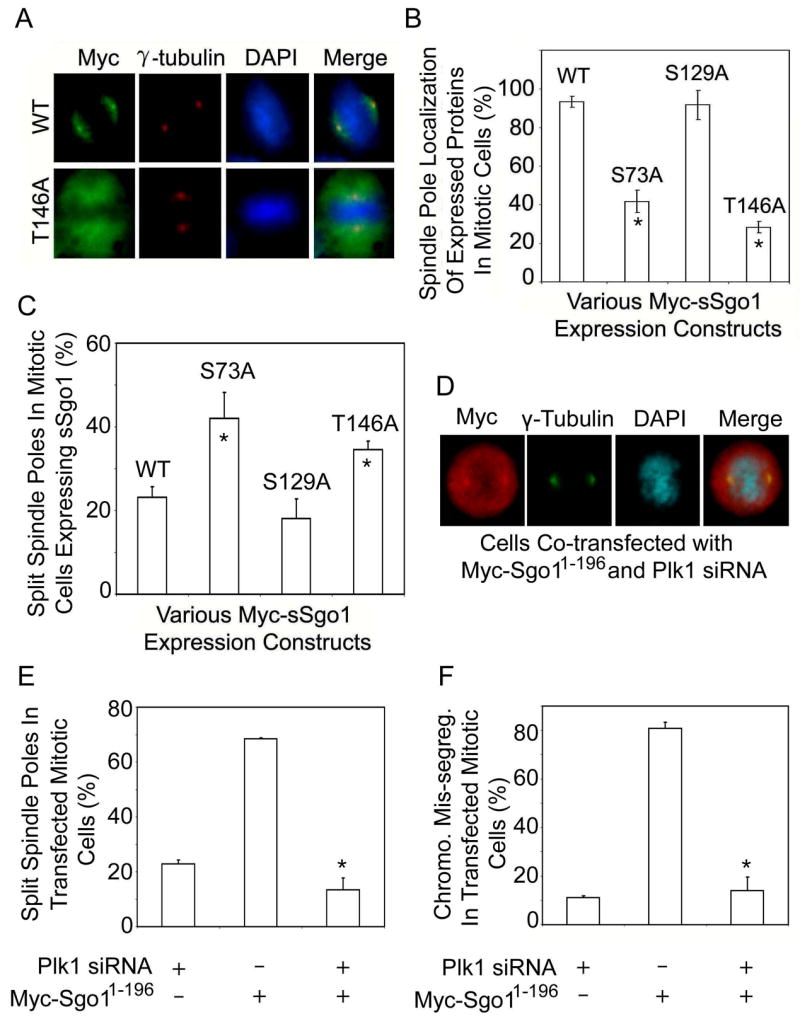
Plk1 regulates Sgo1 function. (A) HeLa cells transfected with Myc-sSgo1 or Myc-sSgo1T146A expression plasmid for 24 h were stained with antibodies to the Myc tag (green) and γ-tubulin (red). (B) HeLa cells transfected with Myc-sSgo1 expression plasmid (WT) or with various Myc-tagged sSgo1 mutant plasmids as indicated for 24 h were stained with antibody to the Myc tag. The data were summarized from three independent experiments (>100 transfected mitotic cells). * denotes significant statistical difference (p<0.01). (C) HeLa cells transfected with Myc-tagged sSgo1 (WT) or with mutant expression plasmids for 24 h were stained with antibodies to the Myc tag and γ-tubulin. The data were summarized from three independent experiments (>300 transfected mitotic cells). * denotes significant statistical difference (p<0.05). (D) HeLa cells co-transfected with Myc-Sgo11-196 expression plasmid and Plk1 siRNA for 24 h were stained with antibodies to the Myc tag (red) and γ-tubulin (green). (E and F) HeLa cells co-transfected with Myc-Sgo11-196 expression plasmid and Plk1 siRNA for 24 h were stained with antibodies to the Myc tag and γ-tubulin. The data were summarized from three independent experiments (>300 transfected mitotic cells). * denotes significant statistical difference (p<0.01).
To further elucidate Plk1 function in regulating spindle pole integrity mediated by sSgo1, we asked whether Plk1 depletion could affect spindle pole abnormalities induced by the dominant negative protein Myc-Sgo11-196. Fluorescence microscopy revealed that whereas expression of Myc-Sgo11-196 alone induced centriole splitting in a majority of mitotic cells, Plk1 silencing by RNAi greatly suppressed Myc-Sgo11-196-induced centriole splitting (Fig. 7D and 7E). Transfection of Plk1 siRNA also suppressed chromosomal missegregation induced by Myc-Sgo11-196 (Fig. 7F). Plk1 depletion confirmed by immunoblotting did not affect the expression of transfected Myc-Sgo11-196 in mitotic cells (Supplement 5C). We also observed some structural alterations in spindle poles in Plk1-depleted cells; however, these changes, characterized mainly by diffused γ-tubulin signals along mitotic spindles (Supplement 5D), were different from that induced by Sgo1-deficiency. Combined, these studies strongly suggest that Sgo1 is involved in controlling spindle pole integrity and that its function is at least partially mediated through phosphorylation by Plk1.
DISCUSSION
This study reveals a new function of Sgo1 in the regulation of spindle pole integrity. Given the tight coordination of DNA replication and centrosome duplication during S phase, it is not surprising to observe crosstalk between chromosomes and spindle poles during mitosis. In fact, important regulatory molecules including Plk1, aurora kinases, and ERKs are present at both the kinetochores and the spindle poles. Recent studies indicate that key components, including PP2A and astrin, involved in mitotic control or sister chromatid cohesion are also present at both the spindle poles and kinetochores (Schlaitz et al., 2007; Thein et al., 2007). Moreover, during late mitosis, the mother centriole repositions itself by migrating to the intercellular bridge, apparently influencing cell cleavage at cytokinesis in certain cell types (Piel et al., 2001). It is conceivable that premature centriole splitting or maternal centriole migration before the completion of chromosome segregation would allow the formation of additional microtubule nucleating foci, a condition favoring chromosome missegregation and genomic instability.
Given the observation that separase, a key enzyme in controlling sister chromatid cohesion, also plays an essential role in centriole disengagement during M phase to facilitate their subsequent duplication (Tsou and Stearns, 2006), it is tempting to speculate that a mechanism similar to that of cohesin cleavage may exist at the centrosomes. Our current study supports the notion that separase may have a centrosomal target(s). The most straightforward model would be based on the concept that sSgo1 at the spindle poles may work in a fashion similar to that of Sgo1 at the kinetochores by protecting cohesin. In fact, Rad21, a cohesin subunit cleavable by separase, is transiently associated with the centrosomes during metaphase and anaphase in Drosophila (Warren et al., 2000). In mammalian cells, the cohesin complex also localizes to the spindle poles during mitosis where it interacts with NuMA, a protein associated with the spindle poles, to play an essential role in spindle pole organization (Gregson et al., 2001). Alternatively, sSgo1 can be a potential substrate of separase because its sequence contains a core separase cleavage consensus site at amino acids 128 to 131 [(D/E)XXR] (Sullivan et al., 2004). This would place sSgo1 downstream of separase. Supporting this notion, depletion of separase by RNAi did not significantly suppress massive centriole splitting induced by Sgo1 depletion (data not shown). The discovery that separase is involved in controlling centriole splitting (Tsou and Stearns, 2006) is significant because this would not only allow the centrosome to duplicate once per cell cycle, but also coordinate efficiently with spindle poles and kinetochores to carry out nuclear and cytoplasmic division and suppress genomic instability during mitosis.
A straightforward explanation for Sgo1’s role in mediating centriole cohesion would be its physical presence at the spindle poles. Indeed, our experiments confirm that centrosome/spindle pole sSgo1 signals are detected in interphase and mitotic cells and that these signals disappear upon transfection with Sgo1 siRNA, which depletes both Sgo1 and sSgo1. The failure to detect spindle pole Sgo1 (sSgo1) signals by others may be due partly to different in situ extraction conditions or the nature of the antibodies because sSgo1 contains no sequences encoded by exon 6. The modified protocol using a more stringent extraction process before staining allows us to detect Sgo1 signals at the centrosomes/spindle poles (which, we believe, were otherwise masked). Spindle pole localization of Sgo1 was also observed in a separate study in male meiotic cells in Drosophila (Lee et al., 2005).
Plk1 is a major protein kinase regulating centrosome maturation and spindle pole function during mitosis. Our current studies indicate that Plk1 plays an important role in regulating centriole cohesion mediated by Sgo1. (i) It physically interacts with sSgo1 and the interaction between sSgo1 (and/or Sgo1) and Plk1 appears to be mitosis-specific. (ii) Functional knockdown of Plk1 abolishes the subcellular localization of sSgo1 at mitotic spindles and spindle poles. (iii) Plk1 depletion greatly suppresses centriole splitting and chromosome missegregation induced by Myc-Sgo11-196, a dominant negative mutant. (iv) sSgo1 mutants (especially, serineS73A and threonineT146A) fail to localize properly to spindle poles due to lack of Plk1 phosphorylation sites, and this is correlated with an increase in split spindle poles. Serine 73 is localized in the region mediating microtubule binding (Salic et al., 2004); it is also in the coiled-coil structure, which is known to mediate protein-protein interactions. It is conceivable that phosphorylation of sSgo1 by Plk1 may directly regulate its interaction with microtubules and spindle pole components such as PP2A. In fact, the N-terminal region of Sgo1 interacts with PP2A (Tang et al., 2006). Structural alterations due to changes in phosphorylation status or due to truncation (Sgo11-196) may disrupt proper interaction with PP2A, thus inducing centriole splitting. This is consistent with the observation that depletion of PP2A via RNAi also compromises the integrity of spindle poles (data not shown) and causes the formation of extra γ-tubulin foci (Kitajima et al., 2006).
The dominant negative protein Myc-Sgo11-196 may interfere with the function of endogenous sSgo1 (and Sgo1 as well) by direct physical association because existing evidence indicates that Sgo1 is capable of forming a dimer (Tang et al., 1998). Plk1-mediated phosphorylation of Sgo1 may be required for efficient dimer formation in vivo. Therefore, in the absence of Plk1, the association between the mutant protein and cellular sSgo1 and Sgo1 can be conceivably compromised, resulting in significant functional rescue of defects in mitotic cells expressing transfected Myc-Sgo11-196. It is not surprising to observe that Plk1 knock-down suppresses both spindle pole splitting and chromosome missegregation because Plk1 exhibits localization at both spindle poles and kinetochores (Arnaud et al., 1998).
Accurate chromosome segregation during mitosis is one of the most fundamental processes that allow cells to faithfully transmit their genetic information from one generation to another. Failures in the maintenance of chromosome stability during mitosis inevitably lead to either mitotic catastrophe or malignant transformation. Our current studies indicate that Sgo1 mediates the integrity of both kinetochores and spindle poles, two major mitotic apparatuses, during cell division and the dysregulated function of Sgo1 may be one of the major underlying causes of chromosomal instability.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLa, U2OS, and A549 cell lines were obtained from American Type Culture Collection (ATCC). These cells were cultured under 5% CO2 in Dulbecco’s Minimum Essential Medium (DMEM), 15% fetal bovine serum (FBS), and antibiotics (100 μg/mL penicillin, 50 μg/mL streptomycin sulfate). WI-38 cells from ATCC were cultured under 5% CO2 in Minimum Essential Medium (MEM) with 10 % FBS and 2 mM L-glutamine.
RNA interference
Sgo1 small interfering RNAs (siRNAs) (mixed pool) were obtained from Dharmacon which correspond to the following sequences: 5′CAUCUUAGCCUGAAGGAUAUU3′ (designated as siRNA-1), 5′UGAAAGAAGCCCAAGAUAUUU3′ (siRNA-2), and 5′AAA CGCAGGUCUUUUAUAGUU3′. The 3′UTR sequence used for designing siRNA for silencing Sgo1 is 5′GAGGAUCUGUAAGAGUACACAUU3′. Plk1 siRNA corresponding sequence is 5′AAGGGCGGCUUUGCCAAGUGC3′. Sgo1 or Plk1 siRNA duplexes were transfected into HeLa or U2OS cells with Lipofectamine2000 (Invitrogen). Briefly, cells seeded at 50% confluency in an antibiotic-free culture medium were transfected with siRNA duplexes at a final concentration of 100 nM for 24 h (unless otherwise specified). Negative controls were cells transfected with 100 nM siRNA duplex targeting firefly (Photinus pyralis) luciferase (5′UUCCTACGCTGAGTACTTCGA3′, GL-3 from Dharmacon). In some experiments, HeLa cells were co-transfected with Sgo1 siRNA and various Sgo1 or sSgo1 expression constructs for 24 or 48 h.
Fluorescence microscopy
Cells fixed in methanol or 4% paraformaldehyde (PFA) were treated with 0.1% Triton X-100 on ice and then washed three times with PBS. After blocking with 2.0% BSA in PBS for 15 min, cells were incubated for 1 h with antibodies to Sgo1, γ-tubulin (Sigma), α-tubulin, NuMA, green fluorescence protein (GFP, Santa Cruz), Myc tag (Cell Signaling), Plk1 (Zymed), Ninein (Abcam), NuMA, and CREST, washed with PBS, and then incubated with appropriate secondary antibodies conjugated with Rhodamine-Red-X or FITC (Jackson Immuno Research). Cells were finally stained with 4′,6-diamidino-2-phenylindole (DAPI, 1 μg/ml, Fluka). For detecting Sgo1 signals at spindle poles/centrosomes, we followed the procedure as described (Gregson et al., 2001). Briefly, HeLa cells washed with a cytoskeleton (CSK) buffer (10 mM Pipes, pH 7.0, 100 mM NaCl, 300 mM sucrose, and 3 mM MgCl2) were extracted by incubation in the CSK buffer containing 0.1% Triton-X-100 at RT for 2 min. After brief washing with the CSK buffer, the extracted cells were fixed with methanol or PFA for 5 min at 4°C and treated with 0.1% Triton-X-100 in PBS for 5 min on ice followed by blocking and antibody incubation as described above. Fluorescence microscopy was performed on a Nikon microscope, and images were captured using a digital camera (Optronics). For confocal imaging, a Leica TCS SP5 and a Bio-Rad MRC-1000 confocal microscopes (MRC-1000, Bio-Rad) were utilized.
Centrosome isolation
Centrosomes were isolated from HeLa cells essentially as described (Blomberg-Wirschell and Doxsey, 1998). Briefly, five confluent 150-mm dishes of HeLa cells were treated with nocodazole (5 μg/ml, Sigma) and cytochalasin B (10 μg/ml, Sigma) for 90 min at 37°C. All subsequent steps were performed on wet ice or at 4oC using solutions that had been pre-chilled on ice. Cell lysates were collected into a 15-ml plastic conical tube and various components were supplemented to achieve a final concentration of 10 mM PIPES, 1 mM EDTA, and 1μg/ml protease inhibitors. Lysates were filtered through a 40 μm nytex membrane and then gently laid on top of 20% (w/v) Ficoll (MW 400,000). The samples were then subjected to centrifugation at 25,000g in a swinging bucket rotor for 20 min at 4°C. After centrifugation, centrosomes were layered about 2 mm on top of the Ficoll cushion. Samples (200 μl per fraction) were collected from the bottom of the centrifuge tubes and placed on ice for subsequent Western blot and fluorescence microscopic analyses.
Cell synchronization
To obtain cells that were arrested at the G1/S boundary, double thymidine block was carried out by first culturing HeLa cells in medium containing 2 mM thymidine overnight. After incubating in fresh medium for 8 h, a second overnight incubation in 2 mM thymidine was performed prior to washing and releasing the block in medium for 8 h. For mitotic arrest, cells were released into medium containing nocodazole (0.5 μg/ml) for 16 h. At the end of block or release, cells were collected for preparation of lysates. Double thymidine block was also employed to enrich mitotic cells expressing transfected Myc-sSgo1. Briefly, HeLa cells were cultured in medium containing 2 mM thymidine overnight. After washing, cells were cultured in fresh medium for 4 h before co-transfection with the Myc-sSgo1 construct and 3′UTR Sgo1 siRNA. Four h after transfection, cells were supplemented with medium containing 2 mM thymidine for 18 h. Cells were then washed and cultured in fresh medium for 10 h before being processed for fluorescent microscopy..
Western blot
HeLa cells were transfected with Sgo1 or Plk1 siRNA for 24 h or treated with nocodazole (0.5 μg/ml) for 16 h. Mitotic cells were collected by shake-off. The remaining cells were also harvested after trypsin treatment. An equal amount of proteins were subjected to immunoblotting with antibodies to Sgo1, Plk1 (Zymed), NuMA (Santa Cruz), PCNA (Sigma), γ-tubulin (Sigma), or β-actin (Sigma). Specific signals were detected with horseradish peroxidase–conjugated goat-anti-rabbit (or anti-mouse) secondary antibodies (Sigma) and enhanced chemiluminescence reagents (Amersham Pharmacia Biotech).
Supplementary Material
Acknowledgments
We thank Kenji Fukasawa and Adrian Salic for the GFP-centrin and mouse Sgo1 plasmids, respectively. We also thank Alexey Khodjakov for HeLa cell line stably expressing GFP-centrin, Duane Compton for the NuMA antibody, Tiansen Li for Rootletin antibody, and Jon Kajstura and Jane Lin for assistance in confocal microscopy. We are also grateful to Yuqiang Fang for technical assistance and to Lisa Buerle for technical and administrative assistance. The work is supported in part by grants from the National Institutes of Health (CA90658 and CA74229) to WD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnaud L, Pines J, Nigg EA. GFP tagging reveals human Polo-like kinase 1 at the kinetochore/centromere region of mitotic chromosomes. Chromosoma. 1998;107:424–429. doi: 10.1007/s004120050326. [DOI] [PubMed] [Google Scholar]
- Blomberg-Wirschell M, Doxsey SJ. Rapid isolation of centrosomes. Methods Enzymol. 1998;298:228–238. doi: 10.1016/s0076-6879(98)98022-3. [DOI] [PubMed] [Google Scholar]
- Clarke AS, Tang TT, Ooi DL, Orr-Weaver TL. POLO kinase regulates the Drosophila centromere cohesion protein MEI-S332. Dev Cell. 2005;8:53–64. doi: 10.1016/j.devcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Dionne MA, Howard L, Compton DA. NuMA is a component of an insoluble matrix at mitotic spindle poles. Cell Motil Cytoskeleton. 1999;42:189–203. doi: 10.1002/(SICI)1097-0169(1999)42:3<189::AID-CM3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Gregson HC, Schmiesing JA, Kim JS, Kobayashi T, Zhou S, Yokomori K. A potential role for human cohesin in mitotic spindle aster assembly. J Biol Chem. 2001;276:47575–47582. doi: 10.1074/jbc.M103364200. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. Two for two: Cdk2 and its role in centrosome doubling. Oncogene. 2002;21:6154–6160. doi: 10.1038/sj.onc.1205826. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Lee JY, Hayashi-Hagihara A, Orr-Weaver TL. Roles and regulation of the Drosophila centromere cohesion protein MEI-S332 family. Philos Trans R Soc Lond B Biol Sci. 2005;360:543–552. doi: 10.1098/rstb.2005.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- Mayor T, Meraldi P, Stierhof YD, Nigg EA, Fry AM. Protein kinases in control of the centrosome cycle. FEBS Lett. 1999;452:92–95. doi: 10.1016/s0014-5793(99)00534-7. [DOI] [PubMed] [Google Scholar]
- McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudjou M, Bordes N, Paintrand M, Bornens M. gamma-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J Cell Sci. 1996;109(Pt 4):875–887. doi: 10.1242/jcs.109.4.875. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Oshimori N, Ohsugi M, Yamamoto T. The Plk1 target Kizuna stabilizes mitotic centrosomes to ensure spindle bipolarity. Nat Cell Biol. 2006;8:1095–1101. doi: 10.1038/ncb1474. [DOI] [PubMed] [Google Scholar]
- Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Sauer K, Lehner CF. The role of cyclin E in the regulation of entry into S phase. Prog Cell Cycle Res. 1995;1:125–139. doi: 10.1007/978-1-4615-1809-9_10. [DOI] [PubMed] [Google Scholar]
- Schlaitz AL, Srayko M, Dammermann A, Quintin S, Wielsch N, MacLeod I, de Robillard Q, Zinke A, Yates JR, III, Muller-Reichert T, Shevchenko A, Oegema K, Hyman AA. The C. elegans RSA complex localizes protein phosphatase 2A to centrosomes and regulates mitotic spindle assembly. Cell. 2007;128:115–127. doi: 10.1016/j.cell.2006.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M, Hornig NC, Porstmann T, Uhlmann F. Studies on substrate recognition by the budding yeast separase. J Biol Chem. 2004;279:1191–1196. doi: 10.1074/jbc.M309761200. [DOI] [PubMed] [Google Scholar]
- Tang TT, Bickel SE, Young LM, Orr-Weaver TL. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes Dev. 1998;12:3843–3856. doi: 10.1101/gad.12.24.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Shu H, Qi W, Mahmood NA, Mumby MC, Yu H. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Tang Z, Sun Y, Harley SE, Zou H, Yu H. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci USA. 2004;101:18012–18017. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein KH, Kleylein-Sohn J, Nigg EA, Gruneberg U. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J Cell Biol. 2007;178:345–354. doi: 10.1083/jcb.200701163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Uhlmann F. The mechanism of sister chromatid cohesion. Exp Cell Res. 2004;296:80–85. doi: 10.1016/j.yexcr.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu T, Fang Y, Xie S, Huang X, Mahmood R, Ramaswamy G, Sakamoto KM, Darzynkiewicz Z, Xu M, Dai W. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103:1278–1285. doi: 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- Wang X, Dai W. Shugoshin, a guardian for sister chromatid segregation. Exp Cell Res. 2005;310:1–9. doi: 10.1016/j.yexcr.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Wang X, Yang Y, Dai W. Differential subcellular localizations of two human Sgo1 isoforms: implications in regulation of sister chromatid cohesion and microtubule dynamics. Cell Cycle. 2006;5:635–640. [PubMed] [Google Scholar]
- Warren WD, Steffensen S, Lin E, Coelho P, Loupart M, Cobbe N, Lee JY, McKay MJ, Orr-Weaver T, Heck MM, Sunkel CE. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr Biol. 2000;10:1463–1466. doi: 10.1016/s0960-9822(00)00806-x. [DOI] [PubMed] [Google Scholar]
- Watanabe Y. Sister chromatid cohesion along arms and at centromeres. Trends Genet. 2005;21:405–412. doi: 10.1016/j.tig.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wang X, Dai W. Human Sgo1 is an excellent target for induction of apoptosis of transformed cells. Cell Cycle. 2006;5:896–901. doi: 10.4161/cc.5.8.2691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.