Summary
We sought to determine if the innate immune response is under circadian regulation and whether this impacts overall health status. To this end, we used infection of Drosophila with the human opportunistic pathogenic bacteria Pseudomonas aeruginosa as our model system [1]. We show that the survival rates of wild-type flies vary as a function of when during the day they are infected, peaking in the middle of the night. Although this rhythm is abolished in clock mutant flies, those with an inactive period gene are highly susceptible to infection, whereas mutants with impairment in other core clock genes exhibit enhanced survival. After an initial phase of strong suppression, the kinetics of bacterial growth correlates highly with time-of-day and clock mutant effects on survival. Expression profiling revealed that night-time infection leads to a clock-regulated transient burst in the expression of a limited number of innate immunity genes. Circadian modulation of survival was also observed with another pathogen, Staphylococcus aureus. Our findings suggest that medical intervention strategies incorporating chronobiological considerations could enhance the innate immune response, boosting the efficacy of combating pathogenic infections.
Results and Discussion
Time-of-day and clock effects on the survival rates of Drosophila infected with P. aeruginosa
The innate immune response [2, 3] and circadian clock mechanisms [4] are both highly conserved between Drosophila and mammals. Pseudomonas aeruginosa is a human opportunistic pathogen commonly found in hospital-acquired infections [1], and studies using Drosophila have revealed numerous insights into understanding the pathogenicity of these gram-negative bacteria [5]. To test if the circadian system modulates the ability of D. melanogaster to combat a pathogenic infection, we first entrained control rhythmic flies (yw) under standard 12 hr light/12 hr dark cycles [LD; where zeitgeber time 0 (ZT0) is defined as lights-on] for 2 days and on the third day inoculated them at different times of day with the PA14-isogenic mutant strain of P. aeruginosa, which is defective in phopholipase C (PA14 plcs) [6] (Fig. 1A and B, and Fig. S1). The PA14 plcs strain is a less virulent strain than PA14 and was chosen in our studies because although infection with this attenuated strain evokes rapid mortality (between 1-2 days) of many flies, a sizable proportion survives throughout an extended post-infection observation period (at least 1 week in our standard experimental setup; termed ‘survivors’) (Fig. 1A and B), as previously reported [6]. By establishing conditions that yielded a mixed population response with individuals that succumbed quickly to the infection and those that survived over an extended period of time, this allowed us to better evaluate whether the clock modulates the ability of flies to successfully combat a pathogenic infection. Adult flies were infected by the standard method of lightly stabbing their abdomens with a fine needle dipped in a concentrated liquid culture containing PA14 plcs. We also included control groups that were contemporaneously mock-treated with needles placed in just the growth media (on average, 90-100% of the flies stabbed with control needles survived to the end of the test period; Table S1).
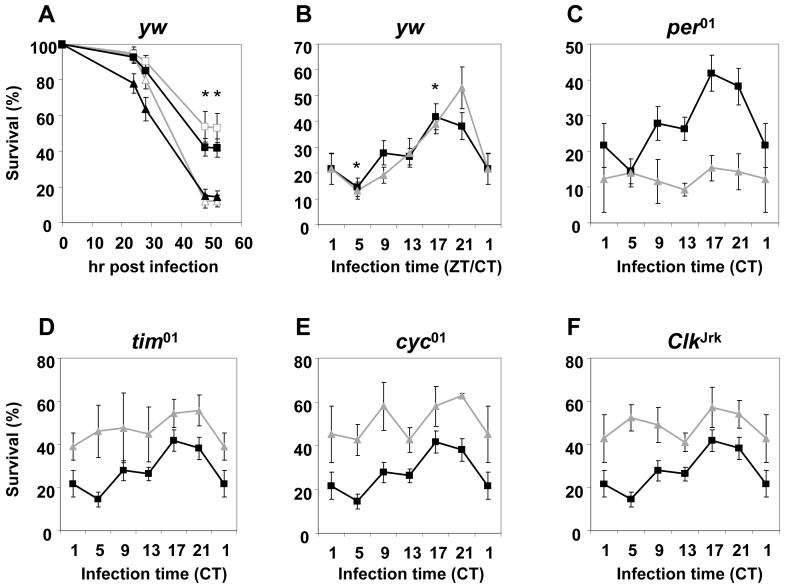
Figure 1. Time of infection during a daily cycle and the clock regulate the survival outcomes of flies infected with P. aeruginosa.
(A) Time course in the proportion of yw flies surviving after infection with PA14 plcs at ZT5 (open triangles, n = 251; indicates total number of flies from several independent experiments that were infected and used to calculate the average survival data shown) and ZT21 (open squares, n = 249) during LD, or CT5 (filled triangles, n = 321) and CT17 (filled squares, n = 321) during DD. Asterisks indicate significantly higher survival rates for the ZT21-or CT17-groups compared to the ZT/CT5-groups (two-tailed Student’s t-test; * denotes p < 0.005). (B) Survival rates of yw flies infected with PA14 plcs at different times of day during either LD (grey triangles, n = 246-253; i.e., indicates the range in the total number of flies from several independent experiments that were infected at the different times in a daily cycle) or DD (black squares, n = 315-323). Survival profiles were evaluated by one-way ANOVA followed by Tukey-Kramer HSD analysis with the following results; (1) in LD, the ZT21-group survived better than the ZT1-, ZT5-, or ZT9- groups (one-way ANOVA, p < 0.0005; Tukey-Kramer HSD, α = 0.01). At α = 0.001, only the ZT5-group died significantly more than the ZT21-group; (2) in DD, the CT17- and CT21-groups have higher survival rates compared to the CT5-group (one-way ANOVA, p < 0.005; Tukey-Kramer HSD, α = 0.05). At α = 0.01, only the CT17-group exhibited significantly higher survival compared to the CT5-group (*). (C-F) Survival rates of clock mutant flies (grey triangles) infected with PA14 plcs during DD compared to control yw flies (black squares). Results reflect the average of at least three independent experiments. Error bars indicate S.E.M.
Control (yw) flies exhibit a diurnal profile in their survival rates (one-way ANOVA, p < 0.0005 for Figs. 1A and B). Tukey-Kramer HSD analysis indicated that control flies infected at ZT21 have significantly higher rates of survival than those infected at ZT1, 5, or 9 (when α = 0.01; see also legend to Fig. 1). Flies infected at ZT21 survived approximately 4-fold better compared to the trough values at ZT5 (two-tailed Student’s t-test, p < 0.005 at 48 hrs post infection and thereafter) (Figs. 1A and B). We observed a similar daily pattern in survival rates when inoculating flies with bacterial titers 5- to 20-fold lower compared to our standard conditions (compare Figs. 1A and B to S1), demonstrating that the time-of-day effects on survival are observed over a broad range of initial bacterial doses.
To determine if the survival rhythm is endogenously driven, flies were entrained by three LD cycles and subsequently maintained in constant darkness (DD) followed by inoculation on the second day of DD. In addition, we also infected the well-characterized per01, tim01, ClkJrk and cyc01 arrhythmic clock mutants that carry inactivated period (per), timeless (tim), clock (Clk) and cycle (cyc) genes, respectively [4]. To minimize genetic background effects, the clock mutants were evaluated in the same yw background as the control strain. The daily profile of the survival rate for control rhythmic flies in DD (Fig. 1B; one-way ANOVA, p < 0.005; statistical analysis summarized in legend to Fig. 1 and Table S2) was almost identical to that observed in LD except that flies infected at circadian time 17 (CT17; in this manuscript we use the term CT as equivalent to ZT, which is a reasonable approximation given the near 24 hr behavioural rhythms of Drosophila) showed the best survival rates (Tukey-Kramer HSD when α = 0.01), indicating the survival rhythm is under circadian regulation. For example, flies infected at CT17 survived approximately 3-fold better compared to the trough values observed at CT5 (two-tailed Student’s t-test, p < 0.005 at 48 hrs post infection and thereafter) (Fig. 1A and B; Table S2). Importantly, similar results whereby survival rates are significantly higher at CT17 compared to CT5 were also observed when using rhythmic flies with different genetic backgrounds, including the Canton-S (CS) and Oregon R (OR) wildtype strains (Fig. 2A; two-tailed Student’s t-test, p < 0.05; and data not shown).
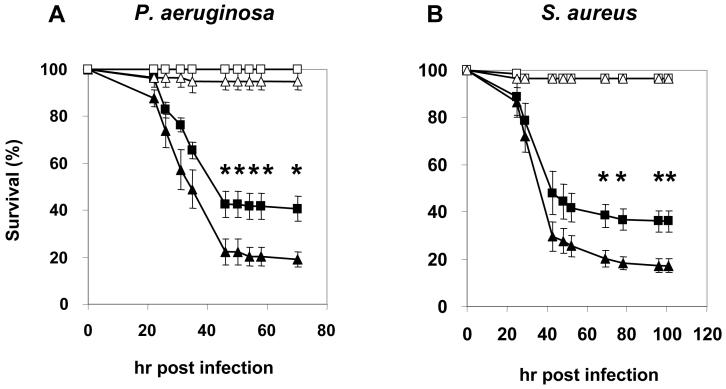
Figure 2. Night-time infections lead to higher survival rates in a variety of wildtype D. melanogaster strains infected with either P. aeruginosa or S. aureus.
(A) Time course depicting the survival rates of wildtype flies (Canton-S and Oregon R) infected with PA14 plcs at CT5 (filled triangles) and CT17 (filled squares) on the second day of DD; also shown, mock-injury groups pricked at CT5 (open triangles) and CT17 (open squares). Very similar survival curves were obtained for Canton-S and Oregon R flies (data not shown), and hence the data were pooled. Results are the average of four independent experiments and indicate significantly higher survival rates for flies infected at CT17 compared to CT5 (two-tailed Student’s t-test; CT5 vs. CT17, *denotes p < 0.05). Error bars indicate S.E.M. The number of flies analyzed is as follows: n = 151 for the infected CT5-group, n = 154 for the infected CT17-group, n = 70 for the mock injury CT5- or CT17- groups. (B) Time course depicting the survival rates of wildtype flies (Canton-S and Oregon R) infected with S. aureus at CT5 (filled triangles) and CT17 (filled squares); also shown, mock-injury groups pricked at CT5 (open triangles) or CT17 (open squares). Results are the average of four independent experiments and indicate significantly higher survival rates for flies infected at CT17 compared to CT5 (two-tailed Student’s t-test; CT 5 vs. CT 17, *denotes p < 0.05). Error bars indicate S.E.M. The number of flies analyzed is as follows: n = 160 for the infected CT5- or CT17-group, n = 80 for the mock injury CT5- or CT17-group.
In contrast to rhythmic control and wildtype strains, a daily survival rhythm was not observed in the four arrhythmic clock mutant strains tested (Fig. 1C-F; one-way ANOVA, p > 0.05, Table S2). More extensive analysis comparing CT5 and CT17 groups did not reveal significant time-of-day differences in the survival rates of per01 or ClkJrk flies (Table S3). Although we cannot rule out the possibility of small but real time-of-day variations in the percent survival of the clock mutants (Tables S2 and S3), our data indicate that the robust daily rhythm observed in control and wildtype flies is either abolished or greatly attenuated in the mutants. In addition, close inspection of the survival patterns of the clock mutants suggests the possibility of low amplitude cycles that peak twice per day (Fig. 1C-F). Intriguingly, in many cases higher frequency ‘ultradian’ rhythms are more readily observed or enhanced when circadian systems are severely compromised [7].
Although we do not observe robust daily rhythms in survival for the different arrhythmic clock mutants analyzed, per01 flies manifested relatively higher mortality rates compared to control flies, whereas tim01, ClkJrk and cyc01 flies showed overall enhanced survival compared to their background controls (Fig. 1C-F). Similar results were also obtained when examining the survival patterns of the clock mutants in LD, except that mortality of all the clock mutants tested was overall slightly higher during the daytime, suggesting that in these mutants the light/dark conditions have direct effects on the ability to survive the infection (data not shown). While our manuscript was under review, Shirasu-Hiza et al. (2007) reported that per01 flies were more susceptible to Streptococcus pneumoniae and Listeria monocytogenes compared to wildtype flies [8], consistent with our findings. However, in that study tim01 flies also succumbed to death faster than wild-type flies when infected with the same pathogens. The possible discrepancy between the two studies with regards to the ability of tim01 flies to survive pathogenic infections is presently unclear and might be due to the use of different bacteria and/or the mode of pathogenicity. Although the reasons underlying the differential effects of clock mutations are not known, the collective findings suggest that per function plays a protective role in Drosophila infected with pathogenic bacteria. Future studies will be required to better evaluate the roles of the different clock genes in innate immunity.
To expand our observations we also inoculated wildtype flies with another human pathogenic bacteria, Staphylococcus aureus (S. aureus), which unlike P. aeruginosa is gram-positive. As with P. aeruginosa, the CT17-group exhibited higher rates of survival compared to the CT5-group (Fig. 2B; two-tailed Student’s t-test, p < 0.05). Taken together, the findings indicate that Drosophila survive night-time infections significantly better than day-time ones.
Bacterial growth kinetics correlate with survival rates in rhythmic and clock mutant flies
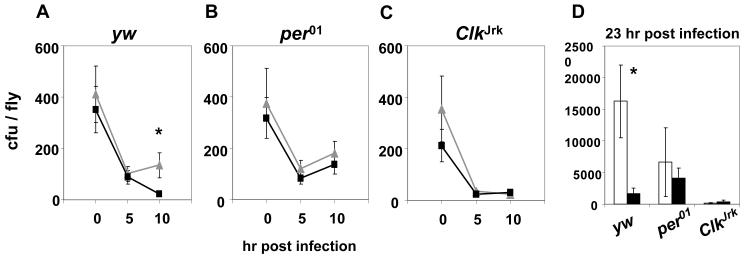
To determine whether the kinetics of bacterial growth correlate with the survival patterns observed, we infected control, per01 and ClkJrk flies on the second day of DD at either CT5 or 17, the trough and peak times for survival rates, respectively. Live flies were collected at several times post-inoculation and bacterial titers measured (Fig. 3). In each experiment, all three genotypes were contemporaneously treated and the results from multiple experiments pooled. Also, a subset of flies were not processed for the bacterial assays but were scored for survival and in rare cases where anomalous survival results were obtained (e.g., little or no mortality) the bacterial data from that experiment was not used. Irrespective of the infection time, all genotypes showed strong decreases in bacterial titers during the first 5 hr post-infection (Fig. 3A-C). However, in control yw flies, the CT5-group had significantly higher bacterial loads at 10 and 23 hr post-infection compared to the CT17-group (two-tailed Student’s t-test, p < 0.05 for 10 hr, p = 0.01 for 23h), whereas per01 and ClkJrk flies did not exhibit significant differences in bacterial loads as a function of infection time (Fig. 3A-D; statistical analysis summarized in Table S4). Nonetheless, the titer of PA14 plcS in per01 flies increased between 5 to 10 hr post-infection but remained very low in ClkJrk flies (Fig. 3B and C). Indeed, pair-wise comparisons indicated significantly higher levels of bacteria in per01 flies at 10 and 23 hr post-infection compared to ClkJrk flies (Table S4), consistent with its relatively lower survival rates (Figs. 1 and S2).
Figure 3. Bacterial growth correlates with time of infection and clock mutant effects on survival rates.
(A-D) Flies of the indicated genotype were infected at either CT5 (grey triangles or open bars) or CT17 (black rectangles or filled bars) and collected at the indicated times. Asterisks indicate significantly higher bacterial titer for the control CT5-group compared to the CT17-group (two-tailed Student’s t-test, p <0.05). Results from at least three independent experiments were combined. Mean ± S.E.M. and values obtained from at least 60 flies per collection time are displayed (n = 60-128, 95-130, 67-90 for yw, per01, and ClkJrk flies, respectively).
To further demonstrate that per01 flies have higher mortality rates compared to ClkJrk flies, we infected per01 flies with approximately half the number of bacteria as ClkJrk flies (Fig. S2A) and compared their survival rates (Fig. S2B). Despite the lower bacterial load used in the infection, significantly more per01 flies died compared to ClkJrk flies (two-tailed Student’s t-test, p < 0.01 after 52 hrs post infection, α = 0.05) Fig. S2B). These results are consistent with the observation that similar survival rhythms are observed in control flies over a wide range of initial bacterial doses (Fig. S1). Thus, the time-of-day differences observed in rhythmic flies, and the enhanced survival of ClkJrk compared to per01 flies cannot be accounted by possible experimental variations in the amount of bacteria used during inoculation.
In summary, after an initial phase of bacterial clearance, there is a tight correlation between bacterial loads and survival outcomes, both for wild-type flies as a function of circadian time and when comparing clock mutants. Indeed, the early bacterial growth pattern in per01 flies mimics that observed in the wild-type CT5-group, increasing after 5 hr post-infection, whereas the ClkJrk response is more similar to the wild-type CT17-group, which declines or remains low at 10 hr post-infection. Thus, our findings suggest that the ability to suppress bacterial growth during the first 10 hr after infection is causally linked to better prognosis for survival.
Clock regulation in the induced profiles of innate immunity genes is highly selective and restricted to the early phase of the infection
The best-studied defense effectors in innate immunity are antimicrobial peptide genes (AMPs) that are rapidly induced following microbial infection [2]. In Drosophila, AMPs are primarily induced via activation of the Toll (mainly responding to fungi or gram-positive bacteria) and/or Imd (mainly responding to gram-negative bacteria) pathways [2]. As an initial attempt to understand the molecular mechanisms underlying the circadian pattern of survival rates and bacterial growth kinetics, we largely focused on the expression patterns of several key players in the Toll and Imd innate immune signalling pathways activated by microbial infection. This included measuring the post-inoculation expression kinetics of several peptidoglycan recognition proteins (PGRP) shown to play key roles as microbial receptors and/or scavengers (e.g., PGRP-SA, -LC and -LB), AMPs [i.e. attacin A (attA), defensin (def), diptericin (dipt), drosocin (drc) and drosomycin (drs)], and some key signalling components such as imd. Control yw, per01 and ClkJrk flies were infected during the second day of DD and collected at different times post-infection. We used real-time quantitative RT-PCR to measure RNA levels in head extracts of adult flies because we noted that the induced levels of many immune relevant genes attain higher values in heads compared to isolated bodies or whole flies (e.g., compare Figs. 3 and S4; data not shown). An immune response in the head has also been described elsewhere [9] (data not shown). For each genotype and gene surveyed we compared the RNA values obtained at the same post-infection time point for the CT5 and CT17 infected groups.
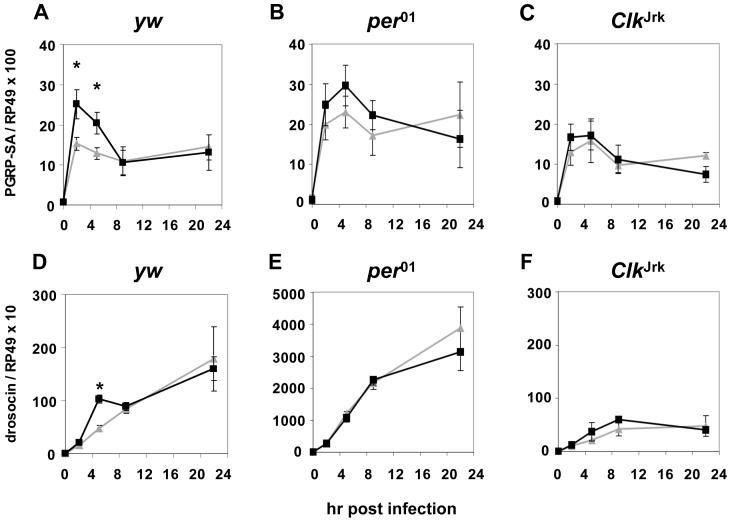
Intriguingly, although many immune relevant genes are induced following infection with Pseudomonas [10], the post-inoculation expression patterns of only PGRP-SA and drc showed differences as a function of infection time in control flies, which were abolished in the clock mutants, indicative of bona-fide circadian regulation (Figs. 4 and S3). For PGRP-SA, its mRNA levels at 2 and 5 hr post-infection are significantly higher in the CT17-group compared to the values obtained at the same post-infection time points in the CT5-group (Fig. 4A), whereas for drc higher mRNA levels were observed at 5 hr post-infection in the CT17-group compared to the same post-infection time point in the CT5-group (Fig. 4D). Similar circadian patterns in the induction profiles for PGRP-SA and drc were also obtained when analyzing extracts prepared from isolated bodies (Fig. S4).
Figure 4. Night-time infection leads to early and transient clock-regulated increases in the mRNA induction profiles of a limited number of immune response genes.
(A-F) Flies were infected at either CT5 (grey triangles) or CT17 (black squares), collected at the indicated times and RNA levels measured. For each genotype we compared the RNA values for the CT5 and CT17 groups that were obtained at the same post-infection time point. Asterisks indicate significantly higher mRNA levels for the yw CT17-group compared to the yw CT5-group (two-tailed Student’s t-test, p < 0.0005 for drosocin, p < 0.05 for PGRP-SA). Results from at least three independent experiments were averaged except that ClkJrk data were derived from two experiments. Error bars indicate S.E.M.
Besides imd, none of the immune relevant genes we probed exhibit circadian fluctuations in basal levels (data not shown; e.g., endogenous levels of imd are higher at CT17 compared to CT5 in control flies, Fig. S3M, compare values at time 0 post-infection). This is consistent with prior work using microarrays to probe daily patterns of expression in head extracts [11]. However, we did not observe time-of-day differences in the induced levels of imd following infection (Fig. S3M). Thus, whether the basal expression of an immune response gene is constitutive or circadian is not necessarily linked to how the clock regulates its expression post-infection. Although it is not clear why the induced profile of drc and not other AMP genes exhibits circadian differences as a function of infection time (compare Figs. 4 and S3), drosocin kills gram-negative bacteria [2] and has been demonstrated to be one of only a few AMPs that when overexpressed can protect flies infected with P. aeruginosa [10]. In this context it is interesting to note that although PGRP-SA has a characterized role as a receptor in the Toll pathway [12], it has recently been implicated in phagocytosis as well [13].
Together, our findings suggest the following scenario for how the clock in Drosophila might influence the progression of an infection with P. aeruginosa. Early in the infection a robust immune response (perhaps both cellular and humoral) is mounted, which is effective in pathogen clearance irrespective of when during a daily cycle the flies are infected, as indicated by the rapid drop in bacterial titer during the first 5 hr post-infection (Fig. 3A). However, the clock regulates the induced levels of a limited subset of innate immunity players, such as PGRP-SA and drc, whereby infections in the mid-night result in a transient burst early during the infection (Fig. 4A and D). Higher levels of a few key immune players over a certain threshold may contribute to keeping the titer of pathogenic bacteria low after the initial rapid declining phase (Fig. 3A). By prolonging the suppression of bacterial growth during a critical window of the infection, this might provide an opportunity to mount or recruit additional host defenses in addition to AMPs, resulting in improved survival (Figs. 1A and B). Thus, our results suggest that the clock modulates the strength or responsiveness of immune activation in a time-of-day dependent manner but only during a critical early phase in the infection process that has physiological consequences on the ability of the host to survive pathogenic infections. Indeed, it is noteworthy that P. aeruginosa eludes host defenses by the early suppression of antimicrobial peptide gene expression [10]. It will be of interest to determine why the post-infection expression profiles of only certain immune response genes exhibit circadian regulation and how this is apparently restricted to a particular phase of the immune response.
Although the time-of-day differences in the levels of induced PGRP-SA and drc are clearly consistent with the survival rates of wild-type flies infected at different times of day, this is not the case for the per01 and ClkJrk mutants where the overall levels of drc are much lower in ClkJrk compared to per01 flies (Fig. 4; a trend observed for other AMP genes surveyed, Fig. S3 and data not shown). Although seemingly paradoxical, this is not unanticipated as there are precedents in the literature showing that flies can be more susceptible to bacterial infection despite elevated levels of AMP expression, indicating that excessive or inappropriate immune activation can be deleterious [14-17]. In this context it is important to consider that besides the production of AMPs, innate immunity in adult Drosophila includes a proteolytic cascade leading to melanization and a cellular immune response characterized by phagocytosis [18]. It is possible that inactivation of per might affect other host defense mechanisms that cannot be compensated by a potentially hypersensitive humoral immune response. Conversely, ClkJrk and other clock mutant flies (Fig. 1) might have a heightened activity of cellular immunity. Presently, our results based on probing the expression profiles of several immune response genes (Figs. 4 and S3) would seem to demand that the molecular mechanisms governing the time-of-day differences in survival for flies with functional clocks are different from those affecting survival rates in per01 and ClkJrk flies. Otherwise stated, it does not appear likely that the survival rates of per01 and ClkJrk flies are simply due to their ‘clocks’ being pegged or held at a phase that is similar to either ZT/CT5 or ZT/CT17 in wildtype flies, respectively. While future work will be required to resolve the molecular underpinnings governing the differential clock mutant effects on survival, these considerations suggest that core clock genes have ‘non-circadian’ related roles (i.e., roles not solely limited to their functions in timekeeping) in fighting microbial infections. Indeed, our findings add to a growing list of physiological and behavioral pathways that are differentially regulated in different clock mutants; e.g., mutations in per but not tim, Clk or cyc play a key role in long-term memory formation in Drosophila [19].
If the ability to evoke a stronger response at night enhances the efficacy of fighting a microbial infection, why restrict it to the night? It is widely thought that maintaining an optimal immune system is metabolically costly, competing for limited metabolic resources with other energetically demanding activities such as foraging or mating [20]. Within this framework we suggest that the clock might function as a temporal sieve to ensure the proper allocation of metabolic resources at biologically desirable times. On a more medical perspective, our results suggest that the innate immune system is a prime target for interventions based on chronobiological considerations in the hopes of boosting the ability to combat pathogenic infections.
Supplementary Material
Acknowledgments
We are grateful to D. Karahentsev and S. Minakhina for helping us set up the infection procedure, L. Rahme for PA14 strains and technical advice, B. Lazzaro for inspiring us to use P. aeruginosa and B. Lemaitre for helpful communication. We also thank A. Sehgal for ywper01 and yw;;tim01 flies, J. Hall for yw;;cyc01 flies, R. Allada for yw;;ClkJrk flies, and S. Kurata for Oregon-R flies. This work was partially supported by an N.I.H. grant (NS-34958) to I.E.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Van Delden C, Iglewski BH. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–38. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 3.Kimbrell DA, Beutler B. The evolution and genetics of innate immunity. Nat Rev Genet. 2001;2:256–267. doi: 10.1038/35066006. [DOI] [PubMed] [Google Scholar]
- 4.Stanewsky R. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J Neurobiol. 2003;54:111–147. doi: 10.1002/neu.10164. [DOI] [PubMed] [Google Scholar]
- 5.Vodovar N, Acosta C, Lemaitre B, Boccard F. Drosophila: a polyvalent model to decipher host-pathogen interactions. Trends Microbiol. 2004;12:235–242. doi: 10.1016/j.tim.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Lau GW, Goumnerov BC, Walendziewicz CL, Hewitson J, Xiao W, Mahajan-Miklos S, Tompkins RG, Perkins LA, Rahme LG. The Drosophila melanogaster toll pathway participates in resistance to infection by the gram-negative human pathogen Pseudomonas aeruginosa. Infect Immun. 2003;71:4059–4066. doi: 10.1128/IAI.71.7.4059-4066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shirasu-Hiza MM, Dionne MS, Pham LN, Ayres JS, Schneider DS. Interactions between circadian rhythm and immunity in Drosophila melanogaster. Curr Biol. 2007;17:R353–355. doi: 10.1016/j.cub.2007.03.049. [DOI] [PubMed] [Google Scholar]
- 9.Libert S, Chao Y, Chu X, Pletcher SD. Trade-offs between longevity and pathogen resistance in Drosophila melanogaster are mediated by NFkappaB signaling. Aging Cell. 2006;5:533–543. doi: 10.1111/j.1474-9726.2006.00251.x. [DOI] [PubMed] [Google Scholar]
- 10.Apidianakis Y, Mindrinos MN, Xiao W, Lau GW, Baldini RL, Davis RW, Rahme LG. Profiling early infection responses: Pseudomonas aeruginosa eludes host defenses by suppressing antimicrobial peptide gene expression. Proc Natl Acad Sci U S A. 2005;102:2573–2578. doi: 10.1073/pnas.0409588102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 12.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 13.Garver LS, Wu J, Wu LP. The peptidoglycan recognition protein PGRP-SC1a is essential for Toll signaling and phagocytosis of Staphylococcus aureus in Drosophila. Proc Natl Acad Sci U S A. 2006;103:660–665. doi: 10.1073/pnas.0506182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukae N, Yokoyama H, Yokokura T, Sakoyama Y, Nagata S. Activation of the innate immunity in Drosophila by endogenous chromosomal DNA that escaped apoptotic degradation. Genes Dev. 2002;16:2662–2671. doi: 10.1101/gad.1022802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsichritzis T, Gaentzsch PC, Kosmidis S, Brown AE, Skoulakis EM, Ligoxygakis P, Mosialos G. A Drosophila ortholog of the human cylindromatosis tumor suppressor gene regulates triglyceride content and antibacterial defense. Development. 2007;134:2605–2614. doi: 10.1242/dev.02859. [DOI] [PubMed] [Google Scholar]
- 17.Tsuda M, Langmann C, Harden N, Aigaki T. The RING-finger scaffold protein Plenty of SH3s targets TAK1 to control immunity signalling in Drosophila. EMBO Rep. 2005;6:1082–1087. doi: 10.1038/sj.embor.7400537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hultmark D. Drosophila immunity: paths and patterns. Curr Opin Immunol. 2003;15:12–19. doi: 10.1016/s0952-7915(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 19.Sakai T, Tamura T, Kitamoto T, Kidokoro Y. A clock gene, period, plays a key role in long-term memory formation in Drosophila. Proc Natl Acad Sci U S A. 2004;101:16058–16063. doi: 10.1073/pnas.0401472101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmid-Hempel P. Evolutionary ecology of insect immune defenses. Annu Rev Entomol. 2005;50:529–551. doi: 10.1146/annurev.ento.50.071803.130420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.