Abstract
Although protein degradation is enhanced in muscle-wasting conditions and limits the rate of muscle growth in domestic animals, the proteolytic system responsible for degrading myofibrillar proteins in skeletal muscle is not well defined. The goals of this study were to evaluate the roles of the calpains (calcium-activated cysteine proteases) in mediating muscle protein degradation and the extent to which these proteases participate in protein turnover in muscle. Two strategies to regulate intracellular calpain activities were developed: overexpression of dominant-negative m-calpain and overexpression of calpastatin inhibitory domain. To express these constructs, L8 myoblast cell lines were transfected with LacSwitch plasmids, which allowed for isopropyl β-d-thiogalactoside-dependent expression of the gene of interest. Inhibition of calpain stabilized fodrin, a well characterized calpain substrate. Under conditions of accelerated degradation (serum withdrawal), inhibition of m-calpain reduced protein degradation by 30%, whereas calpastatin inhibitory domain expression reduced degradation by 63%. Inhibition of calpain also stabilized nebulin. These observations indicate that calpains play key roles in the disassembly of sarcomeric proteins. Inhibition of calpain activity may have therapeutic value in treatment of muscle-wasting conditions and may enhance muscle growth in domestic animals.
Keywords: fodrin/nebulin
Two proteolytic systems have been studied with regard to their role in muscle protein wasting: calpains and the proteasome. Calpains are calcium-activated cysteine proteases, which were originally identified in porcine muscle (1, 2). Two ubiquitous isoforms are well characterized (μ- and m-calpain), and several tissue-specific isoforms have also been reported (3). Calpains are regulated by a variety of factors, including a 30-kDa small subunit (3), calcium and phospholipids (3–6), and calpastatin, a widely distributed calpain-specific inhibitor (7–15). Calpastatin possesses four inhibitory domains that contain a consensus inhibitory sequence (TIPPEYR), which, in itself, is able to inhibit calpain specifically (8, 11–15).
Evidence that calpains mediate degradation of the myofibrillar apparatus is compelling. In vitro, calpains initiate digestion of individual myofibrillar proteins, including desmin, filamen, C-protein, tropomyosin, troponin T, troponin I, titin, nebulin, vimentin, gelsolin, and vinculin (16–25). However, calpains do not degrade α-actin, α-actinin, or myosin heavy chain (23, 24). Calpains tend to be concentrated in the Z disk (26), the site where disassembly begins (27). Calpains are activated by calcium, and treatment of purified myofibrils with calcium causes rapid and complete loss of the Z disk (28). Calpains are also activated in conditions of muscle wasting, including vitamin E deficiency (29), Duchenne muscular dystrophy (30), and fasting (31). Calpain mRNA concentrations are increased markedly during fasting (32). The course of changes in calpain activity over time corresponds to morphological changes in muscle (33). Based on these data, Goll et al. (23, 24) have proposed that calpains play a significant role in myofibrillar protein degradation, especially in the disassembly of the myofibril during early stages of turnover.
Despite this, some reports have shown that the proteasome is also involved in myofibrillar protein degradation (34–39). The proteasome, a large, ubiquitous ATP- and ubiquitin-dependent proteolytic system, is able to degrade actin and myosin in vitro (35). A recent study by Solomon et al. (35) indicated that the proteasome degrades intact monomeric myofibrillar proteins, except when they are associated with other myofibrillar proteins.
Previous studies are limited by being performed in vitro with nonspecific protease inhibitors. To assess the function of calpains in living muscle cells, we developed two genetic strategies to regulate calpain activity: overexpression of dominant-negative (DN) m-calpain and overexpression of calpastatin inhibitory domain (CID). Our expectation was that specific regulation of calpains in living muscle cells would reveal calpain function. Our data indicate that calpains play significant roles in L8 muscle-cell protein degradation and participate in the degradation of nebulin. These data indicate that inhibition of calpains may effectively slow myofibrillar protein digestion in vivo.
MATERIALS AND METHODS
Plasmid Construction.
Site-directed mutagenesis of rat m-calpain. Rat m-calpain cDNA was supplied by John Elce (Queen’s University, Kingston, ON, Canada). Site-directed mutagenesis was performed with the Kunkel method and Muta-Gene T7 Enzyme Refill Pack Version 2 (Bio-Rad; ref. 40) to introduce a codon that encoded alanine in place of cysteine in the m-calpain active site. An oligonucleotide (5′-AGCCAGAAGCCAGGCGCTCCCAAGGGCTCC-3′, synthesized by the Central Services Lab, Oregon State University), was phosphorylated with T4 kinase (Promega) as a primer for synthesis of a cDNA strand. Successful mutation at 105 amino acid (cysteine → alanine) was verified first by HgaI restriction-enzyme digestion (a new restriction site generated after mutation) and sequencing. The DN-m-calpain gene, digested with NcoI and SalI, was subcloned with NotI linkers into pOP13CAT (Stratagene) expression vector after removal of the chloramphenicol acetyltransferase (CAT) gene. This plasmid was designated pOP13DN. Correct orientation of the insert was confirmed by sequencing and restriction-enzyme digestion.
Construction of CID.
The CID nucleotide sequence (Fig. 1A) was deduced from peptide EKLGERDDTIPPEYRELLEKKTGV (8) with standard mammalian codon usage. A translation start codon (ATG), Kozak sequence, and NotI site were added sequentially to the 5′ end. A translation stop codon (TGA) and NotI site were added at the 3′ end (Fig. 1B). This 97-bp full-length single-stranded CID expression construct was synthesized. Double-stranded full-length CID expression construct was generated by PCR with a forward primer P1 (5′-TGCGGCCGCCATGGAGAAGCT-3′) and a reverse primer P2 (5′-TGCGGCCGCTCACACGCCGGT-3′). Double-stranded CID cDNA was subcloned into pOP13CAT expression vector and designated pOP13CID. Correct orientation of the insertion was confirmed by sequencing.
Figure 1.
CID construct. (A) Complete CID nucleotide sequence. (B) CID construct containing CID nucleotide sequence (lowercase letters in the middle), Kozak sequence (bold italic), translation start codon ATG (underlined), translation stop codon TGA (underlined), and NotI sites (5′ and 3′).
Cell Culture.
Rat L8 myoblasts and transfectants were cultured in DMEM (GIBCO) with addition of 1000 mg/liter d-glucose, 584 mg/liter glutamine, 110 mg/liter sodium pyruvate, 4 mg/liter pyridoxine hydrochloride, 3.7 g/liter sodium bicarbonate, 100 units/ml penicillin-streptomycin (GIBCO), and 10% fetal bovine serum (HyClone). When cells reached 90–95% confluence, the medium was changed to complete DMEM containing 2% horse serum medium (HyClone) to obtain fully differentiated myotubes. The medium was changed every other day for either myoblast or myotubule maintenance.
Stable Cotransfection.
Calcium phosphate transfection (40) was used for preparation of cell lines. A day before transfection, one 10-cm plate of myoblasts at 80–90% confluence was split into four 10-cm plates. On the day of transfection, cells were fed with 9.0 ml of fetal bovine serum medium 4 h before calcium phosphate precipitation. Plasmids were purified by CsCl gradient centrifugation (40). Antibiotic selection with hygromycin B (200 μg/ml; GIBCO) was used for selection of cells containing the p3′SS repressor plasmid, and geneticin (400 μg/ml; GIBCO) was used for selection of the cells transfected with pOP13CAT, pOP13CID, or pOP13DN. When the clones were large enough to be visualized without a microscope, each clone was transferred from the original plate and cultured sequentially in a 24-well plate, a 12-well plate, a 6-well plate, a 5-cm plate, and finally, a 10-cm plate. We selected 20 clones of each cotransfectant.
The clones cotransfected with p3′SS (repressor plasmid; Stratagene) and pOP13CAT (expression vector) were named L8/PC (plasmid control cell line). The clones cotransfected with p3′SS and pOP13DN were named L8/DN (dominant negative cell line). The clones cotransfected with p3′SS and pOP13CID were named L8/CID (CID cell line). The ability to differentiate was tested for each transfectant. Only the transfectants that were able to differentiate fully were used for further experiments.
Reverse Transcription–PCR (RT-PCR).
RT-PCR assays were used to study expression of the various constructs in cultured cells. RNA was isolated by using the guanidinium-extraction method (40). RT-PCR was performed in two separate steps: reverse transcription to generate the single-stranded cDNA and PCR to amplify the cDNA. RNA used for the RT reaction was treated with RQ1 DNase (RNase-free, Promega) to minimize genomic DNA contamination. Oligo(dT) 18-mer primers were used for the RT reaction.
PCR.
The m-calpain RT-PCR consisted of 2 μl of RT reaction as a template, 1 μl of forward primer RT-anti-P (5′-GATGAAACTGGCCAAAGA-3′), and 1 μl of reverse primer m-P2 (5′-AGCTCCTCTGGGACCTCATAGATG-3′). The reaction mixture in this and in all other PCRs also contained 1.5 mM MgCl2, 0.2 mM 2′-deoxyribonucleoside 5′-triphosphate, 1× Taq DNA polymerase buffer, and 0.5 μl of Taq DNA polymerase (Promega). The cycle used in this and in other PCRs was 30 cycles at 94°C for 1 min, 55°C for 1.5 min, and 74°C for 1 min. pOP13DN was used as the template in one reaction as a positive control.
The CID RT-PCR included 10 μl of RT reaction as a template, forward primer P1 (5′-CATGGAGAAGCTGGGCGA-3′), and 1 μl of reverse primer P2 (5′-TCACACGCCGGTCTTCTT-3′). pOP13CID was used as the template in one reaction as a positive control.
The CAT RT-PCR (positive control for induction of isopropyl β-d-thiogalactoside; IPTG) included 10 μl of RT reaction as a template, 1 μl of forward primer CAT1 (5′-ATGGAGAAAAAAATCACTGGATAT-3′), and 1 μl of reverse primer CAT2 (5′-TTACGCCCCGCCCTGCCACTCAT-3′). pOP13CAT was used as a template in one reaction as a positive control.
The LacI RT-PCR included 6 μl of RT reaction as a template, 1 μl of forward primer lacP1 (5′-TGTCGATGGTAGAAGGAAG-3′), and 1 μl of reverse primer lacP2 (5′-GTGGTTTTTCTTTTCACCAG-3′). p3′SS was used as the template in one reaction as a positive control. All of the primers were 50 pmol/μl.
Products from each PCR (15 μl) were used for gel electrophoresis. A 1% TAE agarose gel was used for the 1100-bp m-calpain RT-PCR product, the 680-bp CAT gene RT-PCR product, and the 600-bp lacI RT-PCR product. A 2% TAE agarose gel was used for the 80-bp CID RT-PCR product.
Measurement of Total Protein Degradation.
After 6 days of differentiation, cells were supplied with 2 μCi/ml of [3H]tyrosine (NET-127, DuPont/NEN), and 5 mM IPTG was added to one-half of the plates. After 24 h, the plates were washed with DMEM containing 2 mM tyrosine (chase) and refilled with DMEM containing 2 mM tyrosine. At that time, 1.5 ml of medium was taken from each plate, and radioactivity was measured by scintillation counter. This measurement was designated as the radioactivity present at time zero. Time zero plates from cultures not treated with IPTG were described as T0, whereas time zero plates from cultures treated with IPTG were described as T0I. The rest of the plates were cultured for an additional 6 or 12 h. Of these, half continued to be treated with IPTG. After 6 (T6 or T6I) or 12 h (T12 or T12I), 1.5 ml of medium from each plate was taken, and radioactivity was measured. From these measurements, protein degradation (D) at 6 (D6 or D6I) or 12 h (D12 or D12I) and the percentage reduction of protein degradation caused by addition of IPTG were calculated.
Protein Extraction and Western Blotting.
The methods used in this study for recovery of cell protein were similar to those reported by Wang et al. (41). The protein concentration was measured with a protein assay kit (Bio-Rad). Standard techniques (40) were used for electrophoresis and transfer of proteins to Optitran membranes (Schleicher & Schuell) and for blotting. A 4–12.5% gradient gel with a 100:1 ratio of acrylamide to bisacrylamide with no stacking gel was used for nebulin protein separation. A 7.5% separating gel and 5% stacking gel were used for m-calpain and LacI protein separation. A 4–20% gradient gel and 4% stacking gel were used for fodrin protein separation.
The blots were rinsed with 50 mM Tris-base, pH 7.5, and 150 mM NaCl (TBS), blocked by 5% skim milk in TBS plus 0.05% Tween-20 (TTBS) for 1 h, and probed with nebulin (1:5000, A-9891, Sigma), LacI (1:1000, Stratagene), m-calpain (1:2000), and spectrin antibodies (1:1000, Chemicon) for 2.5 h. After antibody probing, the blots were washed with TTBS for 30 min and transferred into a secondary antibody solution with 1:5000 dilution of either anti-rabbit or anti-mouse IgG (Bio-Rad) conjugate for at least 30 min. After another 30-min wash, the blots were developed with either an enhanced chemiluminescence (Amersham) or a LumiGLO (Kirkegaard & Perry Laboratories) detection system.
Production of m-Calpain Antibody.
An antipeptidic antibody against the amino-terminal 18-mer segment of preautolysis m-calpain was developed by using a synthetic peptide (42), AGIAAKLAKDREAAEGLGC, conjugated to keyhole limpet hemocyanin and affinity-purified with Imject Activated Immunogen Conjugation Kits (Pierce). A female, 12-week-old New Zealand White rabbit was injected with the immunogen. Antiserum m-calpain was collected 4 weeks after injection and was purified with Sulfolink coupling gel (Pierce).
Statistical Analysis.
Raw data from a single time point (6 or 12 h) were analyzed with sigma plot 3.0. A Student’s t test or ANOVA was used to evaluate differences among the control and treatments. A significance level of 5% was used for all comparisons (43).
RESULTS
Overexpression of the DN-m-Calpain and CID: Detection at the mRNA Level.
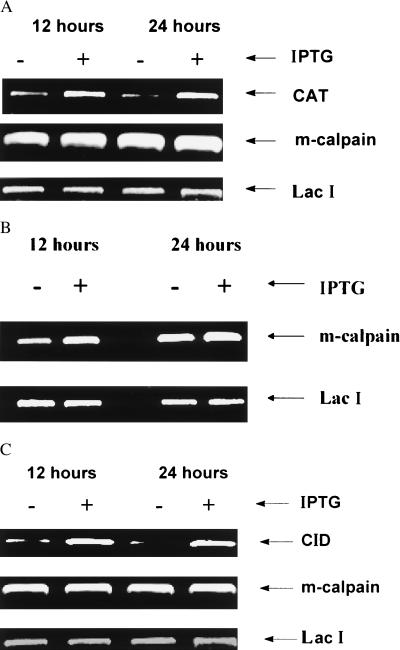
To verify that cell lines expressed constructs as desired, several confirmation studies were completed. In the first of these (Fig. 2), we assessed effects of IPTG treatment (at 12 and 24 h) on CAT, m-calpain, and LacI mRNAs by RT-PCR in the L8/PC (Fig. 2A), L8/DN (Fig. 2B), and L8/CID (Fig. 2C) cell lines. In L8/PC cells, IPTG induced expression of CAT without affecting m-calpain or LacI mRNAs (Fig. 2A). In L8/DN cells, IPTG increased expression of m-calpain mRNA but did not affect LacI (Fig. 2B). The signal for m-calpain shown in Fig. 2B represents both the endogenous and DN-m-calpain; our interpretation is that the increase in signal is caused by synthesis of DN-m-calpain. We also assessed effects of IPTG on CID, m-calpain, and LacI mRNAs (Fig. 2C). IPTG increased CID mRNA but did not affect m-calpain and LacI mRNAs. The signal for CAT (Fig. 2A) and CID mRNAs (Fig. 2C) in the absence of IPTG indicates the “leakiness” of the LacSwitch expression system. These results show that exogenous-gene mRNA was inducible by IPTG and stable for at least 24 h.
Figure 2.
IPTG induction of exogenous gene expression in different cell lines by RT-PCR assay. All of the transfectants were grown to 90–100% confluence, after which IPTG (5 mM) was added to half of the plates and incubated for 12 or 24 h. RT-PCR was performed as described in text. (A) L8/PC cell line. (B) L8/DN cell line. (C) L8/CID cell line.
Effects of Overexpressing DN-m-Calpain on m-Calpain at the Protein Level.
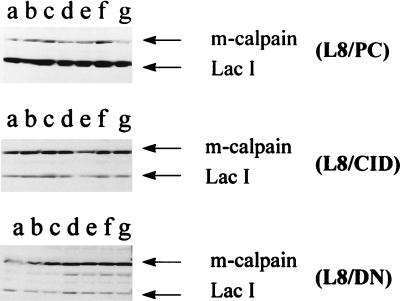
To determine whether DN-m-calpain would be expressed at the protein level, Western blot analysis was performed (Fig. 3). As expected, in L8/PC and L8/CID cell lines (Fig. 3 Top and Middle), addition of IPTG did not affect m-calpain or LacI concentrations. However, in the L8/DN cell line (Fig. 3 Bottom), total m-calpain protein was increased after addition of IPTG at 8 h. This increase indicates that DN-m-calpain was expressed at the protein level.
Figure 3.
Effect of overexpression of various constructs on calpain concentration in transfectants. Myoblasts were cultured to 90–100% confluence, after which IPTG (5 mM) was added to each plate. The plates (except lane a) were cultured for an additional 8 h (lane b), 1 day (lane c), 2 days (lane d), 3 days (lane e), 4 days (lane f), or 5 days (lane g). Western blots were prepared and polyclonal rat m-calpain antibody and polyclonal LacI antibody were used to probe the blots as described in the text. (Top) L8/PC cell line; (Middle) L8/CID cell line; (Bottom) L8/DN cell line.
Effects of Overexpressing CID and DN-m-Calpain on Fodrin Degradation.
Fodrin, a nonerythroid spectrin and a multifunctional protein, is a major component of the cortical cytoskeleton of most eukaryotic cells (44, 45). Calpains cleave fodrin into 150- and 145-kDa fragments after addition of A23187 (Ca2+ ionophore) and maitotoxin; this addition stimulates calcium influx in both excitable and nonexcitable cells in SH-SY5Y neuroblastoma cells and cerebrocortical cultures (41, 46). Because fodrin is a well characterized calpain substrate, we compared its hydrolysis to the hydrolyses of the cell lines developed in this study to determine whether they provided good models for control of calpain activity.
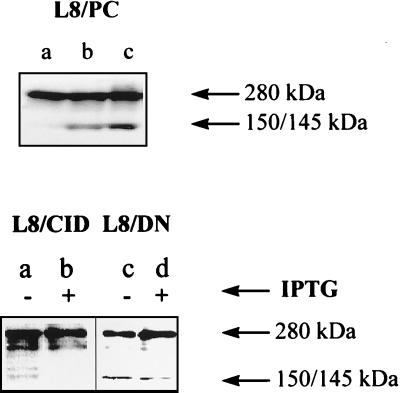
To study the impact of overexpression of CID and DN-m-calpain on endogenous calpain activity, we examined fodrin breakdown after addition of A23187 to culture. In L8/PC cells, fodrin was cleaved into 145- and 150-kDa fragments after addition of 10 μM and 20 μM A23187, respectively, within a 1-h incubation period (Fig. 4 Upper). In L8/CID and L8/DN cell lines, fodrin breakdown, after addition of 20 μM A23187 within a 2-h incubation period, was reduced by addition of IPTG (Fig. 4 Lower, lanes b and d). Specifically, IPTG caused intact fodrin to accumulate and reduced production of fodrin fragments. These results show that endogenous calpain activity is reduced by overexpression of DN-m-calpain or CID.
Figure 4.
Inhibition of fodrin-breakdown product by reduction of calpain activity in various cell lines. (Upper) Fodrin degradation in L8/PC cell line: no A23187 treatment (lane a), A23187 (10 μM) for 1 h (lane b), and A23187 (20 μM) for 2 h (lane c). (Lower) Fodrin degradation in the presence and absence of IPTG in the cell lines L8/CID and L8/DN exposed to A23187 (20 μM) for 2 h. Monoclonal spectrin antibody (nonerythroid) was used for Western blotting.
Contribution of Calpain to Total Protein Degradation.
Total protein degradation after serum withdrawal was measured with a tyrosine labeling experiment. IPTG did not affect total protein degradation after serum withdrawal the in L8/PC cell line. However, in the L8/DN cell line, total protein degradation was reduced by 30% (P < 0.05) by overexpression of the dominant negative m-calpain for 12 h. In the L8/CID cell line, total protein degradation was reduced by 63% (P < 0.05) after overexpression of CID for 12 h. IPTG also caused a 33% reduction in degradation at 6 h in the L8/CID cell line. These data indicate that a large portion of myotube protein degradation is mediated by calpains.
Stabilization of Nebulin by Calpains.
Nebulins, a family of giant proteins with molecular masses of 600–900 kDa in various skeletal muscle tissues (45, 47), are the sole constituent of a set of long, inextensible, longitudinal filaments that span the space between the Z disk and the distal region of a thin (actin) filament (48). An entire nebulin filament consists of a combination of 200-, 180-, 40-, 35-, and 23-kDa subfragments (49). To determine whether the calpains degrade this important structural protein, we assessed its degradation in our cell lines.
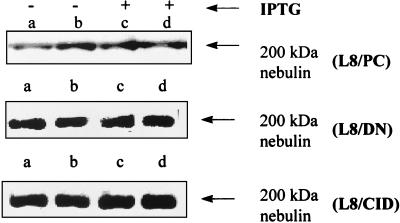
L8/PC, L8/CID, and L8/DN cell lines were exposed to IPTG for 24 h. At the end of treatment, all the cells were changed to serum-free medium and incubated for 12 more hours in the presence (Fig. 5, lanes c and d) or absence (Fig. 5, lanes a and b) of IPTG. Western blotting was performed, and monoclonal nebulin antibody was used as a probe. IPTG did not affect nebulin in L8/PC cells. Based on scanning densitometry, we determined that nebulin was stabilized by 20% after overexpression of DN-m-calpain and by 34.5% after overexpression of CID. These results indicate that the calpain system is involved in degradation of a key myofibrillar protein in living cells.
Figure 5.
Stabilization of nebulin by reduction of calpain activity. The three cell lines (L8/PC, L8/DN, and L8/CID) were exposed to IPTG (5 mM) for 24 h, after which nebulin concentration was assessed by Western blotting. Lanes a and b are duplicates, as are lanes c and d. Nebulin was stabilized by 20% after overexpression of DN-m-calpain and 33% after overexpression of CID.
DISCUSSION
Accumulation and maintenance of muscle mass depends on control of both protein synthesis and protein degradation. In recent years, interest has focused on degradation, because typically this process is accelerated in a wide variety of pathologic conditions. For example, degradation is enhanced in cachexia (34), AIDS wasting (50), various endocrine disorders (51), genetic muscular dystrophies (30), and in extended bed rest (52). Degradation may also be a constraint to growth in domestic animals (53). Reducing the rate of muscle protein degradation in farm animals may represent a strategy that could promote animal growth effectively and enhance efficiency of growth.
The protease(s) that mediates degradation needs to be identified, because it would provide a logical target for intervention. In this regard, two proteolytic systems have been studied: the proteasome and calpain. Several recent studies point to the involvement of the proteasome in myofibrillar protein degradation (34, 35–39); however, it is not clear whether proteasome is the key rate-limiting protease or whether it is simply one protease in the pathway of degradation. A recent study by Solomon et al. (35) indicated that the proteasome can degrade monomeric (free) myofibrillar proteins, except when they are associated with other myofibrillar proteins. These data suggest that another protease must exist upstream of the proteasome to provide it with monomeric myofibrillar proteins as substrate.
Several lines of evidence suggest that calpains may be the rate-limiting proteases; however, this conclusion has not been corroborated in living cells. To assess calpain function, we developed two genetic strategies to manipulate endogenous calpain activities: overexpression of DN-m-calpain and overexpression of CID. Overexpression of DN-m-calpain was expected to inhibit m-calpain activity specifically by competing with endogenous m-calpain for substrate. Overexpression of CID was expected to inhibit both μ- and m-calpain activities, because calpastatin has been shown to inhibit both calpain isoforms (8, 11–15). To assess the utility of these approaches: (i) calpain mRNA concentration was assessed by RT-PCR; (ii) m-calpain concentration was assessed by Western blotting; and (iii) fodrin degradation was assessed. For analysis of calpain function in protein breakdown, we measured total protein degradation and examined the concentration of nebulin, a Z disk- and α-actin-binding protein. Our results showed that exogenous genes (DN-m-calpain and CID) were overexpressed in an inducible manner at the mRNA level (Fig. 2 B and C) and/or at the protein level (Fig. 3 Bottom).
To determine whether expression of DN and CID regulated the calpain system as expected, we evaluated fodrin degradation. Fodrin is a well characterized calpain substrate (41, 46, 54, 55). In fact, specific calpain cleavage sites of fodrin have been identified (56). Our data clearly show that overexpression of our various constructs (Fig. 4) stabilized fodrin. Therefore, these data provide compelling evidence of our ability to control calpain activity in living muscle cells. Previously, the study of protease function in tissues was carried out with the use of protease inhibitors. However, their use has always been accompanied by concerns of nonspecificity. The strategies outlined here successfully control protease activities in living muscle cells and allow unequivocal identification of protease function in muscle tissue.
Our data also indicated that calpains were involved significantly in total protein turnover during conditions of stimulated degradation. The involvement was evaluated by release of radiolabeled tyrosine into cell-culture medium from prelabeled protein. Overexpression of CID caused a 63% reduction in total protein degradation, whereas overexpression of DN caused only a 30% reduction. Our expectation was that the CID construct would inhibit both μ- and m-calpain, whereas the DN construct would specifically inhibit m-calpain by competing with endogenous intact m-calpain for substrate. If this expectation proves to be true, the difference in inhibition caused by these two constructs may approximate the degradation in L8 cells that is mediated by other calpains (i.e., μ-calpain). A more direct means of assessing the role of μ-calpain in muscle cells would be to overexpress a dominant negative μ-calpain construct, similar to the one used in this study.
Many believe that calpains are rate-limiting enzymes in myofibrillar protein degradation, but other compelling data suggest that the proteasome also participates in this process (34–39). A recent article by Solomon et al. (35) indicated that activity of the proteasome could be secondary to another protease, because it was capable of degrading monomeric myofibrillar proteins, not higher structures. We chose to investigate the possibility that calpain degrades myofibrillar protein by investigating nebulin. Whereas intact nebulin approaches 600 kDa, it undergoes autofragmentation into 200-, 180-, 40-, 35-, and 23-kDa subfragments in the presence of 0.1 mM Ca2+ (49, 53). The 200-kDa fragment was the principle nebulin fragment recovered from muscle preparations in our experiment. We found that inhibition of calpain activity, by overexpression of either CID or DN, stabilized the 200-kDa fragment of nebulin. Although nebulin is known to be a calpain substrate in vitro, inhibition of calpains in vivo only has small effects on nebulin degradation. Further studies are needed to examine the role that calpains play in degrading other key structural proteins of the sarcomere.
Earlier studies with L8 cells (57) have documented that serum withdrawal caused an ≈20% increase in total protein degradation. It is interesting to note that stabilization of nebulin and of total proteins by overexpression of the calpain system was detectable only under conditions of serum starvation (i.e., accelerated degradation). These data indicate that calpain may function in degrading proteins under stimulated conditions.
Calpains play substantial roles in protein breakdown in muscle cells. They degrade fodrin, a well characterized calpain substrate in vivo, and account for a large proportion of total protein degradation. Calpains degrade nebulin, an important architectural protein of the sarcomere, and may be particularly important to degradation during pathological conditions. Further studies are needed to discover other substrates for calpains in muscle and to elucidate roles of μ-calpain as well.
Acknowledgments
We thank Dr. John Elce for the gift of rat m-calpain cDNA, Yoji Ueda for the preparation for m-calpain antibody, and Ying-yi Xiao for participation in selecting the cell lines. This work was supported by U.S. Department of Agriculture Grant 94-37206-1098 to N.E.F.
ABBREVIATIONS
- CAT
chloramphenicol acetyltransferase
- CID
calpastatin inhibitory domain
- DN
dominant negative
- IPTG
isopropyl β-d-thiogalactoside
- PC
plasmid control
- RT
reverse transcription
References
- 1. Dayton W R, Goll D E, Zeece M G, Robson R M, Reville W J. Biochemistry. 1976;15:2150–2158. doi: 10.1021/bi00655a019. [DOI] [PubMed] [Google Scholar]
- 2.Dayton W R, Reville W J, Goll D E, Stromer M H. Biochemistry. 1976;15:2159–2167. doi: 10.1021/bi00655a020. [DOI] [PubMed] [Google Scholar]
- 3.Sorimachi H, Saido T, Suzuki K. FEBS Lett. 1994;3443:1–5. doi: 10.1016/0014-5793(94)80595-4. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki K, Saido T C, Hirai S. Ann NY Acad Sci. 1992;674:218–227. doi: 10.1111/j.1749-6632.1992.tb27490.x. [DOI] [PubMed] [Google Scholar]
- 5.Saido T C, Sorimachi H, Suzuki K. FASEB J. 1994;8:814–822. [PubMed] [Google Scholar]
- 6.Saido T C, Nago S, Shiramine M, Tsukaguchi M, Yoshizawa T, Sorimachi H, Ito H, Tsuchiya T, Kawashima S, Suzuki K. FEBS Lett. 1994;346:263–267. doi: 10.1016/0014-5793(94)00487-0. [DOI] [PubMed] [Google Scholar]
- 7.Kawasaki H, Emori Y, Suzuki K. Arch Biochem Biophys. 1993;305:467–472. doi: 10.1006/abbi.1993.1448. [DOI] [PubMed] [Google Scholar]
- 8.Croall D E, McGrody K S. Biochemistry. 1994;33:13223–13230. doi: 10.1021/bi00249a008. [DOI] [PubMed] [Google Scholar]
- 9.Otsuka Y, Goll D E. J Biol Chem. 1987;262:5839–5851. [PubMed] [Google Scholar]
- 10.Ma H, Yang H Q, Takano E, Lee W J, Hatanaka M, Maki M. J Biochem (Tokyo) 1993;113:591–599. doi: 10.1093/oxfordjournals.jbchem.a124088. [DOI] [PubMed] [Google Scholar]
- 11.Demartino G N, Wachendorfer R, McGuire M J, Croall D E. Arch Biochem Biophys. 1988;262:189–198. doi: 10.1016/0003-9861(88)90181-6. [DOI] [PubMed] [Google Scholar]
- 12.Emori Y, Kawasaki H, Imajoh S, Minami Y, Suzuki K. J Biol Chem. 1988;263:2364–2370. [PubMed] [Google Scholar]
- 13.Maki M, Takano E, Osawa T, Ooi T, Murachi T, Hatanaka M. J Biol Chem. 1988;263:10254–10261. [PubMed] [Google Scholar]
- 14.Maki M, Bagci H, Hamaguchi K, Ueda M, Murachi T, Hatanaka M. J Biol Chem. 1989;264:18866–18869. [PubMed] [Google Scholar]
- 15.Emori Y, Kawasaki H, Imajoh S, Minami Y, Suzuki K. J Biol Chem. 1988;263:2364–2370. [PubMed] [Google Scholar]
- 16.Dayton W R, Goll D E, Stromer M H, Reville W J, Zeece M G, Robson R M. In: Proteases and Biological Control, Cold Spring Harbor Conferences on Cell Proliferation. Reich E, Rifkin D B, Shaw E, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1975. pp. 551–577. [Google Scholar]
- 17.Waxman L. In: Protein Turnover and Lysosome Function. Segal H L, Doyle D J, editors. New York: Academic; 1978. pp. 363–377. [Google Scholar]
- 18.Ishiura S, Murofushi H, Suzuki K, Imahori S. J Biochem (Tokyo) 1978;84:225–230. doi: 10.1093/oxfordjournals.jbchem.a132111. [DOI] [PubMed] [Google Scholar]
- 19.Ishiura S, Sugita H, Suzuki K, Imahori K. J Biochem (Tokyo) 1989;86:579–581. doi: 10.1093/oxfordjournals.jbchem.a132558. [DOI] [PubMed] [Google Scholar]
- 20.Toyo-oka T, Shimizu T, Masaki T. Biochem Biophys Res Comm. 1978;82:484–491. doi: 10.1016/0006-291x(78)90900-2. [DOI] [PubMed] [Google Scholar]
- 21.Toyo-oka T, Masaki T, Okamoto J, Tanaka T. J Mol Cell Cardiol. 1979;11:769–786. doi: 10.1016/0022-2828(79)90402-4. [DOI] [PubMed] [Google Scholar]
- 22.Azamza J L, Raymond J, Robin J M, Cottin P, Ducastaing A. Biochem J. 1979;183:339–347. doi: 10.1042/bj1830339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goll D E, Kleese W C, Szpacenko A. In: Animal Growth Regulation. Campion D R, Hausman G J, Martin R J, editors. New York: Plenum; 1989. pp. 141–182. [Google Scholar]
- 24.Tan F C, Goll D E, Otsuka Y. J Mol Cell Cardiol. 1988;20:983–997. doi: 10.1016/0022-2828(88)90576-7. [DOI] [PubMed] [Google Scholar]
- 25.Koohmaraie M. J Anim Sci. 1992;70:3697–3708. doi: 10.2527/1992.70123697x. [DOI] [PubMed] [Google Scholar]
- 26.Kumamoto T, Kleese W C, Cong J, Goll D E, Pierce P R, Allen R E. Anatom Rec. 1992;232:60–77. doi: 10.1002/ar.1092320108. [DOI] [PubMed] [Google Scholar]
- 27.Bird J W C, Carter J H, Triemer R E, Brooks R M, Spanier A M. FASEB J. 1980;39:20–25. [PubMed] [Google Scholar]
- 28.Busch W A, Stromer M H, Goll D E, Suzuki A. J Cell Biol. 1972;52:367–381. doi: 10.1083/jcb.52.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dayton W R, Schollmeyer J V, Chan A C, Allen C E. Biochim Biophys Acta. 1979;584:216–230. doi: 10.1016/0304-4165(79)90266-6. [DOI] [PubMed] [Google Scholar]
- 30.Arahata K, Sugita H. Trends Pharm Sci. 1989;10:437–439. doi: 10.1016/S0165-6147(89)80005-7. [DOI] [PubMed] [Google Scholar]
- 31.Arakawa N, Takashima M, Kurata T, Fujimaki M. Agric Biol Chem. 1983;47:1517–1522. [Google Scholar]
- 32.Ilian M A, Forsberg N E. Biochem J. 1992;287:163–171. doi: 10.1042/bj2870163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahlmann B, Kuehn L, Reinauer H, Kay J, Stauber W T. Contrib Nephrol. 1989;73:127–136. doi: 10.1159/000417385. [DOI] [PubMed] [Google Scholar]
- 34.Temparis S, Asensi M, Taillandier D, Aurousseau E, Larbaud D, Obled A, Bechet D, Ferrara M, Estrela J M, Attaix D. Cancer Res. 1994;54:5568–5573. [PubMed] [Google Scholar]
- 35.Solomon V, Goldberg A L. J Biol Chem. 1996;271:26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- 36.Furuno K, Goodman M N, Goldberg A L. J Biol Chem. 1990;265:8550–8557. [PubMed] [Google Scholar]
- 37.Medina R, Wing S S, Haas A, Goldberg A L. Biomed Biochim Acta. 1991;50:347–356. [PubMed] [Google Scholar]
- 38.Tawa N E, Kettelhut I C, Jr, Goldberg A L. Am J Physiol. 1992;263:E326–E334. doi: 10.1152/ajpendo.1992.263.2.E326. [DOI] [PubMed] [Google Scholar]
- 39.Medina R, Wing S, Goldberg A. Biochem J. 1995;307:631–637. doi: 10.1042/bj3070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ausubel F M, Brent R, Kingston R, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1990. [Google Scholar]
- 41.Wang K K W, Posner A, Hajimohammadreza I. BioTechniques. 1996;20:662–668. doi: 10.2144/19962004662. [DOI] [PubMed] [Google Scholar]
- 42.Locker R H, Wild D J C. J Biochem. 1986;99:1473–1484. doi: 10.1093/oxfordjournals.jbchem.a135617. [DOI] [PubMed] [Google Scholar]
- 43.Steel R G D, Torrie J H. Principles and Procedures of Statistics. New York: McGraw-Hill; 1980. [Google Scholar]
- 44.Martin S J, O’Brien G A, Nishioka W K, McGahon A J, Mahboubi A, Daido T C, Green D R. J Biol Chem. 1995;270:6425–6428. doi: 10.1074/jbc.270.12.6425. [DOI] [PubMed] [Google Scholar]
- 45.Vanags D M, Porn-Ares I, Coppola S, Burgess D H, Orrenius S. J Biol Chem. 1996;271:31075–31085. doi: 10.1074/jbc.271.49.31075. [DOI] [PubMed] [Google Scholar]
- 46.Wang K K W, Nath R, Raser K J, Hajimohammadreza I. Arch Biochem Biophys. 1996;331:208–214. doi: 10.1006/abbi.1996.0300. [DOI] [PubMed] [Google Scholar]
- 47.Hu D H, Kimura S, Maruyama K. J Biochem. 1986;99:1485–1492. doi: 10.1093/oxfordjournals.jbchem.a135618. [DOI] [PubMed] [Google Scholar]
- 48.Wang K, Wright J. J Cell Biol. 1988;107:2199–2212. doi: 10.1083/jcb.107.6.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tatsumi R, Hattori A, Takahashi K. J Biochem. 1993;113:797–804. doi: 10.1093/oxfordjournals.jbchem.a124121. [DOI] [PubMed] [Google Scholar]
- 50.Wrzolek M A, Sher J H, Kozlowski P B, Rao C. Muscle Nerve. 1990;13:508–515. doi: 10.1002/mus.880130607. [DOI] [PubMed] [Google Scholar]
- 51.Kettelhut I C, Wing S S, Goldberg A L. Diabetes Metab Rev. 1988;4:751–772. doi: 10.1002/dmr.5610040805. [DOI] [PubMed] [Google Scholar]
- 52.Tischler M E, Rosenberg S, Satarug S, Henriksen E J, Kirby C R, Tome M, Chase P. Metabolism. 1990;39:756–763. doi: 10.1016/0026-0495(90)90113-q. [DOI] [PubMed] [Google Scholar]
- 53.Bardsley R G, Allcock S M J, Dawson J M, Dumelow N W, Higgins J A, Lasslett Y V, Lockley A K, Buttery P J. Biochimie. 1992;74:267–273. doi: 10.1016/0300-9084(92)90125-x. [DOI] [PubMed] [Google Scholar]
- 54.Blomgren K, Kawashima S, Saido T C, Karlsson J-O, Elmered A, Hagberg H. Brain Res. 1995;684:143–149. doi: 10.1016/0006-8993(95)00399-b. [DOI] [PubMed] [Google Scholar]
- 55.Vanderklish P, Saido T C, Gall C, Arai A, Lynch G. Mol Brain Res. 1995;32:25–35. doi: 10.1016/0169-328x(95)00057-y. [DOI] [PubMed] [Google Scholar]
- 56.Stabach P R, Cianci C D, Glantz S B, Zhang Z, Morrow J S. Biochemistry. 1997;36:57–65. doi: 10.1021/bi962034i. [DOI] [PubMed] [Google Scholar]
- 57.Hong D-H, Forsberg N E. J Anim Sci. 1994;72:2279–2288. doi: 10.2527/1994.7292279x. [DOI] [PubMed] [Google Scholar]